Abstract
Embryonic stem cells (ESCs) are pluripotent cells that can either self-renew or differentiate into many cell types. Oct4 and Sox2 are transcription factors essential to the pluripotent and self-renewing phenotypes of ESCs. Both factors are upstream in the hierarchy of the transcription regulatory network and are partners in regulating several ESC-specific genes. In ESCs, Sox2 is transcriptionally regulated by an enhancer containing a composite sox-oct element that Oct4 and Sox2 bind in a combinatorial interaction. It has previously been shown that Pou5f1, the Oct4 gene, contains a distal enhancer imparting specific expression in both ESCs and preimplantation embryos. Here, we identify a composite sox-oct element within this enhancer and show that it is involved in Pou5f1 transcriptional activity in ESCs. In vitro experiments with ESC nuclear extracts demonstrate that Oct4 and Sox2 interact specifically with this regulatory element. More importantly, by chromatin immunoprecipitation assay, we establish that both Oct4 and Sox2 bind directly to the composite sox-oct elements in both Pou5f1 and Sox2 in living mouse and human ESCs. Specific knockdown of either Oct4 or Sox2 by RNA interference leads to the reduction of both genes' enhancer activities and endogenous expression levels in addition to ESC differentiation. Our data uncover a positive and potentially self-reinforcing regulatory loop that maintains Pou5f1 and Sox2 expression via the Oct4/Sox2 complex in pluripotent cells.
Embryonic stem cells (ESCs) are derived from the inner cell mass (ICM) of the mammalian blastocyst. They are able to undergo self-renewing cell division under specific cell culture conditions for extended periods, thereby maintaining their pluripotency (22, 43). This pluripotency is best displayed by their ability to give rise to all embryonic lineages subsequent to their reintroduction into the blastocyst. In addition, ESCs can also differentiate into a variety of different cell types when cultured in vitro (17, 19, 20, 40, 52). This property of ESCs, particularly for human ESCs, holds great promise for regenerative therapeutic medicine (14, 32).
Several key regulators have been identified that are essential both for the formation of the ICM during mouse preimplantation development and for self-renewal of pluripotent ESCs (3, 10, 24, 26, 41). These regulators include Oct4, Sox2, and Nanog. Oct4 (also known as Oct3 and encoded by Pou5f1) is a POU domain-containing transcription factor that binds to an octamer sequence, ATGCAAAT. During mouse preimplantation development, zygotic Pou5f1 expression is activated at the four-cell stage and is later restricted to the pluripotent cells of the ICM and epiblast. In the mouse postimplantation embryo, Pou5f1 expression is down-regulated upon epiblast differentiation and its expression is maintained only in the primordial germ cells (51). In addition, Oct4 is highly expressed in human and mouse ESCs, and its expression diminishes when these cells differentiate and lose pluripotency (31). Several target genes of Oct4 in ESCs have been identified, and these include Fgf4, Utf1, Opn, Rex1/Zfp42, Fbx15, and Sox2 (5, 6, 8, 12, 27, 46, 47, 53). The regulatory regions of these genes contain an octamer element capable of binding Oct4, at least in an in vitro setting. These sites have been shown to be important for transcriptional activity of their respective genes as indicated by comparisons of octamer mutant and wild-type constructs in reporter assays.
The octamer elements within the enhancers of Fgf4, Utf1, Opn, Fbx15, and Sox2 are found in proximity to Sox2-binding sox elements. Sox2 (SRY-related HMG box 2) is an HMG domain-containing transcription factor essential for pluripotent cell development (3). Sox2 has an expression pattern similar to that of Pou5f1 through mouse preimplantation development, as it is expressed in all blastomeres of the four-cell embryo and becomes restricted to the ICM and epiblast of the blastocyst (3). These two factors are also expressed in ESCs. Of the Sox2-Oct4 target genes, all but Opn have the octamer and sox heptamer elements separated by either 0 or 3 bp. Such proximity suggests that these factors may interact with each other. Indeed, two structures have recently been solved for a POU/HMG ternary complex bound to composite sox-oct elements; one of these is on an element separated by 3 bp (36) and the other is on an element separated by 0 bp (50). Both reveal that the POU and HMG domains mediate specific protein-protein and DNA-protein interactions. In addition, it has also been demonstrated that Sox2 and Oct4 can interact in the absence of DNA and that the HMG and POU domains are involved in this interaction (2). Hence, Oct4 and Sox2 are capable of forming heterodimers both on and off the DNA.
Despite the limited number of Oct4 target genes, we can nevertheless place Oct4 upstream in the hierarchy of the ESC-specific gene regulatory network. Therefore, elucidating the mechanisms behind the transcriptional regulation of Pou5f1 is of considerable interest (33). Previous studies have defined regulatory regions that are important for driving Pou5f1 expression in different cell types of the early mouse embryo through the analysis of lacZ reporter genes under the control of different mouse Pou5f1 genomic fragments (51). These regions include the core promoter which is located within the first 250 bp of the transcription initiation site. A proximal enhancer, located about 1.2 kb upstream (−1524 to −30), is responsible for Pou5f1 expression in the epiblast, and a distal enhancer region (located about 2 kb upstream) drives expression in the morula, ICM, and primordial germ cells. This distal enhancer is also required for ESC-specific expression. Alignment between the upstream sequences of human, bovine, and mouse Pou5f1 promoters revealed four conserved regions (CR1 through CR4) (30). Interestingly, CR4 overlaps with the distal enhancer, suggesting that evolutionarily conserved elements may be regulating the activity of the distal enhancer. To date, the factors that bind and regulate these key regulatory elements have not been identified.
Two regulatory regions (SRR1 and SRR2) in Sox2 are known to confer ESC-specific expression (47). SRR2, located 1.2 kb downstream of the transcription start site, contains the composite sox-oct element. Mutations within this composite element disrupted the in vitro formation of a DNA/protein complex and additionally resulted in the loss of SRR2 enhancer activity. More importantly, the reduction of Pou5f1 abolished the SRR2 activity, indicating that Oct4 can positively regulate SRR2, but whether Oct4 and Sox2 are indeed bound to SRR2 in ESCs remains to be demonstrated. In addition, the necessity of Sox2 binding to the SRR2 region for the transcriptional activity of Sox2 has not been established. Nevertheless, this still strongly suggests that Sox2 is the downstream target of Oct4 (and possibly Sox2 itself). Interestingly, Oct4 has recently been shown by chromatin immunoprecipitation (ChIP) to bind to an octamer site within SRR1 of Sox2 (8).
In this study, we show that Oct4 and Sox2 bind to important regulatory elements of Pou5f1 and Sox2 as a binary complex in living human and mouse ESCs. Using RNA interference (RNAi), we further examine the functional role exerted by this complex on these regulatory elements and the corresponding expression of their endogenous genes. Our data support a model in which the Oct4/Sox2 complex plays a key role in maintaining the expression of essential transcription factors in ESCs through autoregulatory and multicomponent loop network motifs. What emerges is a genetic regulatory network linking the key transcription factors in ESCs.
MATERIALS AND METHODS
Cell culture.
293T cells were cultured in Dulbecco's modified Eagle's medium (GIBCO) supplemented with 10% heat-inactivated fetal bovine serum (GIBCO) and maintained at 37°C with 5% CO2. E14 mouse ESCs, cocultured with mouse primary embryonic fibroblast feeder in 0.1% gelatin-coated flasks, were maintained in Dulbecco's modified Eagle's medium (GIBCO), supplemented with 15% heat-inactivated fetal bovine serum, 0.055 mM 2-β-mercaptoethanol (GIBCO), 2 mM l-glutamine (GIBCO), 0.1 mM minimal essential medium with nonessential amino acids (GIBCO), 5,000-U/ml penicillin-streptomycin (GIBCO), and 1,000 U/ml of leukemia inhibitory factor (LIF) (Chemicon). For certain experiments, the ESCs were cultured under feeder-free conditions. In differentiation experiments, the cells were treated with 0.5 μM of retinoic acid (RA). The human ESC line HUES-6 (obtained from Doug Melton, Harvard University) was passaged according to the method of Cowan et al. (11).
RNAi design and construction of plasmids for shRNA synthesis.
Nineteen-base-pair gene-specific regions for RNA interference were designed based on the work of Reynolds et al. and Ui-Tei et al. (37, 48). Oligonucleotides were cloned into pSUPER.puro (BglII and HindIII sites; Oligoengine), which expresses 19-nucleotide hairpin-type short hairpin RNAs (shRNAs) with a 9-nucleotide loop, as described previously (7). All sequences were analyzed by BLAST search to ensure that they did not have significant sequence similarity with other genes. The oligonucleotides used were as follows: for Gfp RNAi, 5′-GATCCCCGAACGGCATCAAGGTGAACTTCAAGAGAGTTCACCTTGATGCCGTTCTT TTTA-3′ and 5′-AGCTTAAAAAGAACGGCATCAAGGTGAACTCTCTTGAAGTTCACCTTGATGCCGTTCGGG-3′; for Pou5f1 RNAi, 5′-GATCCCC GAAGGATGTGGTTCGAGTATTCAAGAGATACTCGAACCACATCCTTCTTTTTA-3′ and 5′-AGCTTAAAAAGAAGGATGTGGTTCGAGTATCTCTTGAATACTCGAACCACATCCTTCGGG-3′; for Sox2 RNAi, 5′-GATCCCCGAAGGAGCACCCGGATTATTTCAAGAGAATAATCCGGGTGCTCCTTCTTTTTA-3′ and 5′-AGCTTAAAAAGAAGGAGCACCCGGATTATTCTCTTGAAATAATCCGGGTGCTCCTTCGGG-3′. For the Oct4 RNAi target sequence, the Reynolds score and Ui-Tei class value were 6 and class Ib, respectively. For the Sox2 RNAi target sequence, these were 5 and class Ia, respectively.
Luciferase reporter constructs.
For human POU5F1-Luc, a 3-kb fragment of the human POU5F1 promoter was cloned into pGL3-Basic plasmid (BglII and NcoI sites) upstream of the firefly luciferase gene (Promega). The ΔCR4-A construct contained a 27-bp deletion of AGGCCTGCCCCTCCCCCTCCTCTGAGA; the ΔCR4-B construct contained a 22-bp deletion of AGAGAGATGCATGACAAAGGTG (the oct-sox motif is underlined); the ΔCR4-C construct contained a 25-bp deletion of CTGGGGAGGGGCCTCCTCCTGTTCC; the ΔCR4 construct contained a 1,028-bp deletion of XhoI and SalI fragments. All these deletions were introduced into the human POU5F1-Luc plasmid.
