Abstract
Nuclear receptors (NRs) regulate target gene transcription through the recruitment of multiple coactivator complexes to the promoter regions of target genes. One important coactivator complex includes a p160 coactivator (GRIP1, SRC-1, or ACTR) and its downstream coactivators (e.g., p300, CARM1, CoCoA, and Fli-I), which contribute to transcriptional activation by protein acetylation, protein methylation, and protein-protein interactions. In this study, we identified a novel NR coactivator, GAC63, which binds to the N-terminal region of p160 coactivators as well as the ligand binding domains of some NRs. GAC63 enhanced transcriptional activation by NRs in a hormone-dependent and GRIP1-dependent manner in transient transfection assays and cooperated synergistically and selectively with other NR coactivators, including GRIP1 and CARM1, to enhance estrogen receptor function. Endogenous GAC63 was recruited to the estrogen-responsive pS2 gene promoter of MCF-7 cells in response to the hormone. Reduction of the endogenous GAC63 level by small interfering RNA inhibited transcriptional activation by the hormone-activated estrogen receptor. Thus, GAC63 is a physiologically relevant part of the p160 coactivator signaling pathway that mediates transcriptional activation by NRs.
Steroid and thyroid hormones, retinoic acid, and vitamin D play important roles in the regulation of growth, development, and homeostasis. The biological functions of these hormones are mediated by nuclear receptors (NRs), a family of ligand-regulated transcription factors (18, 26). Nuclear receptors share similar structural features, including an N-terminal activation domain of variable length, a central highly conserved DNA-binding domain (DBD), and a C-terminal ligand binding domain (LBD). Most NRs have two transcriptional activation domains, the N-terminal activation function AF-1 and the C-terminal activation function AF-2. Ligand binding causes NRs to undergo conformational changes which potentiate their ability to bind and recruit multiple coactivator complexes to the promoters of target genes (8, 20). Each coactivator complex constitutes a signal transduction pathway which transmits the activating signal from the NRs to specific downstream targets in the transcription machinery. For example, the Swi/Snf complex participates in the remodeling of chromatin conformation around the promoter by means of an ATPase activity; the p160 coactivator complex contains enzymes which contribute to chromatin remodeling by catalyzing posttranslational modifications of histones and other proteins; and the TRAP/DRIP/mediator complex helps to recruit and activate RNA polymerase II.
The p160 coactivator family has three members, SRC-1, GRIP1/TIF-2, and AIB1/ACTR/RAC3/pCIP/TRAM1 (8, 20, 29). The p160 coactivators have a central NR interaction domain consisting of three leucine-rich motifs; a C-terminal region containing two activation domains, AD1 and AD2; and an N-terminal basic helix-loop-helix/Per-Arnt-Sim (bHLH-PAS) domain. The C-terminal activation domains AD1 and AD2 recruit secondary coactivators which lie downstream in the signaling pathway and transmit the activation signal from the enhancer element-bound NR to the transcription machinery. AD1 recruits the related histone acetyltransferases p300 and CBP (6, 25, 30), while AD2 recruits the protein arginine methyltransferases CARM1 and PRMT1 (5, 13). These histone-modifying enzymes are recruited to the target gene promoters through their interaction with p160 coactivators, which bind to hormone-activated NRs (4, 24). They contribute to transcriptional activation by acetylating or methylating histones, coactivators, transcription factors, and presumably other unknown proteins. The N-terminal bHLH-PAS domain is the most conserved region among the p160 coactivators (1). In various proteins, this motif is involved in DNA binding, ligand binding, and protein-protein interactions (9, 10). In p160 coactivators, it functions as an additional activation domain, AD3, by recruiting downstream coactivators, including CoCoA and Fli-I (11, 15). Thus, the p160 coactivators contribute to transcriptional activation by serving as bridges between NRs and downstream secondary coactivators.
The N-terminal bHLH-PAS domain of p160 coactivators is large enough (about 350 amino acids) to have multiple interaction surfaces. Here we report the identification of another protein, GRIP1-associated coactivator 63 (GAC63), as a novel 63-kDa NR coactivator, which interacts with both the GRIP1 N-terminal region and some NRs, cooperates synergistically but selectively with other NR coactivators and is required for optimal transcriptional activation by estrogen receptor α (ERα). GAC63 thus represents a new component of the p160 coactivator signal transduction pathway.
MATERIALS AND METHODS
Yeast two-hybrid screening.
Yeast two-hybrid screening was performed as described previously (11), using the N-terminal bHLH-PAS domain of GRIP1 (amino acids 5 to 479) as bait.
Plasmids.
The following plasmids were used to express GAC63 in mammalian cells and in vitro: pSG5.HA (5) for proteins with an N-terminal hemagglutinin (HA) epitope; pM (Clontech) for fusion proteins with an N-terminal Gal4-DBD; and pcDNA3.1.V5.His (Invitrogen) for proteins with a C-terminal V5 epitope and His6 tag. pGEX-5X1 vector (Amersham Pharmacia) was used to express fusion proteins with an N-terminal glutathione S-transferase (GST) in Escherichia coli. cDNA fragments encoding the indicated GAC63 amino acids were amplified by PCR with primers containing appropriate restriction sites and inserted into vectors as follows: full-length GAC63, GAC63(1-200), GAC63(200-370), GAC63(370-567), GAC63(1-352), and GAC63(184-567) into EcoRI and XhoI sites of pSG5.HA; full-length GAC63, GAC63(1-134), GAC63(1-352), GAC63(130-492), GAC63(130-567), GAC63(382-567), and GAC63(468-567) into EcoRI and SalI sites of pM; and full-length GAC63 into EcoRI and XhoI sites of pGEX-5X1 and pcDNA3.1.V5.His vectors.
