Abstract
Activation of the endothelium by inflammatory cytokines is a key event in the pathogenesis of vascular disease states. Proinflammatory cytokines repress the expression of KLF2, a recently identified transcriptional inhibitor of the cytokine-mediated activation of endothelial cells. In this study the molecular basis for the cytokine-mediated inhibition of KLF2 is elucidated. Tumor necrosis factor alpha (TNF-α) potently inhibited KLF2 expression. This effect was completely abrogated by a constitutively active form of IκBα, as well as treatment with trichostatin A, implicating a role for the NF-κB pathway and histone deacetylases. Overexpression studies coupled with observations with p50/p65 null cells support an essential role for p65. A combination of promoter deletion and mutational analyses, chromatin immunoprecipitation assays, and coimmunoprecipitation studies indicates that p65 and histone deacetylases 4 cooperate to inhibit the ability of MEF2 factors to induce the KLF2 promoter. These studies identify a novel mechanism by which TNF-α can inhibit endothelial gene expression. Furthermore, the inhibition of MEF2 function by p65 and HDAC4 has implications for other cellular systems where these factors are operative.
The vascular endothelium is critically involved in the body's response to inflammation. Proinflammatory stimuli, such as cytokines, induce endothelial dysfunction and confer a proadhesive and prothrombotic phenotype (3). While these events are important in certain physiologic states, such as wound healing, sustained endothelial activation can lead to deleterious consequences, as seen in a number of chronic inflammatory disease states, such as rheumatoid arthritis, inflammatory bowel disease, and atherosclerosis. As such, an understanding of the molecular mechanism regulating endothelial activation by inflammatory mediators is essential (8).
Kruppel-like factors (KLFs) are a member of a multigene family of transcriptional factors having a three-Cys2-His2-zinc-finger domain (4, 7). Previous studies demonstrate a key role for the Kruppel factors in various processes of cell growth and differentiation. For example, KLF1/EKLF (erythroid Kruppel-like factor) is essential for red blood cell maturation, whereas KLF4/GKLF (gut Kruppel-like factor) regulates the differentiation and maturation of dermal and gastrointestinal epithelial cells (15, 24, 26, 28). Furthermore, an emerging literature also supports a role for this family of transcriptional factors in vascular cell biology (7). Lung Kruppel-like factor/KLF2 is highly expressed in the vascular endothelium (17). Systemic knockout of KLF2 results in embryonic death and abnormal vessel formation (17, 18). However, our understanding of the function and targets of KLF2 in endothelial biology have been lacking to date.
We recently identified KLF2 as a novel transcriptional regulator of endothelial proinflammatory activation (19, 29). We found that KLF2 is induced by shear stress and potently inhibited by proinflammatory cytokines. Overexpression of KLF2 induced the key endothelial factors, such as eNOS and thrombomudulin, and inhibited the expression of proadhesive/procoagulant factors, such as VCAM-1 and tissue factor. As a consequence, immune cell adhesion to an endothelial monolayer is strongly attenuated (19, 29). These data support KLF2 as a potent and critical regulator of endothelial proinflammatory activation.
One of the key observations made in this previous study was that proinflammatory cytokines repress KLF2 expression (29). This is an unusual and noteworthy finding, because proinflammatory cytokines can induce hundreds of genes in endothelial cells but inhibition occurs much less frequently. In light of KLF2's potent anti-inflammatory properties, we reasoned that the downregulation of this factor may be an important mechanism by which cytokines activate the endothelium. In this study, we sought to understand the molecular basis underlying the ability of proinflammatory cytokines to inhibit KLF2 expression. Our studies suggest that the tumor necrosis factor alpha (TNF-α)-mediated reduction of KLF2 expression is dependent on NF-κB. Furthermore, we provide evidence that p65 HDAC4 and -5 inhibit the ability of the MADS-box factor MEF2 to regulate KLF2 expression.
MATERIALS AND METHODS
Cell culture and reagents.
Human umbilical vein endothelial cells (HUVEC) were obtained from Cambrex Company (Baltimore, MD). Bovine aortic endothelial cells (BAEC) were from Cell Application (San Diego, CA) and were studied between passages 4 and 7. COS-7 cells were obtained from American Type Culture Collection. p65 and p50 null mouse embryos fibroblasts were a kind gift from Alexander Hoffmann (California Institute of Technology) with permission from David Baltimore and were maintained in Dulbecco's modified Eagle medium. Human TNF-α (R&D systems, Minneapolis, MN) was used at final concentrations of 10 ng/ml. Antibodies recognizing p65, MEF2 (C-21), and HDAC4 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), anti-FLAG from Sigma (St. Louis, CA), and anti-histone H3 from Cell Signaling Tech (Beverly, MA). Control and SR-IκBα adenoviruses were purchased from Gene Therapy Center, The University of North Carolina at Chapel Hill, with permission from Alex Baldwin. Plasmids were obtained from different sources: HDAC1, -4, -5, and -6 were from Stuart Schreiber (Harvard University, Boston, MA), HDAC4 (L175A) mutant from Xiang-Jiao Yang (McGill University Health Center, Quebec, Canada), HDAC2 and -3 from E. Seto (H. Lee Moffitt Cancer Center and Research Institute, University of South Florida), p65 (Y26E33) mutant from G. Natoli (Institute for Research in Biomedicine, Switzerland), MEF2A and MEF2C plasmids from E. N. Olson (University of Texas M.D. Anderson Cancer Center), and the full-length KLF2 promoter from J. Leiden (Abbott Laboratories, Chicago). FLAG-tagged HDAC4 in pCDNA3.1 was cloned from the pBJ construct of HDAC4. All mutation constructs of the KLF2 promoter were generated by PCR and were cloned in pGL2 basic vector (Promega).