For the Sox2-SV40-Luc construct, a 461-bp fragment of genomic DNA containing the SRR2 enhancer was amplified and cloned into a pGL3 promoter plasmid (MluI and BglII sites) containing the simian virus 40 (SV40) promoter upstream of a firefly luciferase gene (Promega). The Luc-Sox2 open reading frame (ORF) construct was generated by cloning the Sox2 open reading frame (NM_011443) into XhoI and NotI sites downstream of the Renilla luciferase gene (psiCHECK-2; Promega). Similarly, the Luc-Pou5f1 ORF construct was generated by cloning the Pou5f1 open reading frame (NM_013633) into XhoI and NotI sites downstream of the Renilla luciferase gene (psiCHECK-2; Promega).
The cloning primers used are as follows: for Sox2-SV40-Luc, 5′-CGCTTTACGCGTCTGCCCTTCAGCCGAGTACCG-3′ and 5′-TTGGCTAGATCTGGAGTTCCGGGAATATCCTCC-3′; for Luc Sox2 ORF, 5′-TGAAAACTCGAGATGTATAACATGATGGAG-3′ and 5′-TTTTCAGCGGCCGCCATGTGCGACAGGGGCAG-3′; for Luc Pou5f1 ORF, 5′-TGAAAACTCGAGATGGCTGGACACCTGGCTTCAG-3′ and 5′-TTTTCAGCGGCCGCACCCCAAAGCTCCAGGTTCTCT-3′.
Luciferase reporter assays.
To assay for knockdown in 293T cells, 5 ng of plasmids containing Renilla luciferase fused to ORF and 100 ng of shRNA plasmids (pSUPER.puro; Oligoengine) were transfected with Lipofectamine 2000 (Invitrogen) into 293FT cells. The cells were seeded at a density of 25,000 to 30,000/well in 96-well plates (Costar).
To study the effects of Oct4 and Sox2 knockdown on the Pou5f1 promoter, 100 ng of human POU5F1-Luc plasmid, 100 ng of shRNA plasmid (pSUPER.puro; Oligoengine), and 5 ng of plasmid containing Renilla luciferase (pRL-SV40; Promega) were transfected into E14 ESCs. The pRL-SV40 plasmid served as an internal control for normalizing the transfection efficiency. The E14 ESCs were cocultured with inactivated mouse primary embryonic fibroblasts.
For the Sox2 enhancer assay, E14 ESCs were transfected with 100 ng of plasmids containing the SRR2 enhancer activating firefly luciferase (see Fig. 7A), 100 ng of shRNA plasmid, and 5 ng of plasmids containing Renilla luciferase (pRL-SV40). Firefly and Renilla luciferase activities were measured 48 to 60 h after transfection with the Dual Luciferase System (Promega) using a Centro LB960 96-well luminometer (Berthold Technologies).
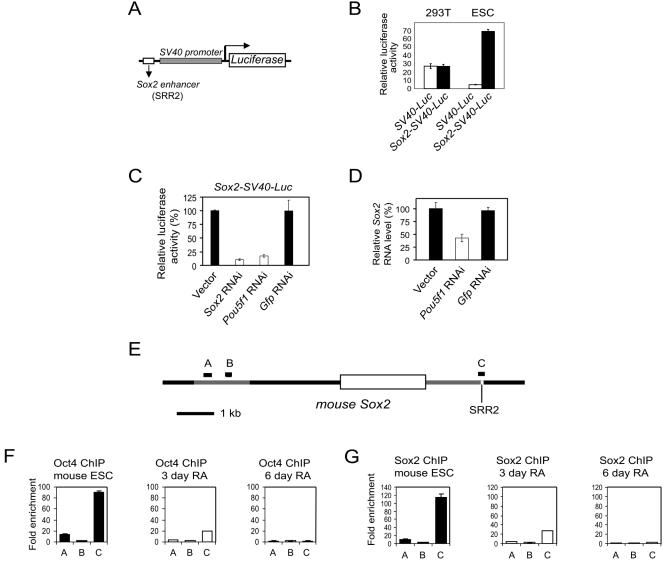
FIG. 7.
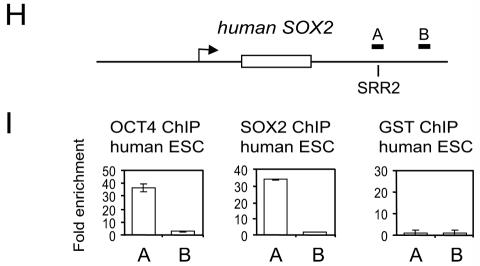
Oct4 and Sox2 bind to the 3′ enhancer of Sox2 and regulate its activity. (A) The reporter construct used to assay for Sox2 enhancer activity consisted of the Sox2 SRR2 site positioned 5′ to the SV40 promoter driving a luciferase reporter. (B) The SRR2 enhancer drives luciferase expression specific to ESC. (C) The Sox2 luciferase reporter construct was cotransfected into mouse ESCs with either empty vector or RNAi constructs directed against Sox2, Pou5f1, or Gfp RNAi vector. The knockdown effect is measured by relative luciferase activity 60 h after transfection, with the empty vector set at 100%. The standard deviations are shown. (D) Endogenous Sox2 mRNA levels in ESCs were measured by real-time PCR after transfection with the respective RNAi constructs containing a puromycin resistance gene. Cells were harvested 2 days posttransfection after continuous puromycin selection. Endogenous Sox2 levels are expressed relative to the empty vector control. Standard deviations are shown. (E) Schematic of mouse Sox2 genomic locus with the single exon represented by an open box and the SRR2 region containing the composite sox-oct element indicated. The relative locations of the amplicons used to detect enriched ChIP fragments are shown (A to C). (F) Measurement by ChIP analysis of Oct4 occupancy in regions of Sox2 in undifferentiated mouse ESCs and those induced to differentiate for 3 and 6 days by RA. Letters correspond to the amplicons indicated in panel E. Standard deviations are shown. (G) Similar to panel F but for Sox2 occupancy. (H) Schematic of human SOX2 genomic locus with the single exon represented by an open box and the SRR2 region containing the composite sox-oct element indicated. The relative locations of the amplicons used to detect enriched ChIP fragments are shown (A and B). (I) Measurement by ChIP analysis of OCT4 and SOX2 occupancies on SOX2 in living human ESCs. A glutathione S-transferase antibody (GST) was used as a negative control. Standard deviations are shown.
RNA preparation/reverse transcription/real-time PCR analysis.
To examine the effects of knocking down endogenous Pou5f1 or Sox2, E14 ESCs were cotransfected with 3.5 μg of shRNA plasmids and 0.5 μg of Gfp plasmids in 60-mm plates at 50% confluency. Selection by puromycin (Sigma) was performed 1 day after transfection for a period of 2 days at a concentration of 1 μg/ml. Total RNA was isolated using Trizol reagent (Invitrogen) and purified with the RNeasy minikit (Qiagen). cDNA synthesis was performed with 500 ng of total RNA and the SuperScript II kit (Invitrogen) according to the manufacturer's instructions. Endogenous mRNA levels were measured by real-time PCR analysis based on SYBR Green detection with the ABI Prism 7900HT real-time PCR machine. Briefly, the real-time PCR mixture contained 2 μl of the reverse transcription reaction product in a total volume of 20 μl, consisting of 1× SYBR Green mix reagent (Applied Biosystems), 50 nM forward primer, and 50 nM reverse primer. Each sample was analyzed in duplicate. The real-time PCR primers used were as follows: for Gapd, 5′-GGTTGTCTCCTGCGACTTCAACAGC-3′ and 5′-CGAGTTGGGATAGGGCCTCTCTTGC-3′; for Pou5f1, 5′-TTGGGCTAGAGAAGGATGTGGTT-3′ and 5′-GGAAAAGGGACTGAGTAGAGTGTGG-3′; for Sox2, 5′-GCACATGAACGGCTGGAGCAACG-3′ and 5′-TGCTGCGAGTAGGACATGCTGTAGG-3′; for Brachury, 5′-CCAACCTATGCGGACAATTCATCTGC-3′ and 5′-GTGTAATGTGCAGGGGAGCCTCGAA-3′; for Nestin, 5′-AGAGGAAGAGCAGCAAGGCCATGAC-3′ and 5′-TCCCTGACTCTGCTCCTTCTTCTTCAT-3′; for Gata6, 5′-TGTGCAATGCATGCGGTCTCTACAGCA-3′ and 5′-TTCATAGCAAGTGGTCGAGGCACCC-3′; for Bmp2, 5′-CCAAGATGAACACAGCTGGTCACAGATAAGGC-3′ and 5′-AGGTGGTCAGCAAGGGGAAAAGGACACTCC-3′; for Cdx2, 5′-CGCAGAACTTTGTCAGTCCTCCGCAGTACC-3′ and 5′-GTATTCGGCGGGGCTGCTGTAGCCCATAGC-3′; for Hand1, 5′-CCTGCCCAAACGAAAAGGCTCAGGACCCAA-3′ and 5′-CGACCGCCATCCGTCTTTTTGAGTTCAGCC-3′.
ChIP assay.