The following plasmids were described previously: pGEX-4T1.GRIP1(5-479), pSG5.HA-GRIP1, pSG5.HA-GRIP1(5-479), pSG5.HA-GRIP1(563-1462), pSG5.HA-CARM1, and pCMV-p300 (11); pSV-AR and pCMV-AR encoding human androgen receptor (AR) and luciferase reporter plasmids MMTV(ERE)-LUC for ER, MMTV-LUC for AR, and GK1-LUC for Gal4 (5); and pSG5-HE0 encoding human ERα, pHE15 encoding ERα(1-281), and pHE19 encoding ERα(179-595) (14, 27).
Protein-protein interaction assays.
GST pull-down assays were performed as described previously (11, 13). Coimmunoprecipitation and immunoblotting were performed as described previously (11), using the following antibodies: anti-His-tag (Invitrogen), anti-GAC63 antiserum 1bg (produced for this study by Zymed), anti-ERα (HC-20; Santa Cruz Biotechnology), normal mouse or rabbit immunoglobulin G (IgG) (Santa Cruz Biotechnology), and anti-HA-tag (3F10; Roche). The GAC63 antiserum was raised against a C-terminal peptide representing GAC63 amino acids 546 to 567 and affinity purified against the same peptide.
Transient transfection assays.
Transient transfection assays were performed with CV-1 cells in 12-well plates as described previously (11). After transfection, cells were cultured with or without 100 nM estradiol (E2) or dihydrotestosterone (DHT) for 48 h before harvesting for luciferase activity assays. The results shown are the means and standard deviations of triplicate points. Instead of using typical constitutive reporter plasmids (which are generally stimulated by coactivators) as internal controls, the results shown are representative of at least three independent experiments.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed with MCF-7 cells treated with or without 100 nM E2 for 60 min as described previously (11, 17), using 1 μg of anti-ERα antibody, 2 μg of anti-GRIP1 antibody A300-025A (Bethyl Laboratories), 2 μg of anti-GAC63 antibody 1bg, or 2 μg of normal rabbit IgG. A total of 0.1 to 5 μl of immunoprecipitated, purified, chromosomal DNA was used for PCR amplification, using the following primers: pS2 (−353/−31), 5′-GGCCATCTCTCACTATGAATCACTTCTGC-3′ (forward) and 5′-GGCAGGCTCTGTTTGCTTAAAGAGCG-3′ (reverse); and β-actin (nucleotides 68 to 327 of GenBank file NM001101), 5′-CTCACCATGGATGATGATATCGC-3′ (forward), and 5′-ATTTTCTCCATGTCGTCCCAGTTG-3′ (reverse).
Quantitative real-time PCR (QPCR) reactions were performed with 2 μl of immunoprecipitated chromosomal DNA on a Stratagene Mx3000P Instrument, using SYBR green to detect PCR products. The following primer pairs were used: pS2 (−353/−31 relative to the transcription start site), 5′-GGCCATCTCTCACTATGAATCACTTCTGC-3′ (forward) and 5′-GGCAGGCTCTGTTTGCTTAAAGAGCG-3′ (reverse); and β-actin (nucleotides 68 to 327 of GenBank file NM001101), 5′-CTCACCATGGATGATGATATCGC-3′ (forward) and 5′-ATTTTCTCCATGTCGTCCCAGTTG-3′ (reverse).
RNA interference.
RNA interference experiments were performed as described previously (11) using Lipofectamine 2000 (Invitrogen). Small interfering RNA (siRNA) oligonucleotides for GAC63 and scrambled oligonucleotides (same nucleotide composition with scrambled sequence) were synthesized by the USC Norris Comprehensive Cancer Center Microchemical Core Laboratory and annealed to form duplexes. The following siRNA sequences were used: siGAC63, 5′-GAACCUCUCCAAGUAAGAGdTdT-3′ (sense) and 5′-CUCUUACUUGGAGAGGUUCdTdT-3′ (antisense); and scrambled siRNA, 5′-UUCUCCGAACGUGUCACAUdTdT-3′ (sense) and 5′-AUGUGACACGUUCGGAGAAdTdT-3′ (antisense).
Reverse transcription-QPCR (RT-QPCR) was also performed. Total MCF-7 cell RNA was extracted with Trizol reagent (Invitrogen) and subjected to reverse transcription by using an iScript cDNA Synthesis kit (Bio-Rad). Two microliters of RT product was subjected QPCR analysis. The primers used were as follows: GAC63, 5′-AAGCCCTGAAGCTCTTGCCAG-3′ (forward) and 5′-CCGAGCATTCCTACGAAGTTC-3′ (reverse); pS2, 5′-CATGGAGAACAAGGTGATCTG-3′ (forward) and 5′-CTTCTGGAGGGACGTCGATGG-3′ (reverse); and GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′-TCTGGTAAAGTGGATATTGTTG-3′ (forward) and 5′-GATGGTGATGGGATTTCC-3′ (reverse).
RESULTS
Isolation of GAC63.
From a yeast two-hybrid screen of a mouse 17-day embryo cDNA library, using the N-terminal fragment of GRIP1 (amino acids 5 to 479) as bait (Fig. 1A), we identified a C-terminal fragment of a mouse protein (amino acids 184 to 567) which is apparently orthologous to HUEL (human embryo lung protein; accession number AF006621). The full-length HUEL sequence is available—but only the C-terminal sequence of its mouse orthologue. Using a 3′ primer from the mouse cDNA sequence and a 5′ primer designed from the mouse genomic sequence (using the translation start site of the human cDNA sequence as a positioning guide), we PCR amplified the full-length mouse cDNA sequence corresponding to HUEL from a mouse 17-day-embryo cDNA library. There are 87% nucleotide identity and 89% amino acid identity (for the complete coding regions) between mouse and human sequences. Because of its binding to GRIP1 and NR coactivator activity (see below), we named it GRIP1-associated coactivator 63 (GAC63; GenBank accession number AY682914).