Northern blot analysis.
HUVECs were treated with TNF-α for the indicated time period and then harvested for RNA. Ten micrograms of RNA was blotted on a membrane and hybridized with various probes as previously described (29). For super-repressor IκBα (SR-IκBα), cells were infected with control or SR-IκBα virus for 48 h and then left untreated or treated with TNF-α for an additional 4 h.
Transient-transfection assays.
BAEC or COS-7 cells were plated at a density of 5 × 104/well in 12-well plates 1 day before transfection. Transient transfection was performed using Fugene6 reagent (Roche Molecular Biochemicals, Indianopolis, IN) according to instructions by the manufacturer. A total of 1 to 2 μg of plasmid DNA was used in transfections, and total DNA was always kept constant. Transfection efficiencies were normalized by cotransfection of cytomegalovirus-β-galactosidase (β-Gal). Cells were harvested 24 h after transfection, assayed for luciferase activity, and normalized to β-galactosidase or total protein as analyzed with a BCA kit (Pierce, Rockford, IL) for each sample. Under our experimental conditions, we did not observe any variation in the internal control (β-Gal values). In some experiments, cells were treated with human TNF-α (10 ng/ml; R&D systems, Minneapolis, MN) for 15 h before harvesting. All transfections were performed in triplicate for at least three independent experiments.
Coimmunoprecipitation assay.
For coimmunoprecipitation, COS-7 cells were cotransfected with Flag-tagged HDAC4, pCMV-MEF2C, and p65 in various combinations using Fugene. The DNA amount was kept constant by adding empty vector. After 48 h of transfection, cells were lysed in radioimmunoprecipitation assay buffer. The presence of trimolecular complex of KLF2, MEF2C, and p65 was detected by immunoprecipitation with anti-MEF2 antibody and immunoblotting with anti-p65 or anti-FLAG antibody (for HDAC4).
Gel shift studies.
Gel shifts were performed as previously described (29). Approximately 5 μg of the nuclear extract from untreated or TNF-α-treated HUVECs was used for the gel shift assay. MEF2 binding sites from the KLF2 promoter were included using the following oligonucleotides: MEF2, wild type, -CCAGGCTTATATACCGCGGCTAAATTTAGGCTGAGCCCGGA; mutant,CCAGGCTTATATACCGCGGCTAtcggTAGGCTGAGCCCGGA (lowercase letters indicate the mutated MEF2 site). Briefly, nuclear extract was incubated with labeled wild-type oligonucleotide for 30 min. Anti-MEF2 or competitor oligonucleotides were preincubated with nuclear extract for the supershift and the competition. Complexes were separated on 6% nondenaturating native gel, dried, and exposed for autoradiography.
siRNA transfection.
Human MEF2-directed small interfering RNA (siRNA) and a nonspecific control siRNA were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). HUVECs were plated 1 day before transfection in antibiotic-free EBM-2 medium. On the day of transfection, 100 nM of specific siRNA was incubated with Lipofectamine 2000 (Invitrogen, CA) at room temperature for 30 min before being added to the HUVECs in OPTI-MEM (Invitrogen, CA). Three hours later the medium was replaced by EBM-2 and cultured for an additional 48 h. Cells were harvested for RNA as well as for protein. MEF2 expression was confirmed by Northern and immunoblot analysis as described earlier.
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was carried out as reported earlier (33). Briefly, HUVECs were left untreated or treated with TNF-α for 4 h. For the super-repressor experiment, HUVECs were infected with control or SR-IκBα adenovirus before TNF treatment. Native protein-DNA complexes were cross-linked by treatment with 1% formaldehyde for 15 min. Equal aliquots of isolated chromatin were subjected to immunoprecipitation with various antibodies. The DNA associated with specific immunoprecipitates or with negative control mouse immunoglobulin G was isolated and used as a template for the PCR to amplify the KLF2 promoter sequences containing the MEF2 site. The primers used were a 5′ primer, CTAGGCAGGCCCCAAACT TCATCC, and a 3′ primer, CTTATAGGCGCGGCAGGCACAG. As a specificity control, the β-actin promoter was amplified from the same templates using the following primers: 5′ primer, GAGCACAGAGCCTCGCCTTT; 3′ primer, AGACAAAGACCCCGCCGGTT. Some ChIP assays were reconfirmed by quantitative real-time PCR, which was performed in triplicate with Brilliant SYBR green mix using the Mx3000P Real-Time PCR system (Stratagene). Data were presented as severalfold change over DNA input.
RESULTS
TNF-α-mediated inhibition of KLF2 expression requires NF-κB.