ChIP assays with feeder-free E14 mouse ESCs were carried out as described previously (9). Briefly, cells were cross-linked with 1% formaldehyde for 10 min at room temperature and formaldehyde was then inactivated by the addition of 125 mM glycine. Chromatin extracts containing DNA fragments with an average size of 500 bp were immunoprecipitated using anti-Oct4 or anti-Sox2 polyclonal antibodies (Santa Cruz Biotechnology). For all ChIP experiments, quantitative PCR analyses were performed in real time using the ABI PRISM 7900 Sequence Detection System and SYBR Green Master Mix as described previously (25). Relative occupancy values were calculated by determining the apparent immunoprecipitation efficiency (ratios of the amount of immunoprecipitated DNA to that of the input sample) and normalized to the level observed at a control region, which was defined as 1.0. All ChIP experiments were repeated at least three times. For all the primers used, each gave a single product of the right size, as confirmed by agarose gel electrophoresis and dissociation curve analysis. These primers also gave no DNA product in the no-template control. The primers used for real-time PCR to quantitate the ChIP-enriched DNA are as follows: Fig. 2, region 1 (mouse Pou5f1), −4003/−3798 relative to translation start site, CAGAACATCTGGATTTGGGAAGAGACGTT and GAGCAGGGAAATCACTCGTGTTAGCATC; Fig. 2, region 2 (mouse Pou5f1), −3383/−3219, CATTATAGGTGTGGCATTCCGCATCTG and TGCCACAAACCACCTGTATTTTAGAACCA; Fig. 2, region 3 (mouse Pou5f1), −2396/−2160, AGGCTAGGGCACATCTGTTTCAAGCTAGT and CCCAGCAGTCCTGTCTGTATTCAATACC; Fig. 2, region 4 (mouse Pou5f1), −2098/−1928 (encompassing CR4-B), GGAACTGGGTGTGGGGAGGTTGTA and AGCAGATTAAGGAAGGGCTAGGACGAGAG; Fig. 2, region 5 (mouse Pou5f1), −1817/−1570, CATGACAGAGTGGAGGAAACGGAAGA and CTGCCCTTGAACTCCTGATAATTCTCCTG; Fig. 2, region 6 (mouse Pou5f1), −1412/−1163 (encompassing CR3), TGCTCTGGGCTTTTTGAGGCTGTGTGATT and TGGCGGAAAGACACTAAGGAGACGGGATT; Fig. 2, region 7 (mouse Pou5f1) −655/−412, TCTACCAACCTGGACAACACAAGATGGAA and GCCACTCCTCAGTTCTTGCTTACCCAC; Fig. 2, region 8 (mouse Pou5f1), −284/−43 (encompassing CR1), AGCAACTGGTTTGTGAGGTGTCCGGTGAC and CTCCCCAATCCCACCCTCTAGCCTTGAC; Fig. 2, region 9 (mouse Pou5f1), −54/+141, GGATTGGGGAGGGAGAGGTGAAACCGT and TGGAAGCTTAGCCAGGTTCGAGGATCCAC; Fig. 2, region 10 (mouse Pou5f1), +1274/+1478 (within intron 1), GGAGTCCCCTAGGAAGGCATTAATAGTTT and GGATTCTCTCGGCTTCAGACAGACTTT; Fig. 3, region A (human POU5F1), −2613/−2396, GGGGAACCTGGAGGATGGCAAGCTGAGAAA and GGCCTGGTGGGGGTGGGAGGAACAT; Fig. 3, region B (human POU5F1), −1779/−1563, CCTGCACCCCTCCACAAATCACTCGC and TGCAATCCCCTCAAAGACTGAGCCTCAGAC; Fig. 3, region C (human POU5F1), −237/−136, GAGGGGCGCCAGTTGTGTCTCCCGGTTTT and GGGAGGTGGGGGGAGAAACTGAGGCGAAGG; Fig. 7E and F, region A (mouse Sox2), −4199/−3964 relative to translation start site, ATTAGTCTGCTCTTCCTCGGAATGGTTGG and TGATGCTTGTTAAAAACGCTTCGCTCCT; Fig. 7E and F, region B (mouse Sox2), −3581/−3421, CCCTGTTCCAAGTCTCTTTCTGCTAGTCA and CACCGATTTCAATCCAACACCATCATAG; Fig. 7E and F, region C (mouse Sox2), +3515/+3664, TTTTCGTTTTTAGGGTAAGGTACTGGGAAG and CCACGTGAATAATCCTATATGCATCACAAT; Fig. 7H, region A (human SOX2), +3662/+3862 relative to translation start site, GGATAACATTGTACTGGGAAGGGACA and CAAAGTTTCTTTTATTCGTATGTGTGAGCA; Fig. 7H, region B (human SOX2), +4464/+4658, CCTCGGGATATTATTCTGCTCAATGA and TAACGCTCTTTATTTAAAGTATGTGGTGGG.
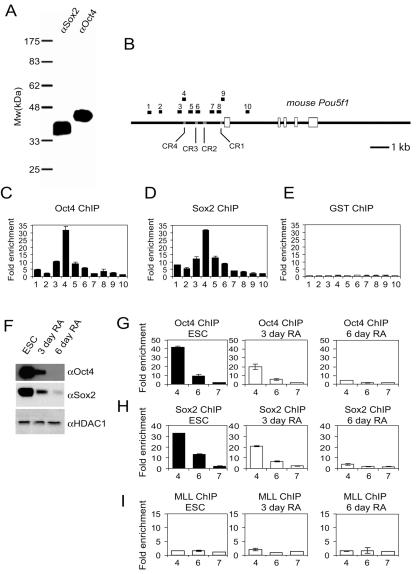
FIG. 2.
Oct4 and Sox2 bind to the distal enhancer of Pou5f1 in living mouse ESCs. (A) Specificity of the antibodies used in the ChIP assays was confirmed by Western blotting of mouse ESC nuclear extracts. (B) The locations of the amplified products (black boxes) of the primer sets used to detect the ChIP-enriched DNA fragments, shown within the context of the genomic structure of mouse Pou5f1. The locations of the conserved regions within the promoter are indicated. The composite sox-oct element is within CR4. Amplicons are numbered in order relative to their sites along the gene. Open boxes represent exons. (C to E) High-resolution mapping of Oct4 (C), Sox2 (D), and control (glutathione S-transferase antibody [GST]) (E) binding sites across the Pou5f1 promoter in mouse ESCs by ChIP analysis. Fold enrichment is the relative abundance of DNA fragments at the indicated regions (see panel B) over a control region as quantified by real-time PCR. Standard deviations are shown. (F to I) A similar analysis of Oct4 (G) and Sox2 (H) occupancy on the Pou5f1 promoter in undifferentiated mouse ESCs and in ESCs induced to differentiate with RA for 3 and 6 days. Oct4 and Sox2 levels are shown to decrease in these differentiating conditions as identified by Western blotting (F) with a histone deacetylase 1 antibody (αHDAC1) used as a loading control. ChIP analysis was used as a subset (4, 6, and 7) of the amplicons described in panel B. (I) A control ChIP assay using an anti-MLL antibody.
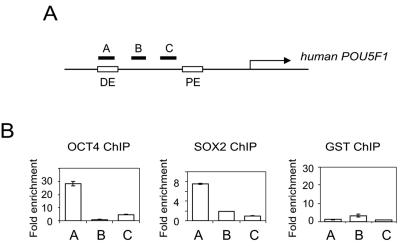
FIG. 3.
OCT4 and SOX2 bind to the distal enhancer/CR4 region of POU5F1 in living human ESCs. (A) Schematic of the location of the amplicons (A, B, and C) used to detect ChIP-enriched fragments in POU5F1 shown relative to the distal enhancer (DE)/CR4 region in which the composite sox-oct element resides, to the proximal enhancer (PE), and to the transcription start site (arrow). (B) Real-time PCR detection of enriched fragments from ChIP assays using OCT4, SOX2, and a control glutathione S-transferase (GST) antibody.
Electrophoretic mobility shift assays (EMSAs).
Nuclear extracts were prepared from E14 mouse ESCs grown under feeder-free conditions using the method of Dignam et al. (13) with modifications as described: cells were treated with trypsin for 1 min and harvested in cold phosphate-buffered saline. After centrifugation, cells were resuspended in cold buffer A (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], protease inhibitor cocktail [Roche]) and incubated on ice for 10 min. Cells were resuspended in buffer A and lysed with a Wheaton homogenizer. Pelleted nuclei were resuspended in buffer B (20 mM HEPES, pH 7.9, 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 1% protease inhibitor cocktail) and incubated on ice and vortexed for 15 s every 10 min, for a total of 30 min. After centrifugation, supernatants were dialyzed against dialysis buffer (20 mM HEPES, pH 7.9, 20% glycerol, 100 mM KCl, 0.83 mM EDTA, 1.66 mM DTT, protease inhibitor cocktail) at 4°C for 1 h. Extracts were then stored at −80°C. The concentrations of the nuclear extracts were determined by a NanoDrop ND-1000 spectrophotometer.
For EMSA double-stranded DNA oligonucleotides (Proligo) labeled with biotin at the 5′ termini of the sense strands were annealed with reverse strands in annealing buffer (10 mM Tris-HCl, pH 8.0, 50 mM NaCl, 1 mM EDTA) and purified with an agarose gel DNA extraction kit (Roche). The sense strand sequence is as follows: CR4-1S-CAGACAGCAGAGAGATGCATGACAAAGGTGCCGTGATGGTTC. The gel shift assays were performed using a LightShift Chemiluminescent EMSA kit (Pierce Biotechnologies). Four microliters (∼15 μg) of nuclear extract was added to a 10-μl reaction mixture (final) containing 3 ng of biotin-labeled oligonucleotide and 1 μg of poly(dG-dC) (Amersham). The final binding buffer composition was 60% dialysis buffer. Binding reaction mixtures were incubated for 10 min at room temperature. Where specified, antibodies (Santa Cruz) were added after the initial incubation for a further 20 min as follows: 1 μl of anti-Oct4 (sc-9081x), anti-Sox2 (sc-17320x), or anti-JunB (sc-46x). For cooperativity binding studies, the probe was labeled with 32P and the indicated amounts of overexpressed Oct4 or Sox2 were added for a further 20 min after the initial incubation. After separation of the different complexes on the gel, the amount of bound DNA for each complex was measured with a PhosphorImager (1). To calculate the predicted noncooperative interaction values, the amount of probe bound by a given concentration of Oct4 (in the absence of Sox2) was multiplied by the amount of probe bound by a given concentration of Sox2 (in the absence of Oct4). For example, the amount of ternary complex expected to form is 1% if 10% of the probe is bound by Oct4 and Sox2 independently. For competitive studies, a specified 600 ng of unlabeled double-stranded competitor was added for a further 20 min after the initial incubation. Binding reaction mixtures were resolved on prerun 5% native polyacrylamide gels in 0.5× Tris-buffered EDTA. Gels were transferred to Biodyne B nylon membranes (Pierce Biotechnologies) using Western blot techniques and detected using chemiluminescence.
RESULTS
Conserved elements within the CR4 region of Pou5f1 promoter.
Previous work comparing the genomic sequences of human, bovine, and mouse Pou5f1 identified four conserved regions within 3 kb upstream of its transcription start site (CR1 to CR4) (30). CR4 is of particular interest as this region maps to the previously characterized distal enhancer of Pou5f1, which has been shown to be responsible for both mouse ESC and early embryonic expression (51).
To enhance the phylogenetic footprint and aid us in identifying specific cis-regulatory elements within this CR4 region, we extended the comparative analysis to include three additional mammals (pig, dog, and rat). The six species in the alignment are eutherian mammals, all of which have an ICM at the blastocyst stage of development and presumably share similar genetic regulatory architecture in regulating their pluripotent phenotypes.
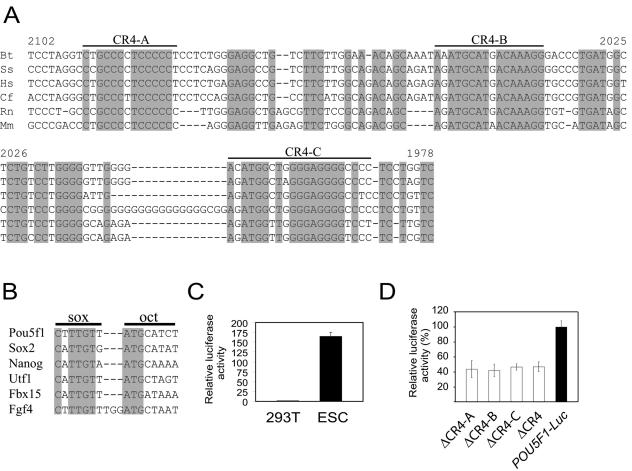
The alignment clearly showed three highly conserved consensus sites (CR4-A, CR4-B, and CR4-C) within the CR4 regions between the six species (Fig. 1A). The CR4-A consensus is C[T,C]GCCC[T,C]TCCCCC[T,C], and the CR4-B consensus is A[A,G]ATGCAT[G,A]ACAAAGG, while the CR4-C consensus is A[C,G]ATGG[C,T]T[G,A]GGGAGGGG[C,T]C[C,T][C,T]. Intriguingly, within the CR4-B region, we identified what clearly resembles the sox-oct composite element which is involved in regulating the transcription of several other ESC-specific genes (Fig. 1B). The sox element in the mouse sequence is identical to that found in Fgf4. Immediately adjacent to this sox element is a strong consensus sequence for an octamer element, differing only in the seventh position. Furthermore, this sox-oct composite element of Pou5f1 varies between species at only 2 of the 15 positions (Fig. 1A), and both of these positions are also the most variable positions between the known sox-oct target genes (Fig. 1B). This evidence suggested to us that Pou5f1 may also be a target of the Oct4/Sox2 complex.