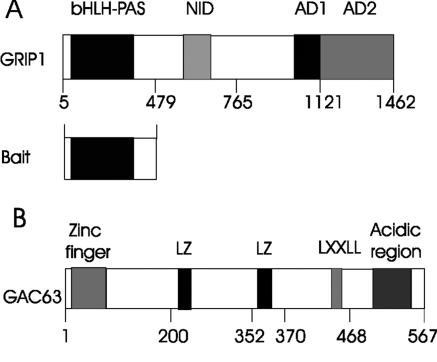
FIG. 1.
Functional domains of GRIP1 and GAC63. The N-terminal bHLH-PAS domain of GRIP1 was used as bait in a yeast two-hybrid screen. Numbers indicate amino acids. NID, nuclear receptor interaction domain; AD1 and AD2, activation domains; LXXLL, leucine-rich motif; LZ, leucine zipper-like motif.
Full-length GAC63 has a continuous open reading frame of 1,704 bp, which encodes a predicted protein of 567 amino acids with a calculated molecular mass of 63 kDa. Affinity-purified antibodies against the predicted C-terminal peptide of GAC63 recognized a 63-kDa endogenous protein in several mammalian cell lines (data not shown). The human orthologue of GAC63, HUEL, is expressed in a wide range of normal human fetal and adult tissues, as well as cancer cell lines (22). GAC63 has several recognizable motifs, including an LXXLL motif reminiscent of an NR binding sequence, a zinc finger-like motif, an acidic region, and two leucine zipper-like domains (Fig. 1B). The C-terminal amino acid sequence 239 to 533 of GAC63 has stretches of homology to cation efflux family proteins (pfam01545.11 in the NCBI Conserved Domains database) (19).
Interaction of GAC63 with GRIP1 and NRs.
To confirm the interaction between GAC63 and GRIP1 in vitro, we performed GST pull-down assays. GST-GRIP1N (amino acids 5 to 479) attached to glutathione-Sepharose-4B beads efficiently bound full-length GAC63, while GST attached to beads did not (Fig. 2A). Conversely, GST-GAC63 bound specifically to full length GRIP1 and SRC-1 synthesized in vitro (data not shown). These results suggest that GAC63 binds directly to the N-terminal region of p160 coactivators.
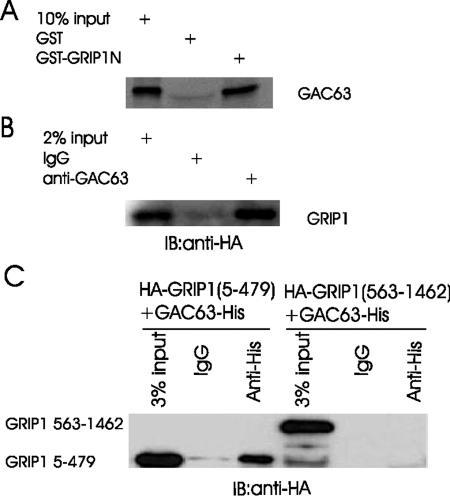
FIG. 2.
GAC63 binds GRIP1 in vitro and in vivo. (A) In GST pull-down assays, GST fusion proteins of GRIP1N (amino acids 5 to 479) were bound to beads and incubated with in vitro-translated GAC63. Bound proteins were eluted and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autofluorography. (B) COS-7 cells were transfected with 2 μg of pcDNA3.1-GAC63-V5.His plasmid and 3 μg of pSG5.HA-GRIP1 plasmid. Cell extracts were immunoprecipitated with anti-GAC63 antibody or normal rabbit IgG. The immunoprecipitated proteins were detected by immunoblotting with anti-HA antibody. (C) COS-7 cells were transfected with 2 μg of pcDNA3.1-GAC63-V5.His plasmid and 2 μg of pSG5.HA-GRIP1(5-479) or pSG5.HA-GRIP1(563-1462). Cell extracts were immunoprecipitated with anti-His-tag antibody. The precipitated proteins were detected by immunoblotting with anti-HA antibody.
To test for binding in vivo, we employed coimmunoprecipitation assays, using COS-7 cells expressing HA-tagged GRIP1 and GAC63 from transiently transfected plasmids. GRIP1, detected by Western blotting with antibodies against HA tag, was precipitated by antibodies against GAC63 but not by normal IgG (Fig. 2B). In addition, GRIP1(5-479) but not GRIP1(563-1462) also coimmunoprecipitated with GAC63 (Fig. 2C). Thus, GAC63 binds to GRIP1 in vivo through the GRIP1 N-terminal region.
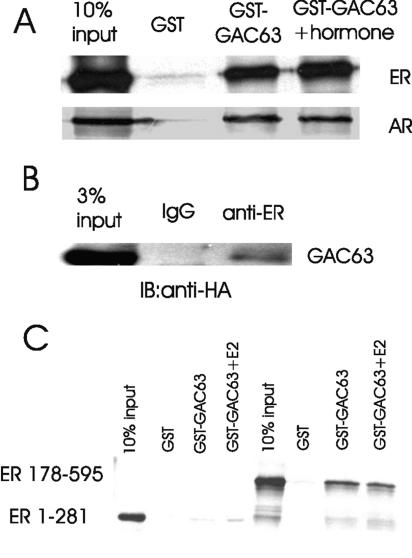
We also tested whether GAC63 binds to NRs. In GST pull-down assays, ERα and AR, translated in vitro with or without the appropriate ligand (E2 for ER and DHT for AR), bound efficiently and specifically to GST-GAC63 (Fig. 3A). The interaction of ER with GST-GAC63 was enhanced modestly by E2 treatment. These findings suggest that GAC63 binds to at least some NRs directly in vitro, in the absence or presence of hormone. In transiently transfected COS-7 cell extracts, weak binding of GAC63 to ER was detected without E2 when antibodies against ER were used for immunoprecipitation (Fig. 3B). No enhancement of binding was detected in the presence of E2. In additional GST pull-down assays, GAC63 bound to a DBD-LBD fragment of ER (in the presence or absence of E2) but bound weakly or not at all to an AF1-DBD fragment of ER, indicating that the ER LBD is the primary binding site for GAC63 (Fig. 3C).