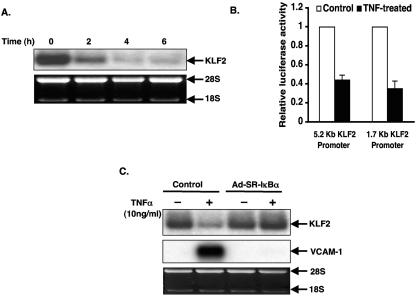
To understand how proinflammatory cytokines like TNF-α regulate KLF2 expression, we treated HUVECs with TNF-α (10 ng/ml) for various time periods and assessed the expression of KLF2 mRNA by Northern blot analysis. As shown in Fig. 1A, maximal inhibition by TNF-α occurred by 4 to 6 h and was sustained for at least 48 h (data not shown). To determine if inhibition occurs at the level of the promoter, we assessed the effect of TNF-α on KLF2 promoter activity in HUVECs. For this purpose, we used luciferase constructs containing the proximal −5.2 kb and −1.7 kb regions of the KLF2 promoter. Consistent with our RNA data, TNF-α substantially reduced the activities of both the promoters (Fig. 1B).
FIG. 1.
TNF-α inhibits KLF2 expression in HUVECs via the NF-κB pathway. (A) 2 × 106 HUVECs were plated in a 10-cm plate. After 24 h, cells were treated with TNF-α (10 ng/ml) for the indicated time period. Cells were harvested, total RNA was extracted, and 10 μg of RNA was subjected to hybridization with human KLF-2 probe. Ethidium bromide staining of 28S and 18S was used as a loading control. (B) HUVECs were transfected with the −5.2 kb or 1.7 kb KLF2 promoter along with β-Gal. After 24 h, cells were left untreated or treated with TNF-α for an additional 15 h. Cells were harvested and assayed for luciferase activities. (C) HUVEC cells were adenovirally infected with control or SR-IκBα virus. After 48 h, cells were left untreated (−) or treated (+) with TNF-α (10 ng/ml) for 4 h. Total RNA was isolated and hybridized with KLF2 probe as described in the legend to panel A.
To investigate whether the TNF-α-mediated inhibition of KLF2 expression is dependent on NF-κB activity, we employed a molecular inhibitor to interrupt this pathway. To more definitively assess whether NF-κB activation is required, we adenovirally overexpressed a nondegradable form of the NF-κB inhibitor IκB (termed super-repressor IκB [SRIκB]). As shown in Fig. 1C, by comparison to control infected cells, the inhibition of KLF2 by TNF-α was completely abolished in HUVECs infected with Ad-SRIκB. Taken together, these data strongly suggest that NF-κB signaling may play a crucial role in TNF-α-mediated inhibition of KLF2 expression.
p65 inhibits KLF2 promoter activity.
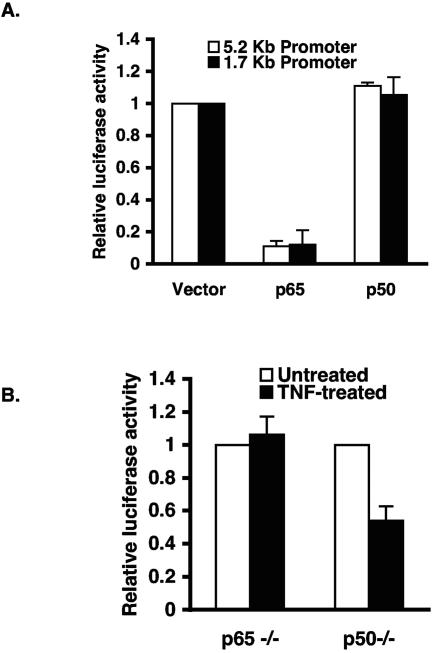
To further understand the molecular mechanism(s) underlying the ability of NF-κB to inhibit KLF2, we assessed the effect of p50 and p65 on KLF2 promoter activity in bovine aortic endothelial cells (BAEC). As shown in Fig. 2A, p65 strongly inhibited KLF2 promoter activity, whereas the p50 subunit alone had no significant effect. The inhibition by p65 was specific, since parallel studies on the VCAM-1 promoter demonstrate that p65 can strongly induce promoter activity (data not shown). To further strengthen this finding, we performed transient-transfection studies in p65 and p50 null mouse embryonic fibroblasts (MEFs) and assessed the effect of TNF-α on KLF2 promoter activity. As shown in Fig. 2B, TNF-α inhibited the −1.7 kb promoter activity in p50−/− cells where p65 expression is intact. However, the effect of TNF-α was completely abrogated in p65−/− MEFs. These observations suggest that p65 (but not p50) is required for TNF-α-mediated inhibition of KLF2 promoter activity.
FIG. 2.
p65 but not p50 inhibits KLF2 promoter activity. (a) BAECs were transiently transfected with 5.2 kb or 1.7 kb KLF2 promoter with or without p65 or p50. Luciferase assays were performed 24 h after transfection. (B) p65 or p50 null mouse embryo fibroblasts were transfected with 1.7 kb KLF2 promoter. After 24 h of transfection, cells were treated with TNF-α (10 ng/ml) for an additional 15 h before harvest for luciferase assays.
p65-mediated inhibition of the KLF2 promoter is independent of DNA binding.