FIG. 1.
(A) Conserved motifs within the distal enhancer of Pou5f1. Alignment of the CR4 region of Pou5f1 from genomic sequence from six eutherian mammals: Bos taurus (Bt), Sus scrofa (Ss), Homo sapiens (Hs), Canus familiaris (Cf), Rattus norvegicus (Rn), and Mus musculus (Mm). Invariant positions within this alignment are indicated by a shaded box. The numbering corresponds to the mouse sequence (Mm) and is relative to the translation start site. CR4-A, -B, and -C are regions discussed in the text. (B) Alignment of the composite sox-oct cis elements from the respective mouse target genes that are known to bind Sox2 and Oct4, including that described in this paper and present in the CR4-B region in panel A (complementary strand). (C) The 3-kb human POU5F1 promoter drives luciferase expression specific to ESC. (D) The effects of deleting the specific CR4-A, CR4-B, and CR4-C regions highlighted in panel A and of the deletion of the entire CR4 region, in the context of the 3-kb human POU5F1 promoter, on luciferase reporter activity in mouse ESCs. Activity is expressed relative to the wild-type construct (POU5F1-Luc).
To test the functional importance of these conserved consensus sites, we cloned a 3-kb promoter fragment from a bacterial artificial chromosome containing POU5F1 (human) and fused it upstream of a luciferase (Luc) gene. This promoter fragment contains all conserved regions (CR1 to CR4). This POU5F1-Luc reporter was then transfected into ES and 293T cells to test for ESC-specific expression. The reporter showed robust expression in ESCs but only background expression in 293T cells (Fig. 1C). Next, we generated stable ESC lines with this promoter fragment driving the enhanced green fluorescent protein gene. The undifferentiated cells were green when observed under the fluorescence microscope, indicating a high level of enhanced green fluorescent protein expression. However, upon differentiation with RA treatment, the level of fluorescence was drastically reduced (data not shown). Hence, consistent with a previous report on mouse Pou5f1 promoter characterization (51), our human reporter is also able to drive ESC-specific expression.
Next, we engineered four reporter constructs harboring deletions of the individual CR4-A, CR4-B, and CR4-C sites as well as one construct (ΔCR4) with deletions in all three CR4 sites. The promoter activities of these constructs were all reduced to 40% of that of the wild type (Fig. 1D). This indicates that each of the three CR4 motifs contributes to the overall CR4 activity. We focused on characterizing the role of DNA binding proteins that regulate Pou5f1 expression through CR4-B, the site in which the composite sox-oct element is contained.
Oct4 and Sox2 bind to the CR4 region in living mouse ESCs.
As the composite sox-oct element in the CR4-B region is conserved throughout eutherian mammals, suggesting functional significance, we examined whether Oct4 and Sox2 bind to this region in ESCs by using a ChIP assay. This assay allows us to investigate the targets of DNA binding proteins in living cells, as formaldehyde preserves the molecular interactions between DNA and protein through covalent cross-linking. We first tested the specificity of antibodies used in this study by Western blotting. The main antibodies used were N19 (anti-Oct4, sc-8628) and Y17 (anti-Sox2, sc-17320) affinity-purified polyclonal antibodies that were raised against peptides. Western blots of ESC nuclear extracts showed that both of these antibodies recognized proteins of the correct sizes (Fig. 2A). More importantly, each antibody recognized only one major polypeptide in the total nuclear extracts.
E14 ESCs were used in our mouse ChIP assays. Undifferentiated ESCs grown in feeder-free conditions were cross-linked with formaldehyde, and the fragmented chromatin lysates were subjected to immunoprecipitation with the Oct4 and Sox2 antibodies. Ten pairs of primers, located sequentially along the entire conserved promoter region and including the first exon, were used to quantify the ChIP-enriched DNA by real-time PCR (Fig. 2B). A peak representing Oct4 and Sox2 binding was observed (approximately 30-fold above background) at the distal enhancer, indicating that this region was specifically bound by the two transcription factors (Fig. 2C and D). A control antibody showed no significant enrichment over the entire surveyed region (Fig. 2E). Identical results were obtained when we used polyclonal antibodies raised against other epitopes of Oct4 and Sox2, further confirming the specificity of this binding (data not shown). Thus, we conclude that the Pou5f1 distal enhancer is a bona fide target of Oct4 and Sox2 in undifferentiated mouse ESCs.
RA treatment overrides the self-renewal properties of ESCs and induces their differentiation. There is a corresponding decrease in endogenous levels of Oct4 and Sox2 (Fig. 2F). We analyzed the binding profiles of Oct4 and Sox2 on Pou5f1 in this RA-induced differentiation model, as this also allowed us to determine the specificity of the antibodies by analyzing the binding in cells deficient in these factors. ESCs at different periods of RA treatment were cross-linked with formaldehyde, and the occupancies of Oct4 and Sox2 on the Pou5f1 promoter were analyzed by ChIP. Upon differentiation, the binding of Oct4 and Sox2 to the Pou5f1 distal enhancer was reduced in close correlation with the degree of differentiation (Fig. 2G and H). By day 3 of RA treatment, the level of Oct4 binding was reduced by almost 50%, and by day 6, no significant enrichment was detected. A mock ChIP using a control antibody did not result in enrichment of the enhancer sequences (Fig. 2I). This indicated that the antibodies recognize specific complexes found in ESCs but not in their differentiated derivatives. From this, we conclude that Oct4 and Sox2 bind to the Pou5f1 enhancer when this gene is being actively transcribed in undifferentiated, pluripotent mouse ESCs.
OCT4 and SOX2 bind to the CR4 region in human ESCs.
The conservation of the composite sox-oct element within the eutherian mammals suggests that OCT4 and SOX2 also bind to the POU5F1 enhancer in human ESCs. Gene expression differences between mouse and human ESCs have previously been described (15, 45, 49), although both are derived from the ICM of the blastocyst and both express Sox2 and Pou5f1 (38). These differences extend to cellular responsiveness to cytokines, as human ESCs are not responsive to LIF and lack the LIF receptor while LIF is crucial for the maintenance of mouse ESCs. To determine if at least part of the underlying genetic regulatory network of mouse and human ESCs is conserved, we extended our investigation on the regulation of Pou5f1 to include human ESCs.
To determine if OCT4 and SOX2 are bound to the CR4 region of the human POU5F1 promoter, we performed ChIP on human ESCs (HUES-6) propagated and passaged on inactivated mouse embryonic fibroblasts. The growth of the human ESCs on a mouse feeder cell layer did not complicate our analysis, as these mouse cells do not express Oct4 or Sox2. Moreover, the primers used to quantify the ChIP-enriched DNA were specific to human sequences (Fig. 3A). Both the OCT4 and SOX2 ChIP assays on human ESCs showed enrichment of DNA fragments in the distal enhancer of POU5F1 in which the composite sox-oct element is located (Fig. 3B). A control antibody did not show any enrichment of these enhancer sequences (Fig. 3B). In summary, we have clearly shown that Oct4/OCT4 and Sox2/SOX2 bind to the distal enhancer of Pou5f1/POU5F1 in living mouse and human ESCs.
Oct4 and Sox2 bind to the composite sox-oct element in vitro, and the binding sites are important for POU5F1 promoter activity.
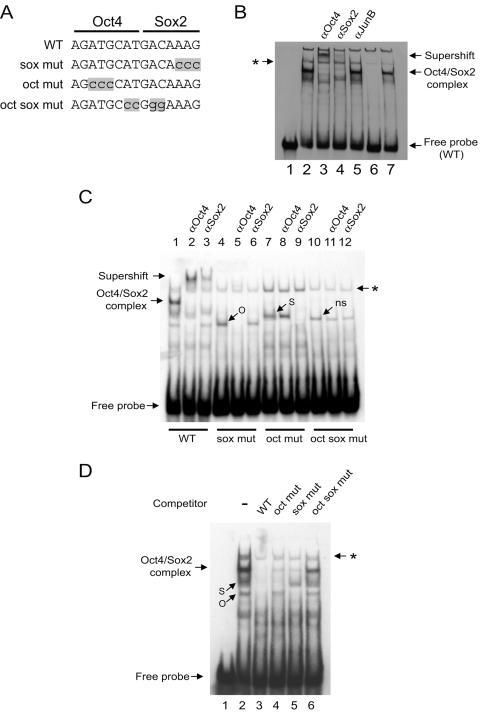
We examined whether native Oct4 and Sox2 bind to the CR4-B region in vitro by an EMSA. A 42-bp CR4-B biotin-labeled double-stranded DNA containing the composite sox-oct element was incubated with 15 μg of nuclear extract from undifferentiated feeder-free mouse ESCs. A major complex between the probe and nuclear proteins was detected (Fig. 4B, lane 2). The addition of Oct4 or Sox2 antibodies led to a supershift of this complex (Fig. 4B, lanes 3 and 4). Conversely, the control anti-JunB antibody did not affect the mobility of this complex. Therefore, this indicated that the DNA duplex was bound specifically by Oct4 and Sox2. The anti-Sox2 supershifted complex was weaker than that formed by the Oct4 antibody (Fig. 4B, lanes 3 and 4), suggesting that the Sox2 antibody interfered with the protein-DNA interaction. The addition of a 200-fold excess of unlabeled probe successfully competed for binding to the Sox2/Oct4 complex (Fig. 4B, lane 6), while an unrelated probe had no effect (Fig. 4B, lane 7). Thus, we conclude that, in vitro, Oct4 and Sox2 bind specifically to the CR4-B sequences as a ternary complex.
FIG. 4.