FIG. 3.
GAC63 binds NRs in vitro and in vivo. (A) GST pull-down assays were performed as in Fig. 2A. Full-length ERα and AR were translated in vitro and incubated with GST or GST-GAC63 in the absence or presence of E2 or DHT. (B) COS-7 cells were transfected with 2 μg of pSG5.HA-GAC63 plasmid and 2 μg of pHE0 plasmid (encoding human ERα). Cell extracts were immunoprecipitated with anti-ERα antibody or normal rabbit IgG. The precipitated proteins were detected by immunoblotting with anti-HA antibody. (C) In GST pull-down assays, in vitro-translated ERα 1-281 (containing AF-1 and DBD) or 178-595 (containing DBD and LBD) was incubated with GST-GAC63 in the absence or presence of E2.
GAC63 is an NR coactivator which cooperates selectively with other NR coactivators.
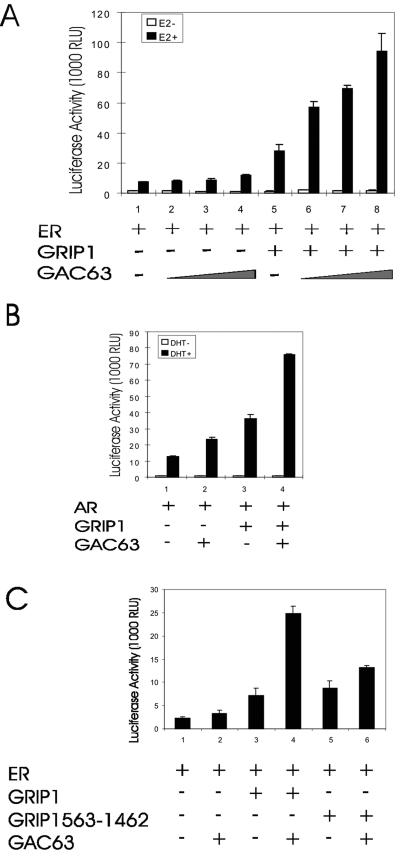
Since GAC63 binds to NRs and the NR coactivator GRIP1, we tested whether GAC63 is involved in NR-regulated transcriptional activation. In transiently transfected CV-1 cells, GRIP1 enhanced the hormone-dependent activity of ERα (Fig. 4A, assays 1 and 5). Increasing amounts of GAC63 expression plasmid enhanced ER activity only slightly (assays 2 to 4) but cooperated synergistically with GRIP1 as a coactivator (assays 6 to 8). Similar results were obtained with AR (Fig. 4B). In addition to GRIP1, GAC63 also cooperated synergistically with the other two p160 family members, SRC-1e and ACTR (data not shown). The synergistic coactivator function between GRIP1 and GAC63 was not observed when the N-terminal 562 amino acids of GRIP1 (containing the GAC63 binding site) were deleted (Fig. 4C).
FIG. 4.
GRIP1-dependent NR coactivator function of GAC63. (A) CV-1 cells were transfected with 200 ng of mouse mammary tumor virus MMTV(ERE)-LUC reporter plasmid; 10 ng of pHE0; 50 ng of pSG5.HA-GRIP1; and 50, 100, or 200 ng of pSG5.HA-GAC63 and grown with or without E2. After 48 h, cell extracts were tested for luciferase activity. RLU, relative light units. (B) CV-1 cells were transfected with 200 ng of MMTV-LUC reporter plasmid, 10 ng of pSV-AR, 50 ng of pSG5.HA-GRIP1, and 200 ng of pSG5.HA-GAC63 and grown with or without DHT before luciferase assays. (C) CV-1 cells were transfected with 200 ng of MMTV(ERE)-LUC reporter plasmid, 10 ng of pHE0, 100 ng of pSG5.HA-GRIP1, or 50 ng of pSG5.GRIP1(563-1462) and 200 ng of pSG5.HA-GAC63 and grown with E2. After 48 h, cell extracts were tested for luciferase activity.
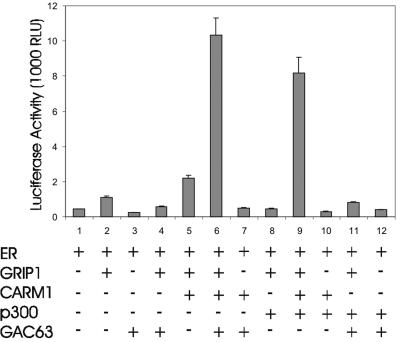
We employed transient transfection assays with low levels of NR expression vector, since these low-NR conditions are characterized by a stringent requirement for multiple coactivators and facilitate the observation of synergistic interaction among multiple coactivators (13, 16). As shown previously under these conditions (16), GRIP1 and the combination of GRIP1 and CARM1 slightly enhanced reporter gene activity, while the combination of GRIP1, CARM1, and p300 caused a dramatic synergistic enhancement of ER-dependent reporter gene activity (Fig. 5, assays 1, 2, 5, and 8 to 10). The combination of GRIP1, CARM1, and GAC63 also produced a dramatic synergy of coactivator function, which was entirely dependent on the presence of GRIP1 (assays 6 and 7). However, the combination of GRIP1, p300, and GAC63 produced no synergy (assay 11). Thus, GAC63 cooperates selectively with other specific coactivators to enhance ER function.