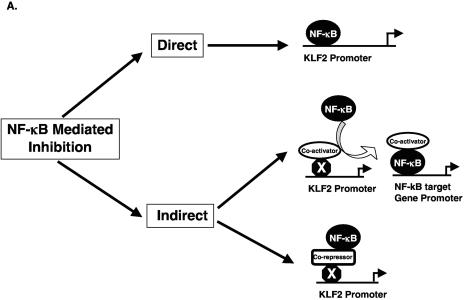
We considered several mechanisms that may account for the p65-mediated inhibition of KLF2 promoter activity. For example, as shown schematically in Fig. 3A, inhibition may occur through direct DNA binding by p65 (12). Alternatively, inhibition may be indirect perhaps by affecting the function of a factor that normally sustains KLF2 promoter activity (labeled X in Fig. 3A). This indirect inhibition may occur through p65-mediated sequestration of a coactivator (e.g., p300/CBP) away from the KLF2 promoter (16) or by recruitment of transcriptional corepressors (e.g., HDACs) to the KLF2 promoter. Indeed, previous studies do support the ability of p65 to interact with p300/CBP as well as HDACs (13) (10).
FIG. 3.
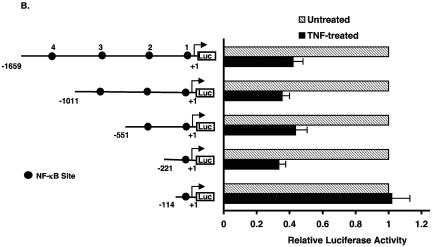
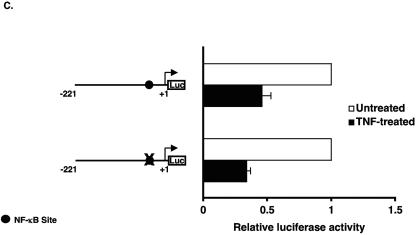
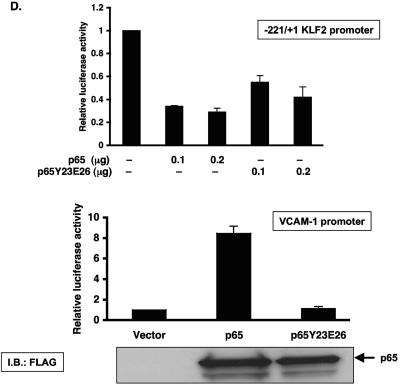
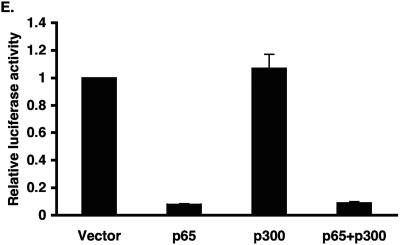
Inhibition of KLF2 promoter activity by p65 is a non-DNA-binding effect. (A) Schematic representation of possible mechanism for TNF-α-mediated inhibition of KLF2. (B) Various deletion constructs of KLF2 promoter were transfected in HUVECs in 12-well plates. Twenty-four hours after transfection, cells were untreated or treated with TNF-α for an additional 15 h before harvest for luciferase assay. (C) HUVECs were transfected with the −221/+1 KLF2 promoter containing wild-type or mutated NF-κB. After 24 h of transfection, cells were treated with vehicle or with TNF-α. Cells were harvested, and the luciferase activities were measured as described for panel A. (D) BAECs were transfected with −221/+1 KLF2 promoter with p65 or p65 mutant (Y23E26). Twenty-four hours posttransfection, cells were harvested for luciferase assay. (E) Indicated plasmids were transiently transfected in BAECs. Cells were lysed in reporter lysis buffer after 24 h of transfection. Luciferase activities were assayed as described above.
To address the first possibility (direct inhibition), we examined the sequence of the −1.7 kb KLF2 promoter and identified four potential NF-κB sites. As shown in Fig. 3B, promoter deletion constructs excluding NF-κB sites 2, 3, and 4 did not alter the ability of TNF-α to inhibit the KLF2 promoter. However, deletion from the −221 bp to −114 bp luciferase construct resulted in complete loss of inhibition. Because the −114 bp construct still retains NF-κB site 1, this region is unlikely to mediate inhibition in context of the full-length promoter. However, to verify this, we mutated NF-κB site 1 in the context of the −221bp-Luc promoter construct and assessed the TNF-α-mediated effect on KLF2 promoter. As expected, TNF-α-mediated inhibition was unaltered (Fig. 3C). Similar results were noted in cotransfection experiments assessing the effect of p65-mediated inhibition on KLF2 promoter activity (data not shown). Furthermore, a point mutant of p65 (Y23E26) that is defective in DNA binding (27) was also able to inhibit KLF2 promoter activity to an extent similar to that for wild-type p65 (Fig. 3D, upper panel). In contrast, this DNA-binding defective mutant of p65 was unable to transactivate a luciferase reporter containing the NF-κB consensus site derived from the VCAM-1 promoter. Taken together, these data suggest that TNF-α/p65-mediated inhibition occurs through a 107-bp region (−221 → −114) of the KLF2 promoter and is not dependent on binding to an NF-κB site.
p65-mediated repression is indirect and involves HDACs.
We next considered the possibility that the p65-mediated repression occurs indirectly—either through sequestering of coactivators, such as p300/CBP, or through recruitment of repressors, such as HDACs (Fig. 3A). To address the former possibility, we performed cotransfection studies. Because p300/CBP are found in rate-limiting amounts, we reasoned that overexpression of this coactivator should abrogate the p65-mediated inhibition of the KLF2 promoter. Indeed, this has been shown in other contexts, such as the rescue of Smad3-mediated repression of proinflammatory target genes (32). As shown in Fig. 3E, p65 strongly inhibited the KLF2 promoter, and this inhibition was maintained in the presence of cotransfected p300. Similar to p300, another coactivator, p/CAF, was also unable to rescue p65's inhibitory effect (data not shown).