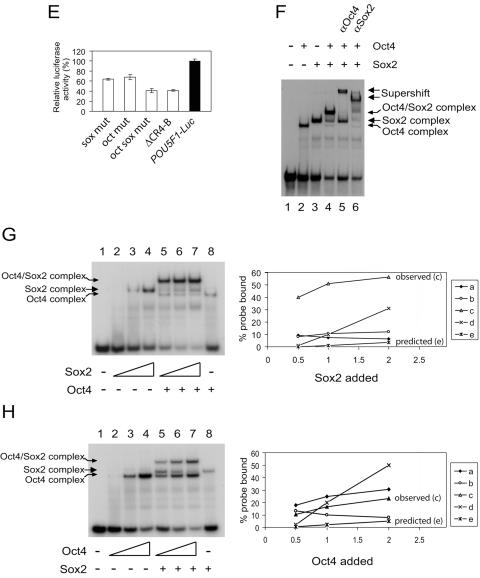
Oct4 and Sox2 bind to the composite sox-oct element in POU5F1. EMSAs were used to analyze the interactions between native Oct4 and Sox2 with a 42-bp double-stranded DNA probe containing the composite sox-oct element. (A) Sequence of the composite element and corresponding mutations (lowercase and shaded) used in this study. (B) EMSA with the wild-type probe detected a specific Oct4 and Sox2 complex. Lane 1 is without nuclear extract; all other lanes are with 15 μg nuclear extract; lanes 3 through 5 are with the respective antibodies added; lane 6 is with 200-fold excess cold probe; lane 7 is with 200-fold excess cold nonspecific probe. The asterisk denotes nonspecific complex associated with all four probes which cannot be competed out by an excess of nonspecific probe. (C) EMSA with probes containing mutations as shown in panel A. O indicates Oct4/DNA binary specific complex, while S indicates Sox2/DNA binary complex. The asterisk denotes nonspecific complex associated with all four probes which cannot be competed out by an excess of nonspecific probe. “ns” denotes a complex which cannot be supershifted with either Oct4 or Sox2 antibodies. (D) EMSAs with the wild-type probe were performed in the presence of excess cold probes (WT, oct mut, sox mut, or oct sox mut). WT, wild type. O, S, and the asterisk are as defined for panel C. (E) The same mutations described in panel A were tested for promoter activity within the context of the 3-kb POU5F1 promoter driving a luciferase reporter. These constructs were transfected into mouse ESCs and tested for luciferase activity 2 days later. Activity is expressed relative to the wild-type promoter (POU5F1-Luc). The CR4-B-deleted construct (Fig. 1) is included for comparison. (F) EMSA using overexpressed Oct4 or Sox2. Whole-cell lysates from 293T cellstransfected with Pou5f1 or Sox2 expression constructs were used in EMSAs. Oct4 and Sox2 are capable of forming specific complexes as confirmed by supershift analysis. (G) Analysis of the cooperativity of Sox2 and Oct4 binding to the composite sox-oct element. An increasing concentration of Sox2 was added to the probe in the absence or presence of a fixed amount of Oct4 (left panel). Quantitative representation of the DNA binding data is presented in the right panel (a, Oct4/DNA complex in the presence of Sox2, lanes 5 to 7; b, Sox2/DNA complex in the presence of Oct4, lanes 5 to 7; c, Oct4/Sox2/DNA complex, lanes 5 to 7; d, Sox2-DNA complex in the absence of Oct4, lanes 2 to 4; e, predicted amount of Oct4/Sox2/DNA complex if Oct4 and Sox2 bind independently of each other and show no cooperativity). The x axis represents the different amounts (microliters) of Sox2 added to the EMSA reaction mixtures. The amount of probe present was determined by PhosphorImager analysis and expressed as the percentage of total probe for each sample (y axis, percent probe bound). (H) Analysis of the cooperativity of Oct4 and Sox2 binding to the composite sox-oct element. Oct4 was titrated with a fixed amount of Sox2. Quantitative representation of the DNA binding data is presented in the right panel (a, Oct4/DNA complex in the presence of Sox2, lanes 5 to 7; b, Sox2/DNA complex in the presence of Oct4, lanes 5 to 7; c, Oct4/Sox2/DNA complex, lanes 5 to 7; d, Oct4-DNA complex in the absence of Sox2, lanes 2 to 4; e, predicted amount of Oct4/Sox2/DNA complex).
Next, we examined the requirement for the oct and sox elements within CR4-B in the formation of the DNA-protein complex. Mutations within the sox element resulted in the formation of a complex of higher mobility than the Oct4/Sox2 complex (Fig. 4A and C, lanes 4 to 6). This complex was disrupted by the addition of the Oct4 antibody but not the Sox2 antibody, indicating that the protein-DNA complex contained Oct4 but not Sox2. The Oct4-DNA complex can be competed out by an excess of specific unlabeled probe but not by an unlabeled nonspecific probe (data not shown). Mutations within the oct element similarly led to the formation of a specific complex containing Sox2 and lacking Oct4, as demonstrated by antibody interference assay and competition experiment with unlabeled probes (Fig. 4C, lanes 7 to 9; data not shown). EMSA with a double-stranded DNA probe containing mutations in both the oct and sox elements indicated that neither Oct4 nor Sox2 could bind to this probe (Fig. 4C, lanes 10 to 12). A complex did form with the oct-sox double mutant probe, but this was not due to Oct4 or Sox2 binding, since addition of the corresponding antibodies to the EMSA reaction mixtures did not disrupt this (Fig. 4C, lanes 11 and 12). From these results, it is evident that Oct4 and Sox2 are capable of binding independently of one another to the oct and sox elements of CR4, respectively. The specificities of the binding were also verified by a competition assay using an excess of unlabeled single or double mutant probes (Fig. 4D). The addition of probes mutated in the sox element reduced the Oct4-containing complexes, while the probes mutated in the oct element competed with the Sox2-containing complexes. These individual mutations in the oct and sox elements, as well as the double mutation, were introduced into our previously constructed POU5F1-Luc vector and transfected into mouse ESCs to test for their effect on promoter activity. The single oct and sox mutations resulted in promoter activities of approximately 60% of wild type whereas the sox-oct double mutation further dropped the activity to 40% of wild type and was equivalent to the activity found when the entire CR4-B region was deleted (Fig. 4E). We conclude that maximal transcriptional activity of POU5F1 in ESCs requires the binding of both Oct4 and Sox2 to the composite sox-oct element.
It has previously been shown that Sox2 can enhance the binding of Oct4 to Utf1 and Fbx15 (36, 46). To address whether these transcription factors also show cooperative binding to the composite sox-oct element in POU5F1, we overexpressed Oct4 and Sox2 in 293T cells which do not normally express these genes. With a 32P-labeled probe containing the composite sox-oct element, EMSA allowed us to investigate cooperative binding using an approach described previously (1, 36). Oct4 and Sox2 were each able to bind to the probe independently of one another (i.e., form a binary complex; Fig. 4F, lanes 2 and 3). When both factors were present in the same EMSA reaction mixture, a Sox2/Oct4/DNA ternary complex was formed as confirmed by supershifts (Fig. 4F, lanes 4 to 6). As the mobilities of the binary and ternary complexes were different, we were able to resolve them in our assay. We carried out the reciprocal experiments of adding an increasing amount of Sox2 to a fixed amount of Oct4 (Fig. 4G, lanes 5 to 7) and an increasing amount of Oct4 to a fixed amount of Sox2 (Fig. 4H, lanes 5 to 7). With the increase in Oct4 or Sox2, there was a corresponding increase in the respective binary complex as well as the Sox2/Oct4/DNA ternary complex. Most significantly, at the highest concentration of Oct4 or Sox2, the intensity of the ternary complex was greater than that of the reciprocal Sox2 or Oct4 binary complex in the absence of Oct4 and Sox2, respectively (Fig. 4G, compare lanes 7 and 8 for Oct4, and Fig. 4H, compare lanes 7 and 8 for Sox2). Supplementing these EMSAs with additional nuclear extract from mock-transfected 293T cells did not enhance the formation of the Sox2/Oct4/DNA ternary complex (data not shown), thus excluding the possibility that an unknown factor in the 293T extract enhanced the formation of this complex. Furthermore, we quantitated the levels of the ternary complexes formed under these different conditions and compared them with the calculated levels based on noncooperative interaction. The amount of the ternary complex expected to form as a result of noncooperative binding of the Oct4 and Sox2 proteins was calculated as the product of the amount of probe bound by Oct4 in the absence of Sox2 and the amount of probe bound by Sox2 in the absence of Oct4. By multiplying the values obtained from the bound complexes of Sox2 (Fig. 4G, lanes 2 to 4) with bound complex of Oct4 (Fig. 4G, lane 8), a graph representing the predicted noncooperative interaction was obtained (Fig. 4G, plot “e” in right panel). The levels of ternary complex formed when the two proteins were mixed together were also quantitated (Fig. 4G, plot “c” of the right panel). The data showed that the actual amount of the ternary complex detected for each point was greater than the amount predicted (Fig. 4G, compare the profiles of “c” and “e”). A similar result was obtained for the reciprocal experiment with a constant level of Sox2 (Fig. 4H). Hence, the increased binding efficiency to the composite sox-oct element by Sox2 and Oct4 when both transcription factors are present suggests the cooperative nature of this interaction.
Regulation of the POU5F1 promoter.
Having established the interaction between Oct4 and Sox2 with the POU5F1 distal enhancer, we sought to understand by a genetic approach the functional role of these factors in the enhancer's activity. Although we showed that the binding events occurred specifically in undifferentiated ESCs when POU5F1 is known to be most transcriptionally active, it was still unclear whether Oct4 or Sox2 was exerting a positive role on POU5F1 expression. To answer this question, we chose a genetic approach that employed RNAi technology.
RNAi has emerged as a powerful tool to study gene function by down-regulating the expression of a gene of interest. We used the pSUPER vector containing the polymerase III H1-RNA gene promoter for directing the synthesis of short interfering RNAs of 19 nucleotides (7). When we expressed a short hairpin RNA from this vector directed against Gfp, it efficiently silenced Gfp expression derived from a cotransfected plasmid (data not shown). Precautions were taken to make sure that the RNAi effect was specific. For each gene, we screened at least six regions for RNAi and used the best construct for subsequent studies (see Materials and Methods). These regions were selected based on their Reynolds scores and Ui-Tei class values (37, 48). To ensure specificity, we also selected regions that showed minimal sequence similarity to other mRNAs by screening with BLAST searches against public databases. Only targets that did not share more than 15 bases of identity with another mRNA were considered. All of the regions we targeted were located within the open reading frame.