FIG. 5.
Synergy among GRIP1, CARM1, and GAC63. CV-1 cells were transfected with 200 ng of MMTV(ERE)-LUC reporter plasmid, 0.2 ng of pHE0, 50 ng of pSG5.HA-GRIP1, 200 ng of pSG5.HA-CARM1, 200 ng of pCMV-p300, and 200 ng of pSG5.HA-GAC63 and treated with E2 before harvest and luciferase assays. RLU, relative light units.
Functional domains of GAC63.
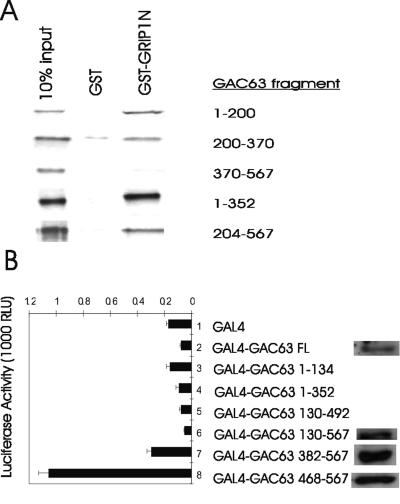
Most coactivators have two types of functional domains: signal input domains to receive the signal from the upstream pathway components with which they interact and signal output or activation domains to transmit the signal to downstream components of the pathway (24). To look for the signal input domains, we employed GST pull-down assays to map the GRIP1 binding domain. Among several GAC63 fragments synthesized in vitro, fragments of the N-terminal region (amino acids 1 to 200 and 200 to 370) bound to GST-GRIP1(5-479) but a GAC63 C-terminal fragment (amino acids 370 to 567) failed to bind (Fig. 6A). Thus, GAC63 has two independent binding sites for GRIP1, located in the N-terminal and central regions of GAC63, which serve as signal input domains.
FIG. 6.
GRIP1 binding and activation domains of GAC63. (A) The indicated GAC63 fragments were translated in vitro and tested in GST pull-down assays for binding to GST or GST-GRIP1(5-479). (B) CV-1 cells were transfected with 200 ng of GK1-LUC reporter plasmid and 200 ng of the indicated pM plasmid encoding Gal4 DBD or Gal4 DBD fused to GAC63 or the indicated GAC63 fragment. Cell extracts were tested for luciferase activity and also by immunoblotting using anti-GAC63 antibodies. RLU, relative light units.
Plasmids encoding GAC63 fragments fused to Gal4-DBD were cotransfected with the Gal4-responsive GK1-LUC (luciferase) reporter plasmid to look for GAC63 domains with autonomous activation activity. Full-length GAC63 and the N-terminal and central fragments of GAC63 did not show any autonomous transcription activity. However, the C-terminal fragment of GAC63(468-567), containing the acidic region, exhibited a weak autonomous activation activity (Fig. 6B). Gal4 DBD fused to GAC63 fragments 130 to 567, 382 to 567, and 468 to 567, all expressed at similar protein levels, while expression of Gal4 DBD fused to full length GAC63 was four to five times lower (Fig. 6B). The Gal4 DBD fusions of full-length GAC63 and GAC63(130-567) failed to activate the reporter gene even when the amount of plasmid encoding these proteins was increased twofold or fourfold (data not shown). The lack of activity of full-length GAC63 and the longer GAC63 C-terminal fragments in this assay suggests that the C-terminal activation domain is regulated by other domains of GAC63. The C-terminal activation domain of GAC63 may serve as a signal output domain.
Recruitment of GAC63 to an estrogen-regulated promoter.
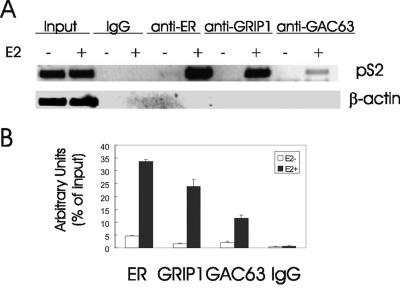
GAC63 binds to some NRs and to NR coactivator GRIP1 (Fig. 2 and 3) and functions as an NR coactivator when GAC63 is overexpressed in transient transfection assays (Fig. 4 and 5). To test whether endogenous GAC63 is recruited to a native, chromosomally integrated, estrogen-regulated promoter in response to E2, we employed ChIP assays to examine the pS2 promoter in MCF-7 breast cancer cells. As previously reported (11, 21), ERα and GRIP1 were recruited to the pS2 promoter region after a 60-min E2 treatment. Similarly, endogenous GAC63 was also recruited to this promoter in a hormone-dependent manner (Fig. 7A). Normal IgG served as a negative control, and the input chromatin levels from hormone-treated and untreated cells were equivalent. The recruitment of ER, GRIP1, and GAC63 was specific for the pS2 promoter, since PCRs with primers for the β-actin coding region failed to produce a signal from the same immunoprecipitated chromatin fractions. Real-time QPCR analysis confirmed the hormone-dependent recruitment of endogenous GAC63 to the native, chromosomally integrated pS2 gene promoter (Fig. 7B).
FIG. 7.
Recruitment of endogenous GAC63 to an endogenous E2-regulated promoter. (A) MCF-7 cells were grown with or without E2 for 60 min and analyzed by ChIP assays, using the indicated antibodies for immunoprecipitation and primers flanking the estrogen response element in the pS2 promoter for PCR analysis of the immunoprecipitated DNA. The results shown are representative of five independent experiments. (B) Real-time QPCR analysis of immunoprecipitated MCF-7 chromosomal DNA using primers for the pS2 promoter was performed with 2 μl of 1:10-diluted samples. The results are shown as percentage of input, are the mean and standard deviation from triplicate reactions, and are representative of two independent experiments.