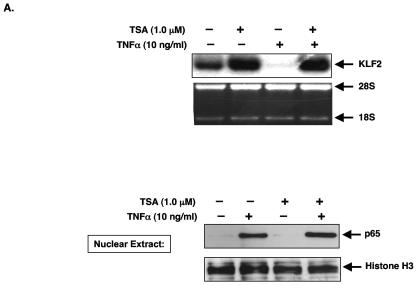
Next, we initiated studies to determine if recruitment of HDACs may be involved, given that these factors often repress transcriptional activity and can interact with p65. As a first step, we assessed whether the HDAC inhibitor trichostatin A (TSA) could affect the TNF-α-mediated inhibition of KLF2 expression. As shown in Fig. 4A, treatment of HUVECs with TNF-α for 4 h potently inhibited KLF2 expression. However, pretreatment with TSA completely abrogated this inhibitory effect, thereby supporting a role for HDACs. To demonstrate that TSA-mediated effect is not due to a reduction in p65 nuclear accumulation, we harvested nuclear extracts in the presence and absence of TSA and TNF-α. As shown in Fig. 4A (lower panel), we observed the expected nuclear accumulation of p65 following TNF-α treatment. Importantly, the accumulation of p65 was not significantly altered in the presence of TSA. Furthermore, we did not observe any significant change in the TNF-mediated induction of p65-DNA binding in the presence of TSA (data not shown).
FIG. 4.
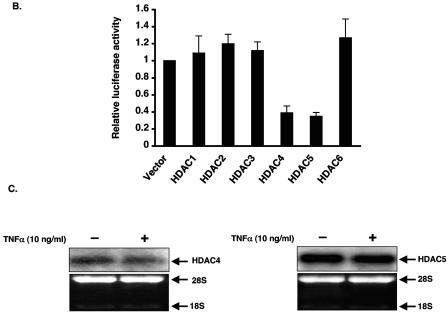
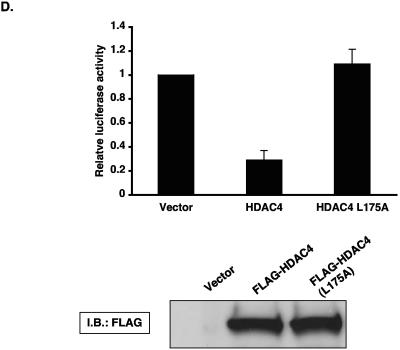
TNF-mediated inhibition of KLF2 involves histone deacetylases. HUVECs were treated with vehicle or TSA (1.0 μM) for 15 h and then were stimulated with TNF-α (10 ng/ml) for an additional 4 h. Cells were harvested for RNA and hybridized with human KLF2 probe as mentioned in the legend to Fig. 1A. In a similar manner, nuclear extracts were harvested from HUVECs and assessed for p65 accumulation. (B) HDAC1 to -6 were transiently transfected in BAECs along with the −221/+1 KLF2 promoter. Cells were harvested and assayed for luciferase activity as described in Materials and Methods. (C) HUVECs were stimulated with TNF-α for 4 h, and total RNA was extracted. Ten micrograms of RNA was hybridized with respective probes as described in the legend to Fig. 1A. Ethidium bromide staining of 28S and 18S was used as a loading control. (D) Wild-type HDAC4 or MEF2-binding-defective mutant of HDAC4 was transiently transfected in BAECs along with the −221/+1 KLF2 promoter. Luciferase activities were determined as described in Materials and Methods. Luciferase lysates were also immunoblotted with anti-FLAG for the expression of wild-type and mutant HDAC4 (lower panel).
To determine which HDACs may be operative, we assessed the effects of various HDACs (1-6) on activity of the −221bp-Luc KLF2 promoter. As shown in Fig. 4B, HDAC1, -2, -3, and -6 did not alter promoter activity. On the other hand, HDAC4 and HDAC5 significantly reduced KLF2 promoter activity. We also verified that these two HDACs are expressed in HUVECs by Northern blot analysis. As shown in Fig. 4C, both HDAC4 and -5 are expressed in endothelial cells. Taken together, these data suggest recruitment of HDACs may be operative in the TNF-α-mediated inhibition of KLF2 expression.
Interestingly, HDAC4 and -5 are distinguished by the presence of an amino-terminal extension that contain an 18-amino-acid motif that mediates binding to MEF2 transcription factor (22). Furthermore, MEF factors have recently been implicated as regulators of endothelial cell biology (10, 31). To gain insight into the importance of MEF2 in the context of HDAC4-mediated inhibition of the KLF2 promoter, we used a point mutant of HDAC4 (L175A) that is defective in binding to MEF2 (30) and assessed the effect of this mutant on KLF2 promoter activity. As shown in Fig. 4D, HDAC4 potently inhibited the KLF2 promoter, whereas the HDAC4 mutant was completely defective in repressing promoter activity (Fig. 4D). These data suggest that HDACs 4 and 5 can repress KFL2 promoter activity. Furthermore, these studies suggest that interaction of HDAC4 with MEF2 factors may be important for this inhibitory effect.
MEF2 factors regulate KLF2 promoter activity.