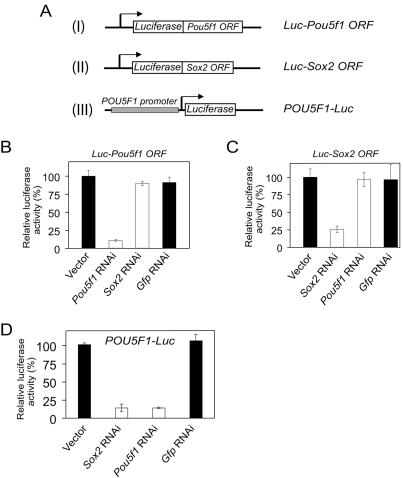
We first assayed the ability of the RNAi constructs to knock down the specific target mRNA in a heterologous system. To this end, we cloned the Pou5f1 or Sox2 open reading frames downstream of the luciferase (Luc) gene (Fig. 5A, constructs I and II). These fusion constructs were then cotransfected with the respective RNAi constructs into 293T cells. If the shRNA was effective in targeting the Luc fusion transcript, it would lead to a selective degradation of the target RNA, resulting in a reduced luciferase activity. In this assay, we showed that our RNAi constructs could mediate efficient knockdown of the coexpressed Luc-Pou5f1 or -Sox2 fusion transcripts and that they were specific for the respective genes. The results for the most effective RNAi constructs are shown (Fig. 5B and C). Using the Pou5f1 RNAi construct, the level of luciferase activity was reduced to less than 10% when measured 48 h after transfection. With the Sox2 RNAi construct, the activity was reduced to approximately 25% of the controls.
FIG. 5.
Regulation of POU5F1 promoter activity by Oct4 and Sox2. (A) Schematic of the luciferase reporter constructs used to measure the effects of RNAi. Construct I contains the Pou5f1 ORF fused downstream of the luciferase reporter, while construct II contains the Sox2 ORF fused downstream of the luciferase reporter. Construct III comprises the POU5F1 promoter containing the sox-oct composite element driving a luciferase reporter. (B and C) Specificity of Pou5f1 (B) and Sox2 (C) RNAi was tested by cotransfection of these constructs with their respective Luc-ORF reporter constructs into 293T cells. (D) Effect of Pou5f1 or Sox2 RNAi on POU5F1 promoter activity was tested by cotransfecting each RNAi plasmid along with the POU5F1-Luc construct into mouse ESCs. In all, luciferase activity was analyzed 2 days after transfection and was expressed relative to the empty RNAi vector control. A Gfp RNAi was used as a nonspecific control. Standard deviations are shown.
Having determined the efficiency and specificity of the RNAi constructs, we proceeded to test the effect of these constructs on a cotransfected POU5F1-Luc reporter (Fig. 5A, construct III) in mouse ESCs. ESCs were harvested 60 h after transfection and measured for luciferase activity. Knockdown of either Pou5f1 or Sox2 suppressed the expression of the transfected POU5F1-Luc vector with an 85% reduction in luciferase activity compared to the empty RNAi or Gfp RNAi vector control (Fig. 5D). Similar results were obtained when we used a mouse Pou5f1-Luc reporter containing the CR4 distal enhancer region (data not shown) (19), indicating a functional conservation between the mouse and human enhancers.
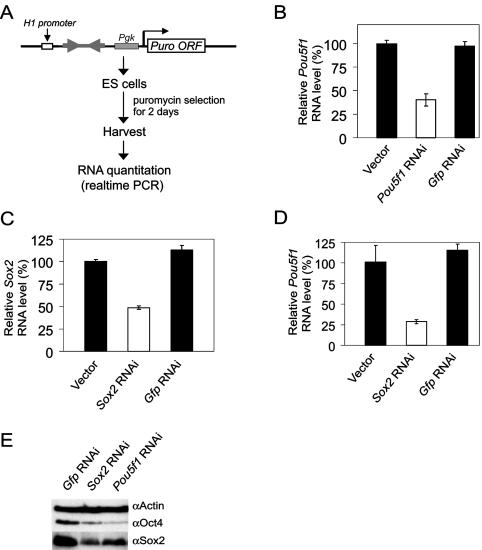
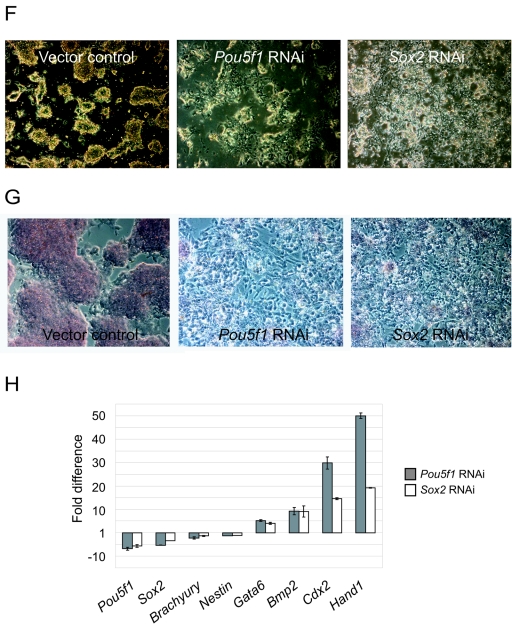
To determine whether our shRNAs designed against Pou5f1 and Sox2 silenced endogenous transcripts in ESCs, we transfected these RNAi constructs in a vector containing a puromycin selection cassette (Fig. 6A). Twenty-four hours after transfection, cells were cultured in medium containing puromycin for 2 days to selectively remove nontransfected ESCs. Approximately 70% of the nontransfected cells were killed after 2 days of selection. Total RNAs were purified from these transfected cells, and the levels of Pou5f1 or Sox2 transcripts were quantified by reverse transcription followed by real-time PCR. We observed a 60% reduction of Pou5f1 transcripts when RNAi was used against Pou5f1 (Fig. 6B). Similarly with Sox2 RNAi, the level of Sox2 transcripts was reduced by 50% relative to the control cells (Fig. 6C), and with this same RNAi there was a corresponding reduction of Pou5f1 transcripts (Fig. 6D). In all these experiments, a significant level of Pou5f1 and Sox2 transcripts remained; these were likely derived from the residual nontransfected ESCs or due to incomplete silencing of the respective genes. Despite this, there was a clear reduction in both Sox2 and Oct4 protein levels when either the Sox2 or Pou5f1 RNAi construct was used, as validated by Western blotting (Fig. 6E). We further assessed whether the reduction in the transcripts and proteins of these factors led to differentiation. After selection, both the Pou5f1 and the Sox2 RNAi-transfected cells had altered morphology compared to transfection controls and undifferentiated ESCs, with the formation of flattened epithelial-cell-like cells (Fig. 6F). We also performed alkaline phosphatase staining on these cells (Fig. 6G). In contrast to the control cells, the epithelial-cell-like cells after either Pou5f1 or Sox2 knockdown showed drastically reduced alkaline phosphatase staining. The RNAs isolated from these cells were reverse transcribed and analyzed by real-time PCR to detect the presence of differentiated cell markers (Fig. 6H). Gene expression profiles for both the Pou5f1 and Sox2 knockdowns were very similar, and both showed an up-regulation of the endoderm and trophectoderm markers such as Gata6, Bmp2, Cdx2, and Hand1 (22, 29). In contrast, mesoderm marker Brachyury and the neuroectoderm marker Nestin were not induced. Our data on Pou5f1 knockdown in ESCs were consistent with previous reports analyzing the gene expression changes when the Pou5f1 level was reduced (18, 24, 29). These results indicates that knockdown of either Pou5f1 or Sox2 caused ESCs to differentiate. In summary, we showed that the knockdown of both Sox2 and Pou5f1 mRNAs caused a reduction in the POU5F1 distal enhancer activity and in endogenous Pou5f1 transcript levels. This indicates that both Sox2 and Oct4 are positively regulating Pou5f1.
FIG. 6.
Regulation of endogenous Pou5f1 expression by Sox2 in ESCs. (A) Schematic of the RNAi vector and a flow chart of the corresponding experiments. The vector contains a puromycin selection cassette. Location of the RNAi sequence is indicated by arrowheads. (B to D) Effects of Pou5f1 (B) and Sox2 (C and D) RNAi on endogenous levels of Pou5f1 (B and D) and Sox2 (C) mRNA in ESCs. Endogenous mRNA levels are expressed relative to the empty RNAi vector control. A nonspecific control Gfp RNAi was included. (E) Knockdown of the protein was confirmed by Western blotting. Actin served as a loading control. (F) Morphological changes of Pou5f1 or Sox2 knockdown cells. Note the presence of flattened epithelial-cell-like cells in knockdown cells not seen at all in vector control ESCs. (G) Alkaline phosphatase staining of Pou5f1 or Sox2 knockdown cells. Note that, in the knockdown cells, almost all the cells stain negatively for alkaline phosphatase. (H) Changes in gene expression following Pou5f1 or Sox2 knockdown by RNAi. cDNAs were prepared from the knockdown cells and analyzed by real-time PCR with fold differences measured against vector control ESCs.
Regulation of the enhancer element in Sox2.
The importance of the enhancer element at the 3′ end of Sox2 has been demonstrated by reporter assays and mutagenesis (47). This element, also known as SRR2 (Sox regulatory region 2), contains a composite sox-oct element. Although Oct6 and Oct4 have been implicated to bind to SRR2 along with Sox2, it is not clear if Oct4 and Sox2 are indeed bound to the SRR2 site in living ESCs.
We further dissected the role of Oct4 and Sox2 in the enhancer activity of SRR2 by genetic manipulation through RNAi. First, the SRR2 element was cloned upstream of an SV40 promoter driving the Luc gene (Fig. 7A); the activity of this construct was then tested in ESCs. This element enhanced the activity of the SV40 promoter in ESCs but not in 293T cells (Fig. 7B). The data are consistent with the finding that SRR2 functioned as an enhancer element when grafted to a heterologous promoter (47).
We then cotransfected our Pou5f1 or Sox2 RNAi constructs along with this Sox2-SV40-Luc reporter into mouse ESCs and assayed for luciferase activity 60 h after transfection. The activity of this reporter was diminished with the knockdown of either Sox2 or Pou5f1 mRNA, while control RNAi using an empty RNAi vector or Gfp RNAi vector showed no effects on this SRR2 reporter (Fig. 7C). In addition to demonstrating that Oct4 and Sox2 positively regulate the SRR2 activity, we showed by reverse transcription-real-time PCR that endogenous Sox2 transcripts were also diminished in mouse ESCs transfected with the Pou5f1 RNAi construct (Fig. 7D). Furthermore, we confirmed by ChIP experiments that Sox2 and Oct4 bound to the SRR2 region of Sox2 in both mouse and human ESCs (Fig. 7E to I) and that these interactions, at least in the mouse, are specific to the undifferentiated ESCs. A control antibody showed no significant enrichment over the entire surveyed region (data not shown). Taken together, our data demonstrate that the SRR2 region containing the composite sox-oct element in Sox2 is a bona fide target for Oct4 and Sox2.
DISCUSSION
During early embryogenesis, Oct4 and Sox2 are found to be coexpressed in several pluripotent cells such as the morula, ICM, epiblast, and germ cells. Gene knockout studies revealed that the primary defect for both the Pou5f1- and Sox2-null animals is in the pluripotent epiblast, though there are slight differences between the two null phenotypes. There is no epiblast development in the Pou5f1-null blastocyst, and the fate of all cell types is towards trophectoderm lineage (26). On the other hand, Sox2-null animals are capable of giving rise, at least transiently, to the epiblast, as epiblast-derived extraembryonic endoderm is detected (3). This transient epiblast formation was suggested to result from maternally derived Sox2 protein. Both Pou5f1- and Sox2-null blastocysts are incapable of giving rise to pluripotent ESCs.