GAC63 is required for efficient gene activation by ER.
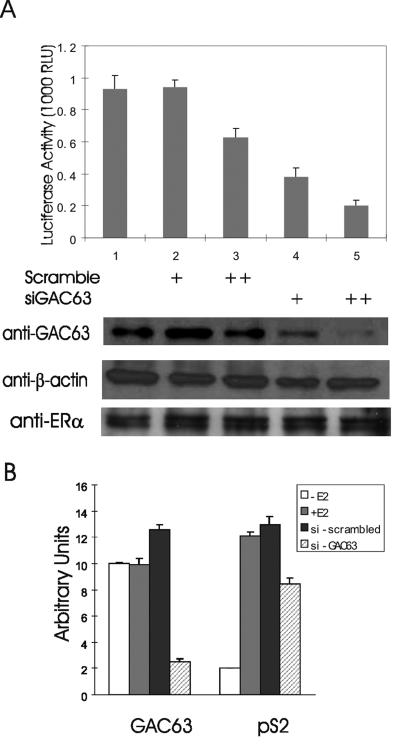
Overexpression of GAC63 enhances ER-regulated reporter gene expression (Fig. 4). To test for a physiological role of endogenous GAC63 in the process of transcriptional activation by endogenous ER, we employed siRNA experiments to decrease the expression of endogenous GAC63. MCF-7 cells were transfected with an ER-responsive reporter plasmid, with or without GAC63-specific siRNA or scrambled-sequence siRNA, and treated with E2. The GAC63 siRNA specifically reduced the level of endogenous GAC63 protein but had no detectable effect on the level of endogenous β-actin or ERα protein (Fig. 8A, lower panels). The scrambled-sequence siRNA did not affect the level of GAC63, β-actin, or ERα protein. The expression of the transiently transfected estrogen-dependent reporter gene was inhibited by 60 to 80% by two different amounts of the GAC63-directed siRNA but only 0 to 40% by equivalent amounts of the scrambled-sequence siRNA (Fig. 8A, upper panel). Reduction of endogenous GAC63 levels by siRNA also compromised the induction of endogenous pS2 gene expression by E2, as observed by RT-QPCR (Fig. 8B). The effect on endogenous pS2 gene expression was specific, since the results shown are normalized to the level of GAPDH transcripts. These data indicate that endogenous GAC63 makes important contributions to the ability of endogenous ER and coactivators to activate transcription of target genes.
FIG. 8.
Requirement of endogenous GAC63 for ER-mediated gene expression. (A) MCF-7 cells were transfected with 200 ng of MMTV(ERE)-LUC reporter plasmid and 90 (+) or 120 (++) pmol of siRNA duplex against GAC63 or scrambled-sequence siRNA duplex. After 48 h, cells were treated with 100 nM E2 and then harvested after an additional 24 h for luciferase assays (upper panel). Cell extracts were also tested by immunoblotting using anti-GAC63 antibody; the membrane was stripped and reprobed with antibody against β-actin (lower panels). In a similar experiment, in which GAC63 expression was dramatically reduced, ERα protein levels were measured (lower panels). The results shown are representative of four independent experiments. RLU, relative light units. (B) MCF-7 cells were transfected with 120 pmol of siRNA duplex against GAC63 or scrambled-sequence siRNA duplex. After 72 h, cells were treated with 100 nM E2 or left untreated for an additional 24 h before harvest. mRNAs were analyzed by RT-QPCR to measure the levels of GAC63, pS2, and GAPDH transcripts. Results shown for GAC63 and pS2 transcripts are normalized by the levels of GAPDH transcripts and are representative of four independent experiments.
DISCUSSION
GAC63 is a novel NR coactivator.
Hormone-activated NRs bound to their specific enhancer elements transmit their activating signal through a complex array of coactivator proteins. Each coactivator complex functions as a branched signal transduction pathway to conduct the activating signal to multiple specific targets in the chromatin and transcription machinery (8, 20, 24). The number of putative nuclear receptor coactivators discovered over the last decade has now exceeded 200 (B. W. O'Malley and R. M. Evans, Nuclear Receptor Signaling Atlas; http://www.nursa.org/index.cfm), suggesting that the coactivator machinery responsible for this signal transduction is extremely large and complex. While the specific molecular contributions and physiological relevance of most of these proteins remain unknown, analysis of the more well-studied coactivators indicates that each complex of coactivators and each component within a specific coactivator complex have specific tasks to perform. For example, the Swi/Snf and p160 coactivator complexes both contribute to chromatin remodeling, but by different mechanisms; and the TRAP/DRIP/mediator complex apparently helps to recruit and activate RNA polymerase II.
In this study, we have identified GAC63 as a new NR coactivator which associates with and cooperates synergistically with the p160 coactivators. Overexpressed GAC63 enhances NR function in transient transfection assays (Fig. 4 to 5). In addition, endogenous GAC63 is recruited, along with ERα and GRIP1, in an E2-dependent manner to the estrogen-responsive pS2 promoter (Fig. 7). Reduction of the endogenous GAC63 level compromises the ability of hormone-activated ERα to activate gene transcription (Fig. 8). By these criteria, GAC63 is a physiologically relevant NR coactivator.