The observations shown in Fig. 4 coupled with the fact that the 107-bp region of the KLF2 promoter essential for TNF-α's inhibitory effect contains a single consensus MEF site led us to further assess the role of MEF factors in the context of the KLF2 promoter. Indeed, we considered the possibility that MEF proteins may be the “X” factor described in the schema outlined in Fig. 3A.
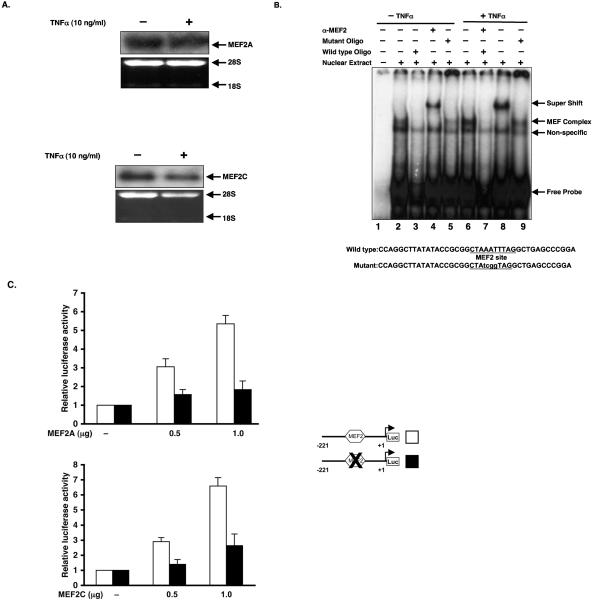
We first verified that MEF2A and MEF2C are expressed in HUVECs. As shown in Fig. 5A, both factors are expressed, and expression is not significantly affected by treatment with TNF-α. We next assessed the ability of the single A/T-rich MEF site in the KLF2 promoter to bind endogenous MEF2 factors using nuclear extract from HUVECs treated with or without TNF-α. As shown in Fig. 5B, MEF2 bound to the oligonucleotide-containing MEF2 site, and this binding was completely abolished by a wild-type competitor. In contrast, mutation of the MEF site resulted in a mild effect on MEF2 binding. Finally, supershift studies verified the presence of MEF2 in this complex. No significant effect was seen on MEF binding to DNA after TNF-α treatment.
FIG. 5.
MEF2 binds and transactivates KLF2 promoter. (A) Total RNA from vehicle or TNF-treated HUVECs was hybridized with a MEF2A or MEF2C probe as described in Materials and Methods. Ethidium bromide of RNA was used as a loading control. (B) Double-stranded labeled oligonucleotides with sequence of the MEF2 site from the KLF2 promoter (−221/+1) were incubated with 5 μg of protein of nuclear extracts of untreated or TNF-treated HUVECs for 30 min. Supershift or competition experiments were carried out by preincubations of the nuclear extract with anti-MEF2 antibody (lanes 4 and 7) or with 100-fold molar excess of competitor oligonucleotides for 30 min. (C) COS-7 cells were seeded in 12-well plates 24 h prior to transfection. Cells were transfected with wild-type or MEF2 site-mutated −221/+1 promoter and an increasing amount of MEF2A or MEF2C. Total DNA was kept constant by adding empty vector. Luciferase activities were assessed as described in Materials and Methods.
To further evaluate the relationship between MEF2 and KLF2, we assessed the effect of MEF2A and MEF2C on KLF2 promoter activity. As shown in Fig. 5C, both MEF2A and MEF2C induce KLF2 promoter (−221bp-Luc) activity in a dose-dependent manner (Fig. 5C). Importantly, mutation of the MEF2 site significantly attenuated MEF2-mediated induction (Fig. 5C).
p65, MEF2, and HDAC4 can form a trimolecular complex.
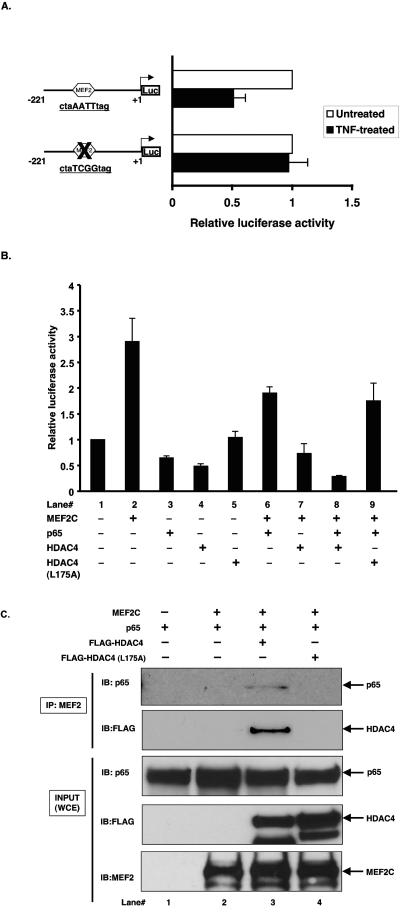
The data in Fig. 4D and Fig. 5 raise the possibility that MEF2 factors may be the “X” factor regulating KLF2 promoter activity. Indeed, we hypothesized that following TNF-α treatment, p65 and HDAC4 may cooperate to repress the MEF2-mediated activation of the KLF2 promoter. If this scenario were true, one would anticipate that the MEF site ought to be essential for the TNF-α-mediated inhibition of the KLF2 promoter. Indeed, as shown in Fig. 6A, mutation of the MEF2 site completely abrogated TNF-α-mediated inhibition of the KLF2 promoter. To test whether p65 and HDAC4 may cooperate to repress MEF2's ability to induce the KLF2 promoter, we performed cotransfection studies. As shown in Fig. 6B, MEF2C transactivated the KLF2 promoter approximately threefold (lane 2). Furthermore, p65 and HDAC4 both inhibited the MEF2C-mediated induction of the KLF2 promoter (lanes 6 and 7). This inhibition was more pronounced with the combination of p65 and HDAC4, as noted in lane 8. In contrast, no cooperative inhibition was seen with the HDAC mutant (L175A; lane 9) that is unable to interact with MEF2 factors.