In this study, we use ESCs as a model for understanding the role of Oct4 and Sox2 in the genetic regulatory network of pluripotent cells. We first established the conditions under which Oct4 and Sox2 interact with the Pou5f1 and Sox2 enhancers in order to understand the function of these regulators at these cis-regulatory target sites. Using specific antibodies against Oct4 and Sox2, we showed by ChIP that these two transcription factors bind to both enhancer elements in mouse and human ESCs (Fig. 2, 3, and 7). Hence, these enhancer elements are the direct targets of their respective gene products and are reciprocally bound by the other regulator. As we detected the highest level of binding in undifferentiated cells when these genes are known to be most transcriptionally active, we conclude that they are associated with the transcriptional activation of these genes and may play a role in positively regulating expression. This was further confirmed through a functional analysis of the interactions in genetic studies.
Using RNAi with reporter gene constructs, we showed that the silencing of Pou5f1 or Sox2 led to the down-regulation of Pou5f1 and Sox2 enhancer activities (Fig. 5, 6, and 7). More importantly, the endogenous transcripts and proteins were also reduced by RNAi in a manner that was consistent with the reporter studies. The effects of RNAi on the reporters may have been indirect, as the silencing of endogenous Pou5f1 or Sox2 led to the differentiation of ESCs and further caused the level of endogenous Oct4 and Sox2 to decrease. However, coupled with the observation that Oct4 and Sox2 directly bind to these regulatory elements when the genes are active, our results indicate that Oct4 and Sox2 play positive roles in the expression of Pou5f1 and Sox2.
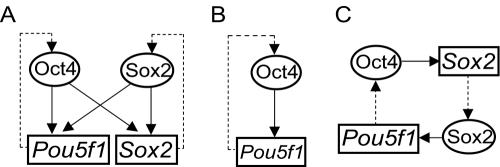
The interactions of this Sox2-Oct4 complex with the respective genes can be described as a transcriptional regulatory network consisting of autoregulatory and multicomponent loops (Fig. 8A) (21). A network motif is a fundamental unit within a complex transcriptional regulatory network. In an autoregulation model, the gene product binds to its own regulatory element (Fig. 8B). This may allow for self-perpetuation and enhanced stability of gene expression. An additional relationship between Oct4 and Sox2 is depicted by a multicomponent loop motif whereby a regulator binds to the regulatory elements of another regulator in a closed loop (Fig. 8C). Such a closed-circuit loop can efficiently generate a bistable system with the ability to switch between two different states. For ESCs, the two states may be the decision to undergo self-renewal or to differentiate and exit from symmetrical division.
FIG. 8.
Oct4-Sox2 regulatory circuitry in ESCs. Transcription factors are represented by ovals, and regulatory elements of the genes are represented by rectangles (gene names are printed in italics). (A) The relationships between Oct4 and Sox2 and their respective genes. The links are based on evidence derived from ChIP and RNAi experiments. A solid arrow indicates a transcription factor positively regulating a gene via direct binding of a cis element. Dashed arrows denote the synthesis of transcription factors by their respective genes. (B) An autoregulation motif for Pou5f1. Although not shown, Sox2 has a similar configuration. (C) A multicomponent loop showing the link between Pou5f1 and Sox2.
This model also requires that the concentrations of the two factors remain relatively constant, as any slight change in the abundance of one protein will destabilize the circuitry. Interestingly, it has been shown that mouse ESCs are exquisitely sensitive to the level of Oct4 (29). Increasing the level of Oct4 by 50% is sufficient to induce differentiation of ESCs into primitive endoderm and mesoderm. We show that mutating either the oct or sox site in the distal enhancer does not completely abolish its enhancer activity, with 60% of the activity still being retained (Fig. 4E). It is conceivable that the modest reduction in Pou5f1 expression may have a significant cellular effect. It would also be of interest to determine whether the alteration of Sox2 levels gives a similar phenotype.
Differential occupancy of the Oct4/Sox2 complex on the various regulatory elements was suggested by the EMSA results. For instance, a direct comparison of ESC nuclear extracts binding to Nanog and Fgf4 at the sox-oct composite elements indicated clear differences in binary and ternary formation between the two (39). In addition, the EMSA indicated that Oct4 was incapable of binding by itself on the Fbx15 composite element (46). This differential binding is likely attributed to variations in the sox-oct composite element, given that the Fgf4 element contains an intervening 3 bp while the Fbx15 element contains an A rather than a C in the otherwise invariant fourth position in the octamer motif (Fig. 1B). Whether the differential binding (as observed in the EMSA) alters transcriptional activity of the Oct4/Sox2 complex itself remains to be seen, but gene-specific sequence conservation within these sox-oct composite elements suggests functional significance.
Although ESCs grown in culture may not mimic the physiological conditions of cells within the ICM of the blastocyst, they provide a good model for understanding the transcriptional regulatory networks in pluripotent cells. Oct4 and Sox2 are key regulators for pluripotency in ESCs. We have recently identified Nanog as a downstream target of Oct4 and Sox2 (39). This expands the list of genes (Fgf4, Utf1, Opn, and Fbx15) which are potentially regulated by both Oct4 and Sox2. It is also important to note that direct binding of these regulators on the regulatory elements of Fgf4, Utf1, and Fbx15 has not been demonstrated by ChIP. Nevertheless, it is apparent that Oct4 and Sox2 are at the top of the hierarchy of transcriptional regulators in ESCs. The cooperative binding of Oct4 and Sox2 may thus be instrumental in recruiting various other interacting protein partners. It should be emphasized that not all Oct4 and Sox2 sites on the same regulatory region are synergistic in transcriptional activation. For example, in the Opn intron, a sox site 39 bp away from an inverted pair of Oct4 sites acts antagonistically in transactivation by Oct4 (6). Repression by Sox2 was shown to require DNA binding and a carboxy-terminal transactivation domain. For Fgf4, Utf1, and Fbx15, the regulatory elements contain Oct4 and Sox2 sites in proximity (either 0- or 3-bp separation) and the Oct4/Sox2 complex is implicated in transactivation. This raises the interesting possibility that perhaps Oct4 and Sox2 collaborate to globally control ESC-specific gene expression through the sox-oct motifs. To address this possibility, the direct targets of both regulators have to be identified. One can then compare these two factors and identify the commonality between them. It is also plausible that the Oct4/Sox2 complex interacts with another factor(s) to activate the network of ESC-specific genes. For example, the Pou5f1 distal enhancer contains three distinct regions (CR4-A to -C) that contribute to expression. Does the Oct4/Sox2 complex communicate with the regulatory protein(s) bound to CR4-A and CR4-C? Are they ubiquitous or ESC-specific factors? These questions require further characterization of the regulators bound to CR4-A and CR4-C. It is also important to note that there are other key regulators for maintenance of the undifferentiated state of ESCs. The LIF/Stat3 pathway is essential for self-renewal of mouse ESCs (23, 28, 35). The removal of LIF leads to the inactivation of Stat3 and induces differentiation. The other key regulator is Nanog. The removal of Nanog via gene targeting or RNAi leads to differentiation of mouse ESCs (10, 24, 39). Intriguingly, overexpression of Nanog is sufficient to bypass the LIF/Stat3 requirement. However, how Stat3 and Nanog interact with the Oct4/Sox2 pathway remains to be studied.
An autoregulation mode can generate a self-perpetuating cycle to maintain stable gene expression through a positive feedback loop. The question is, how can this cycle be broken? In the case of Pou5f1, one key answer is the involvement of a repressor(s) that is induced in differentiated cells. The transcription of Pou5f1 is down-regulated when ESCs differentiate in vitro and when the epiblast differentiates during embryogenesis. There exists a mechanism to break out of the loop of Pou5f1 expression. The promoter of Pou5f1 contains negative regulatory elements which are required for repression when embryonal carcinoma cells differentiate (4, 34, 42, 44). Interestingly, germ cell nuclear factor (Gcnf) has been shown to mediate repression of the Pou5f1 proximal promoter (16). The expression of Gcnf is inversely correlated with the Pou5f1 expression in embryonal carcinoma cells. More importantly, in Gcnf-knockout mouse embryos, the Pou5f1 expression is no longer confined to the germ cell lineage and novel Pou5f1 expression domains are detected. The loss of Pou5f1 expression may subsequently extinguish Sox2 transcription and the expression of other downstream target genes. It is not clear whether Sox2 is similarly subjected to repression when ESCs differentiate. Nevertheless, the finding highlights the importance of an active mechanism to shut down a key regulator(s) upon differentiation.
Acknowledgments
We are grateful to the Biomedical Research Council (BMRC) and the Agency for Science, Technology and Research (A*STAR) for funding. J.-L.C. is supported by the Singapore Millennium Foundation scholarship. Y.-H.L and W.-L.T. are supported by the A*STAR graduate scholarships. X.C. is supported by the NUS Graduate Research Scholarship and President's Graduate Fellowship. B.L. is partially supported by a grant from NIH (DK47636).
We are grateful to Yunhan Hong (National University of Singapore) for the mouse Pou5f1 reporter construct; Doug Melton (Harvard University) for human ESCs; and Sylvia Lim, Kelvin Tan, and Hwee-Goon Tay for technical assistance. Dave Rodda is acknowledged for his advice on EMSA.