The human orthologue of GAC63, HUEL, is expressed in a wide range of human fetal and adult tissues and cancer cell lines (22). The presence of highly homologous cDNA sequences in Drosophila suggests that GAC63/HUEL is conserved over large evolutionary distances. GAC63/HUEL has several potential nuclear localization signals and is primarily cytoplasmic in interphase cells, but it undergoes translocation to the nucleus during the S phase of the cell cycle (23). The mouse chromosomal location is 5C3.1 (from LocusLink on the NCBI website), and the human HUEL gene is located at chromosome 4p12-13, a common site of loss of heterozygosity in a variety of cancers (22). The N-terminal region of HUEL (amino acids 127 to 196) has substantial homology with the minimal DNA binding region (amino acids 139 to 219) of the XPA protein (23), which is involved in DNA repair and is one of the proteins mutated in xeroderma pigmentosum. While the importance of this homology remains to be investigated, the highly conserved nature of this region between GAC63/HUEL and XPA, along with the highly conserved nature of this region in XPA proteins from diverse eukaryotic species (23), suggests an important function for this domain.
Mechanism of GAC63 coactivator function.
While GAC63 bound directly to ERα and AR (Fig. 3), its functional dependence on GRIP1 and on the N terminus of GRIP1 which binds GAC63 (Fig. 4 and 5) suggests that GAC63 acts as a secondary coactivator; i.e., the interaction of GAC63 with NRs appears insufficient for its coactivator function, while the interaction with GRIP1 is essential. The interaction of GAC63 with the N-terminal region of GRIP1 is mediated by two different regions of GAC63, the N-terminal and central regions, which can independently bind to GRIP1 (Fig. 6A). However, an N-terminal deletion mutant of GAC63 lacking amino acids 1 to 183 still functions in cooperation with GRIP1 as a coactivator for ER (data not shown). Thus, the GRIP1 binding site in the N-terminal region of GAC63, which contains a zinc finger-like motif, is not essential for the coactivator function of GAC63; this implies that the GRIP1 binding site in the central region of GAC63 is sufficient to anchor GAC63 to GRIP1 and thus indirectly to the DNA-bound NR. Perhaps the two GRIP1 binding domains represent redundant functions in this particular assay system. An LXXLL motif in the C-terminal region of GAC63 (amino acids 461 to 465) and other related sequences in the central region could represent an NR binding site(s). The importance of these for GAC63 coactivator activity remains to be determined.
The fact that reduction of the endogenous level of GAC63 compromises the activity of ERα (Fig. 8) suggests that GAC63 activates unique downstream targets that are important for transcriptional activation. This conclusion is also supported by the specific coactivator synergy between GAC63 and CARM1 (Fig. 5), which would not be expected if GAC63 and CARM1 shared the same downstream targets. The weak activation domain at the C terminus of GAC63 (Fig. 6B) may function as a signal output domain which contacts a downstream component of the signaling pathway or the transcription machinery.
Multiple contributions of the p160 coactivator complex to the transcriptional activation process.
The large size and high degree of conservation of the bHLH-PAS domains of p160 coactivators (350 amino acids with 60% amino acid sequence identity among the three family members) suggest that this domain has many different interaction surfaces. In fact, several different proteins have already been reported to interact physically and functionally with p160 bHLH-PAS domains. Among these, proteins such as MEF-2C and TEF4 are DNA-binding transcriptional activator proteins which use p160 proteins as primary coactivators (3, 7); the sites they bind to on the p160 coactivators serve as signal input domains. In contrast, the requirement for the N-terminal bHLH-PAS domain of GRIP1 for its coactivator function demonstrates that this region also serves as a third activation or signal output domain (designated AD3) for p160 coactivators (11); AD3 complements the two C-terminal activation domains, AD1 and AD2, which bind CBP/p300 and CARM1/PRMT1, respectively. The secondary coactivators that mediate downstream signaling for the AD3 domain include CoCoA (11), Fli-I (15), and GAC63, which are apparently recruited to the promoter through their interaction with the bHLH-PAS domains of p160 coactivators. Three other proteins with properties of coactivators also bind to the N-terminal bHLH-PAS regions of p160 coactivators: hMMS19, an ER AF-1 coactivator (28); cyclin T1, a component of the transcription elongation factor p-TEFb (12); and BAF57, a subunit of the Swi/Snf complex (2). ANCO-1, which represses the activity of hormone-activated NRs, has also recently been reported to bind p160 bHLH-PAS domains (31). Thus, the bHLH-PAS domains of p160 coactivators are versatile protein interaction domains containing many interaction surfaces which serve diverse functions.
The fact that CoCoA, Fli-I, and GAC63 have no obvious sequence homology and completely different structural elements—CoCoA has a large coiled-coil domain, Fli-I contains gelsolin-like motifs and leucine-rich repeats, and GAC63 has a zinc finger-like motif and leucine zipper-like motifs—suggests that they recognize different surfaces or epitopes of the p160 N-terminal domain and contribute as coactivators by activating different downstream targets in the chromatin and transcription machinery. The nature of the downstream targets and the specific components of the transcription machinery which are regulated by CoCoA, Fli-I, and GAC63 are currently under investigation in our lab.
Acknowledgments
We thank Daniel Gerke for expert technical assistance and Peter Kushner (University of California, San Francisco) for the plasmids encoding ERα and its fragments.
This work was supported by grant DK43093 to M.R.S. from the U.S. National Institutes of Health. Y.-H.C. and J.H.K. were supported by predoctoral fellowships from the University of Southern California/Norris Breast Cancer Research Training Program.