FIG. 6.
p65 interacts with MEF2 in the presence of HDAC4 to associate with the KLF2 promoter. (A) HUVECs were transiently transfected with the proximal −221/+1 KLF2 promoter containing the wild-type or mutated MEF2 binding site. Cells were treated with TNF-α for 15 h before harvest in reporter lysis buffer. Luciferase activities were determined as described in Materials and Methods. (B) COS7 cells were transfected with indicated plasmids alone or in combination. Total DNA was kept constant by adding empty vector. Twenty-four hours after transfection, cells were harvested in 1× reporter lysis buffer and assayed for luciferase activities. Values were corrected with total protein and were expressed as severalfold activation over vector. (C) COS7 cells were transfected with 2 μg of each of p65, MEF2C, HDAC4, or HDAC4 (L175A) alone or in combination. Total DNA was kept constant by adding empty vector. Forty-eight hours posttransfection, cells were harvested in radioimmunoprecipitation assay buffer. Cell lysates were immunoprecipitated with anti-MEF2 followed by immunoblotting with anti-p65 or anti-FLAG (for HDAC4). Ten percent of whole-cell extract (WCE) was used as an input and was directly immunoblotted with respective antibodies.
To determine if p65 and HDAC4 can form a trimolecular complex with MEF2 and thereby inhibit its function, we performed coimmunoprecipitation studies using FLAG-HDAC4, FLAG-HDAC4 (L175A), p65, and MEF2C in various combinations. As shown in Fig. 6C, immunoprecipitation with MEF2 followed by immunoblotting with FLAG showed the association of MEF2C with HDAC4. Consistent with previous reports, the HDAC4 mutant was unable to bind MEF2C (upper second panel; compare lane 3 and 4). Most strikingly, p65 association with MEF2 was observed only in presence of HDAC4 (upper panel; compare lane 2 and 3).
TNF-α-mediated recruitment of p65-HDAC4 complex to the KLF2 promoter.
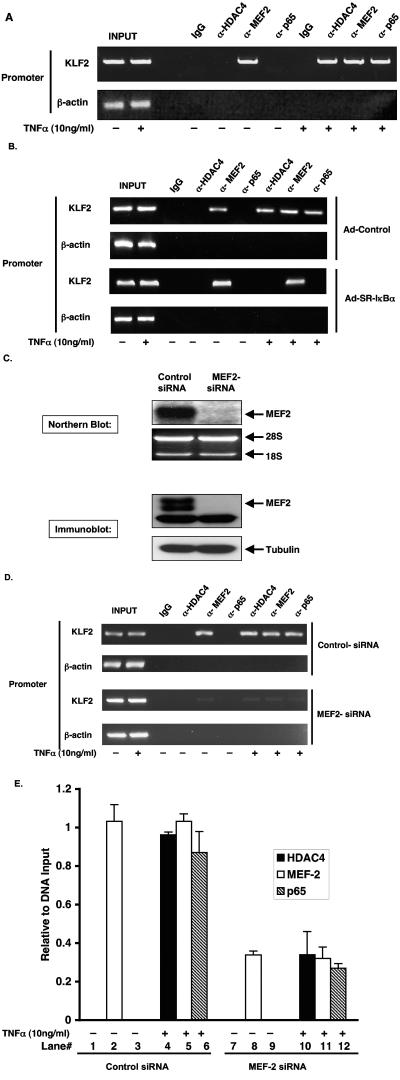
Finally, we performed ChIP assays to look for the association of p65, HDAC4, or MEF2 to the native KLF2 promoter in HUVECs in the presence or absence of TNF-α treatment. As demonstrated in Fig. 7A, we observed that without TNF-α treatment, MEF2 but not p65 or HDAC4 was associated with the KLF2 promoter. Importantly, TNF-α treatment resulted in the association of p65 and HDAC4 along with MEF2 to the KLF2 promoter (upper panel). We further demonstrated that inhibition of the NF-κB pathway by SR-IκBα completely blocks the TNF-α-mediated recruitment of p65 and HDAC4 to the KLF2 promoter, as shown in Fig. 7B. However, MEF2 association with the KLF2 promoter was unaltered.
FIG. 7.