REFERENCES
- 1.Ambrosetti, D. C., C. Basilico, and L. Dailey. 1997. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol. 17:6321-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambrosetti, D. C., H. R. Scholer, L. Dailey, and C. Basilico. 2000. Modulation of the activity of multiple transcriptional activation domains by the DNA binding domains mediates the synergistic action of Sox2 and Oct-3 on the fibroblast growth factor-4 enhancer. J. Biol. Chem. 275:23387-23397. [DOI] [PubMed] [Google Scholar]
- 3.Avilion, A. A., S. K. Nicolis, L. H. Pevny, L. Perez, N. Vivian, and R. Lovell-Badge. 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17:126-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Shushan, E., H. Sharir, E. Pikarsky, and Y. Bergman. 1995. A dynamic balance between ARP-1/COUP-TFII, EAR-3/COUP-TFI, and retinoic acid receptor:retinoid X receptor heterodimers regulates Oct-3/4 expression in embryonal carcinoma cells. Mol. Cell. Biol. 15:1034-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Shushan, E., J. R. Thompson, L. J. Gudas, and Y. Bergman. 1998. Rex-1, a gene encoding a transcription factor expressed in the early embryo, is regulated via Oct-3/4 and Oct-6 binding to an octamer site and a novel protein, Rox-1, binding to an adjacent site. Mol. Cell. Biol. 18:1866-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botquin, V., H. Hess, G. Fuhrmann, C. Anastassiadis, M. K. Gross, G. Vriend, and H. R. Scholer. 1998. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev. 12:2073-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 8.Catena, R., C. Tiveron, A. Ronchi, S. Porta, A. Ferri, L. Tatangelo, M. Cavallaro, R. Favaro, S. Ottolenghi, R. Reinbold, H. Scholer, and S. K. Nicolis. 2004. Conserved POU binding DNA sites in the Sox2 upstream enhancer regulate gene expression in embryonic and neural stem cells. J. Biol. Chem. 279:41846-41857. [DOI] [PubMed] [Google Scholar]
- 9.Cawley, S., S. Bekiranov, H. H. Ng, P. Kapranov, E. A. Sekinger, D. Kampa, A. Piccolboni, V. Sementchenko, J. Cheng, A. J. Williams, R. Wheeler, B. Wong, J. Drenkow, M. Yamanaka, S. Patel, S. Brubaker, H. Tammana, G. Helt, K. Struhl, and T. R. Gingeras. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116:499-509. [DOI] [PubMed] [Google Scholar]
- 10.Chambers, I., D. Colby, M. Robertson, J. Nichols, S. Lee, S. Tweedie, and A. Smith. 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113:643-655. [DOI] [PubMed] [Google Scholar]
- 11.Cowan, C. A., I. Klimanskaya, J. McMahon, J. Atienza, J. Witmyer, J. P. Zucker, S. Wang, C. C. Morton, A. P. McMahon, D. Powers, and D. A. Melton. 2004. Derivation of embryonic stem-cell lines from human blastocysts. N. Engl. J. Med. 350:1353-1356. [DOI] [PubMed] [Google Scholar]
- 12.Dailey, L., H. Yuan, and C. Basilico. 1994. Interaction between a novel F9-specific factor and octamer-binding proteins is required for cell-type-restricted activity of the fibroblast growth factor 4 enhancer. Mol. Cell. Biol. 14:7758-7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donovan, P. J., and J. Gearhart. 2001. The end of the beginning for pluripotent stem cells. Nature 414:92-97. [DOI] [PubMed] [Google Scholar]
- 15.Draper, J. S., C. Pigott, J. A. Thomson, and P. W. Andrews. 2002. Surface antigens of human embryonic stem cells: changes upon differentiation in culture. J. Anat. 200:249-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuhrmann, G., A. C. Chung, K. J. Jackson, G. Hummelke, A. Baniahmad, J. Sutter, I. Sylvester, H. R. Scholer, and A. J. Cooney. 2001. Mouse germline restriction of Oct4 expression by germ cell nuclear factor. Dev. Cell 1:377-387. [DOI] [PubMed] [Google Scholar]
- 17.Geijsen, N., M. Horoschak, K. Kim, J. Gribnau, K. Eggan, and G. Q. Daley. 2004. Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature 427:148-154. [DOI] [PubMed] [Google Scholar]
- 18.Hay, D. C., L. Sutherland, J. Clark, and T. Burdon. 2004. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells 22:225-235. [DOI] [PubMed] [Google Scholar]
- 19.Hong, Y., T. Liu, H. Zhao, H. Xu, W. Wang, R. Liu, T. Chen, J. Deng, and J. Gui. 2004. Establishment of a normal medakafish spermatogonial cell line capable of sperm production in vitro. Proc. Natl. Acad. Sci. USA 101:8011-8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubner, K., G. Fuhrmann, L. K. Christenson, J. Kehler, R. Reinbold, R. De La Fuente, J. Wood, J. F. Strauss III, M. Boiani, and H. R. Scholer. 2003. Derivation of oocytes from mouse embryonic stem cells. Science 300:1251-1256. [DOI] [PubMed] [Google Scholar]
- 21.Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph, G. K. Gerber, N. M. Hannett, C. T. Harbison, C. M. Thompson, I. Simon, J. Zeitlinger, E. G. Jennings, H. L. Murray, D. B. Gordon, B. Ren, J. J. Wyrick, J. B. Tagne, T. L. Volkert, E. Fraenkel, D. K. Gifford, and R. A. Young. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799-804. [DOI] [PubMed] [Google Scholar]
- 22.Loebel, D. A., C. M. Watson, R. A. De Young, and P. P. Tam. 2003. Lineage choice and differentiation in mouse embryos and embryonic stem cells. Dev. Biol. 264:1-14. [DOI] [PubMed] [Google Scholar]
- 23.Matsuda, T., T. Nakamura, K. Nakao, T. Arai, M. Katsuki, T. Heike, and T. Yokota. 1999. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 18:4261-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitsui, K., Y. Tokuzawa, H. Itoh, K. Segawa, M. Murakami, K. Takahashi, M. Maruyama, M. Maeda, and S. Yamanaka. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113:631-642. [DOI] [PubMed] [Google Scholar]
- 25.Ng, H. H., F. Robert, R. A. Young, and K. Struhl 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 26.Nichols, J., B. Zevnik, K. Anastassiadis, H. Niwa, D. Klewe-Nebenius, I. Chambers, H. Scholer, and A. Smith. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95:379-391. [DOI] [PubMed] [Google Scholar]
- 27.Nishimoto, M., A. Fukushima, A. Okuda, and M. Muramatsu. 1999. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol. 19:5453-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niwa, H., T. Burdon, I. Chambers, and A. Smith. 1998. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12:2048-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niwa, H., J. Miyazaki, and A. G. Smith. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24:372-376. [DOI] [PubMed] [Google Scholar]
- 30.Nordhoff, V., K. Hubner, A. Bauer, I. Orlova, A. Malapetsa, and H. R. Scholer. 2001. Comparative analysis of human, bovine, and murine Oct-4 upstream promoter sequences. Mamm. Genome 12:309-317. [DOI] [PubMed] [Google Scholar]
- 31.Palmieri, S. L., W. Peter, H. Hess, and H. R. Scholer. 1994. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev. Biol. 166:259-267. [DOI] [PubMed] [Google Scholar]
- 32.Pera, M. F., B. Reubinoff, and A. Trounson. 2000. Human embryonic stem cells. J. Cell Sci. 113:5-10. [DOI] [PubMed] [Google Scholar]
- 33.Pesce, M., and H. R. Scholer. 2001. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells 19:271-278. [DOI] [PubMed] [Google Scholar]
- 34.Pikarsky, E., H. Sharir, E. Ben-Shushan, and Y. Bergman. 1994. Retinoic acid represses Oct-3/4 gene expression through several retinoic acid-responsive elements located in the promoter-enhancer region. Mol. Cell. Biol. 14:1026-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raz, R., C. K. Lee, L. A. Cannizzaro, P. d'Eustachio, and D. E. Levy. 1999. Essential role of STAT3 for embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. USA 96:2846-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Remenyi, A., K. Lins, L. J. Nissen, R. Reinbold, H. R. Scholer, and M. Wilmanns. 2003. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 17:2048-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reynolds, A., D. Leake, Q. Boese, S. Scaringe, W. S. Marshall, and A. Khvorova. 2004. Rational siRNA design for RNA interference. Nat. Biotechnol. 22:326-330. [DOI] [PubMed] [Google Scholar]
- 38.Robson, P. 2004. The maturing of the human embryonic stem cell transcriptome profile. Trends Biotechnol. 22:609-612. [DOI] [PubMed] [Google Scholar]
- 39.Rodda, D. J., J. L. Chew, L. H. Lim, Y. H. Loh, B. Wang, H. H. Ng, and P. Robson. Transcriptional regulation of Nanog by Oct4 and Sox2. J. Biol. Chem., in press. [DOI] [PubMed]
- 40.Schmitt, T. M., R. F. de Pooter, M. A. Gronski, S. K. Cho, P. S. Ohashi, and J. C. Zuniga-Pflucker. 2004. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat. Immunol. 5:410-417. [DOI] [PubMed] [Google Scholar]
- 41.Scholer, H. R., S. Ruppert, N. Suzuki, K. Chowdhury, and P. Gruss. 1990. New type of POU domain in germ line-specific protein Oct-4. Nature 344:435-439. [DOI] [PubMed] [Google Scholar]
- 42.Schoorlemmer, J., A. van Puijenbroek, M. van Den Eijnden, L. Jonk, C. Pals, and W. Kruijer. 1994. Characterization of a negative retinoic acid response element in the murine Oct4 promoter. Mol. Cell. Biol. 14:1122-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, A. G. 2001. Embryo-derived stem cells: of mice and men. Annu. Rev. Cell Dev. Biol. 17:435-462. [DOI] [PubMed] [Google Scholar]
- 44.Sylvester, I., and H. R. Scholer. 1994. Regulation of the Oct-4 gene by nuclear receptors. Nucleic Acids Res. 22:901-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomson, J. A., J. Itskovitz-Eldor, S. S. Shapiro, M. A. Waknitz, J. J. Swiergiel, V. S. Marshall, and J. M. Jones. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282:1145-1147. [DOI] [PubMed] [Google Scholar]
- 46.Tokuzawa, Y., E. Kaiho, M. Maruyama, K. Takahashi, K. Mitsui, M. Maeda, H. Niwa, and S. Yamanaka. 2003. Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol. Cell. Biol. 23:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomioka, M., M. Nishimoto, S. Miyagi, T. Katayanagi, N. Fukui, H. Niwa, M. Muramatsu, and A. Okuda. 2002. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 30:3202-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ui-Tei, K., Y. Naito, F. Takahashi, T. Haraguchi, H. Ohki-Hamazaki, A. Juni, R. Ueda, and K. Saigo. 2004. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 32:936-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei, C. L., T. Miura, P. Robson, S. K. Lim, X. Q. Xu, M. Y. Lee, S. Gupta, L. Stanton, Y. Luo, J. Schmitt, S. Thies, W. Wang, I. Khrebtukova, D. Zhou, E. T. Liu, Y. J. Ruan, M. Rao, and B. Lim. 2005. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells 23:166-185. [DOI] [PubMed] [Google Scholar]
- 50.Williams, D. C., Jr., M. Cai, and G. M. Clore. 2004. Molecular basis for synergistic transcriptional activation by Oct1 and Sox2 revealed from the solution structure of the 42-kDa Oct1.Sox2.Hoxb1-DNA ternary transcription factor complex. J. Biol. Chem. 279:1449-1457. [DOI] [PubMed] [Google Scholar]
- 51.Yeom, Y. I., G. Fuhrmann, C. E. Ovitt, A. Brehm, K. Ohbo, M. Gross, K. Hubner, and H. R. Scholer. 1996. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 122:881-894. [DOI] [PubMed] [Google Scholar]
- 52.Ying, Q. L., M. Stavridis, D. Griffiths, M. Li, and A. Smith. 2003. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 21:183-186. [DOI] [PubMed] [Google Scholar]
- 53.Yuan, H., N. Corbi, C. Basilico, and L. Dailey. 1995. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 9:2635-2645. [DOI] [PubMed] [Google Scholar]