REFERENCES
- 1.Anzick, S. L., J. Kononen, R. L. Walker, D. O. Azorsa, M. M. Tanner, X.-Y. Guan, G. Sauter, O.-P. Kallioniemi, J. M. Trent, and P. S. Meltzer. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965-968. [DOI] [PubMed] [Google Scholar]
- 2.Belandia, B., R. L. Orford, H. C. Hurst, and M. G. Parker. 2002. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 21:4094-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belandia, B., and M. G. Parker. 2000. Functional interaction between the p160 coactivator proteins and the transcriptional enhancer factor family of transcription factors. J. Biol. Chem. 275:30801-30805. [DOI] [PubMed] [Google Scholar]
- 4.Chen, D., S.-M. Huang, and M. R. Stallcup. 2000. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J. Biol. Chem. 275:40810-40816. [DOI] [PubMed] [Google Scholar]
- 5.Chen, D., H. Ma, H. Hong, S. S. Koh, S.-M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174-2177. [DOI] [PubMed] [Google Scholar]
- 6.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 7.Chen, S. L., D. H. Dowhan, B. M. Hosking, and G. E. Muscat. 2000. The steroid receptor coactivator, GRIP-1, is necessary for MEF-2C-dependent gene expression and skeletal muscle differentiation. Genes Dev. 14:1209-1228. [PMC free article] [PubMed] [Google Scholar]
- 8.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 9.Gu, Y. Z., J. B. Hogenesch, and C. A. Bradfield. 2000. The PAS superfamily: sensors of environmental and developmental signals. Annu. Rev. Pharmacol. Toxicol. 40:519-561. [DOI] [PubMed] [Google Scholar]
- 10.Huang, Z. J., I. Edery, and M. Rosbash. 1993. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature 364:259-262. [DOI] [PubMed] [Google Scholar]
- 11.Kim, J. H., H. Li, and M. R. Stallcup. 2003. CoCoA, a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol. Cell 12:1537-1549. [DOI] [PubMed] [Google Scholar]
- 12.Kino, T., O. Slobodskaya, G. N. Pavlakis, and G. P. Chrousos. 2002. Nuclear receptor coactivator p160 proteins enhance the HIV-1 long terminal repeat promoter by bridging promoter-bound factors and the Tat-P-TEFb complex. J. Biol. Chem. 277:2396-2405. [DOI] [PubMed] [Google Scholar]
- 13.Koh, S. S., D. Chen, Y.-H. Lee, and M. R. Stallcup. 2001. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J. Biol. Chem. 276:1089-1098. [DOI] [PubMed] [Google Scholar]
- 14.Kumar, V., S. Green, G. Stack, M. Berry, J.-R. Jin, and P. Chambon. 1987. Functional domains of the human estrogen receptor. Cell 51:941-951. [DOI] [PubMed] [Google Scholar]
- 15.Lee, Y.-H., H. D. Campbell, and M. R. Stallcup. 2004. Developmentally essential protein Flightless I is a nuclear receptor coactivator with actin binding activity. Mol. Cell. Biol. 24:2103-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, Y.-H., S. S. Koh, X. Zhang, X. Cheng, and M. R. Stallcup. 2002. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol. Cell. Biol. 22:3621-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma, H., Y. Shang, D. Y. Lee, and M. R. Stallcup. 2003. Study of nuclear receptor-induced transcription complex assembly and histone modification by chromatin immunoprecipitation. Methods Enzymol. 364:284-296. [DOI] [PubMed] [Google Scholar]
- 18.Mangelsdorf, D. J., and R. M. Evans. 1995. The RXR heterodimers and orphan receptors. Cell 83:841-850. [DOI] [PubMed] [Google Scholar]
- 19.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 21.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 22.Sim, D. L., and V. T. Chow. 1999. The novel human HUEL (C4orf1) gene maps to chromosome 4p12-p13 and encodes a nuclear protein containing the nuclear receptor interaction motif. Genomics 59:224-233. [DOI] [PubMed] [Google Scholar]
- 23.Sim, D. L., W. M. Yeo, and V. T. Chow. 2002. The novel human HUEL (C4orf1) protein shares homology with the DNA-binding domain of the XPA DNA repair protein and displays nuclear translocation in a cell cycle-dependent manner. Int. J. Biochem. Cell Biol. 34:487-504. [DOI] [PubMed] [Google Scholar]
- 24.Stallcup, M. R., J. H. Kim, C. Teyssier, Y. H. Lee, H. Ma, and D. Chen. 2003. The roles of protein-protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators. J. Steroid Biochem. Mol. Biol. 85:139-145. [DOI] [PubMed] [Google Scholar]
- 25.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear receptor function. Nature 387:677-684. [DOI] [PubMed] [Google Scholar]
- 26.Tsai, M.-J., and B. W. O'Malley. 1994. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 63:451-486. [DOI] [PubMed] [Google Scholar]
- 27.Webb, P., G. N. Lopez, R. M. Uht, and P. J. Kushner. 1995. Tamoxifen activation of the estrogen receptor/AP-1 pathway: potential origin for the cell-specific estrogen-like effects of antiestrogens. Mol. Endocrinol. 9:443-456. [DOI] [PubMed] [Google Scholar]
- 28.Wu, X., H. Li, and J. D. Chen. 2001. The human homologue of the yeast DNA repair and TFIIH regulator MMS19 is an AF-1-specific coactivator of estrogen receptor. J. Biol. Chem. 276:23962-23968. [DOI] [PubMed] [Google Scholar]
- 29.Xu, J., and Q. Li. 2003. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol. Endocrinol. 17:1681-1692. [DOI] [PubMed] [Google Scholar]
- 30.Yao, T.-P., G. Ku, N. Zhou, R. Scully, and D. M. Livingston. 1996. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc. Natl. Acad. Sci. USA 93:10626-10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, A., P. L. Yeung, C. W. Li, S. C. Tsai, G. K. Dinh, X. Wu, H. Li, and J. D. Chen. 2004. Identification of a novel family of ankyrin repeats containing cofactors for p160 nuclear receptor coactivators. J. Biol. Chem. 279:33799-33805. [DOI] [PubMed] [Google Scholar]