TNF-mediated recruitment of p65 and HDAC4 to KLF2 promoter is dependent on MEF2. (A) HUVECs were either vehicle(−) or treated (+) with TNF-α for 4 h and formaldehyde fixed to cross-link the DNA to native chromatin-associated protein complexes, and chromatin lysates were prepared as described in Materials and Methods. Equal aliquots of chromatin lysates were subjected to immunoprecipitation with antibodies against p65, HDAC4, and MEF2. Mouse immunoglobulin G was used as a control. The DNA associated with immunoprecipitates was isolated and used as templates to PCR amplify the KLF2 promoter region containing the MEF2 site. PCR amplification of the β-actin promoter was used as a control. (B) HUVECs were infected with control or SR-IκBα adenovirus. After 48 h of infection, cells were treated with TNF-α for an additional 4 h and assessed for ChIP as described above for panel A. (C) HUVECs were transfected with control or MEF2 siRNA by Lipofectamine 2000 (Invitrogen, CA) according to the manufacturer's protocol. Forty-eight hours posttransfection, cells were harvested for RNA or for protein. Ten micrograms of RNA was blotted on a nylon membrane and hybridized with human MEF2 probe. Ethidium bromide staining of 28S and 18S was used for a loading control. Protein expression was assessed by immunoblotting with MEF2 (C-21) antibody, and tubulin was used as a loading control. (D) Control or MEF2 siRNA was transfected in HUVECs as described for panel C. Forty-eight hours later, cells were treated with TNF-α (+) for 4 h and processed for ChIP assay as described for panel A. (E) Quantitative real-time PCR was performed on DNA isolated from ChIP assay from MEF2 siRNA experiment. DNAs were analyzed in triplicate with Brilliant SYBR Green mix (Stratagene, CA), using the Mx3000P real-time PCR system. Values were presented as relative to DNA input.
Next, we assessed the importance of the MEF2 protein in the context of TNF-α-mediated association of the p65-HDAC4 complex. As shown in Fig. 7C, MEF2-specific siRNA strongly reduced MEF2 mRNA (upper panel) and protein expression (lower panel). We next performed ChIP assays after treatment of HUVECs with control and MEF2-specific siRNA. As shown in Fig. 7D, by comparison to control siRNA-treated cells, knockdown of MEF2 strongly reduced MEF2 binding in the absence or presence of TNF-α. Furthermore, after knockdown, the association of p65 and HDAC4 to the KLF2 promoter was strongly attenuated. To gain the greater insight, we validated these ChIP data by quantitative real-time PCR. As shown in Fig. 7E, knockdown of MEF2 resulted in an approximately 60% reduction in promoter-associated MEF2 (compare lanes 2 and 8) in the absence of TNF-α. Following cytokine stimulation, binding of p65, MEF2, and HDAC4 to the KLF2 promoter was significantly reduced (compare lanes 4, 5, and 6 versus 10, 11, and 12). These results suggest that TNF-α treatment recruits the p65-HDAC4 complex to associate with MEF2 on the KLF2 promoter and that this recruitment is MEF2 dependent.
DISCUSSION
As the interface with blood and tissue, the endothelium is critically involved in the biological response to inflammation. In general, proinflammatory stimuli confer proadhesive and prothrombotic features to the endothelium through the induction of hundreds of target genes, such as VCAM-1 or tissue factor. Less well understood (but equally important) is the fact that these noxious stimuli can reduce the expression of “protective” factors, such as eNOS, thereby rendering the endothelium more vulnerable to activation (3). In this regard, we recently identified KLF2 as a novel factor that inhibits endothelial proinflammatory activation. KLF2 potently induces “protective” factors, such as eNOS procoagulant factors (19). As such, the reduction of KLF2 by proinflammatory stimuli may be a key upstream event leading to endothelial activation, and an understanding of the molecular basis for inhibition of KLF2 is of considerable importance.
One of the first key findings from our work is that the reduction in KLF2 is dependent on NF-κB. Indeed, while the literature is replete with examples of how NF-κB activates target genes, how it inhibits gene expression is less well understood. NF-κB is composed of homo- and heterodimeric complexes of members of the Rel family of proteins, consisting of p65 (RelA), c-Rel, RelB, p50, and p52 (11). The best-studied and most abundant of these complexes is the p50-p65 heterodimer. This heterodimer is normally maintained in the cytoplasm by inhibitor molecules, such as IκB. Typically, following cytokine stimulation, the IKK signalosome is activated and phosphorylates the inhibitor IκB, resulting in its degradation. As a consequence, the p50/p65 complex is liberated, moves to the nucleus, binds to the promoter region of target genes, and induces gene expression (11). In the present study, we found, using a molecular inhibitor (SR-IκB), that inhibition of NF-κB prevented the reduction in KLF2 expression by TNF-α (Fig. 1C). This effect is specific, as evidenced by the fact that VCAM-1, a classic NF-κB-inducible target in endothelial cells, exhibited an antithetical expression pattern (Fig. 1C). Furthermore, studies overexpressing p50 or p65 (Fig. 2) coupled with observations with p50 and p65 null cells implicate p65 as necessary for this inhibitory effect. The ability of p65 to inhibit target genes has also been noted in several recent publications (5, 13). However, our data using null cells (Fig. 2B) provides cogent evidence of a clear distinction between the abilities of these two factors to inhibit transcriptional activity (2) (12).
We considered several plausible mechanisms by which p65 may inhibit KLF2 expression and/or promoter activity (Fig. 3A). For example, it is possible that p65 may bind DNA as a homodimer and inhibit gene expression (12). Indeed, direct binding of NF-κB to DNA has been implicated in acetaldehyde-mediated inhibition of the collagen promoter (23). Other reports suggest that NF-κB can inhibit target gene expression independently of DNA binding—either through sequestration of coactivators (16) or recruitment of corepressors (5, 13). In either case, the function of a factor “X” that normally regulates the target gene promoter is attenuated, resulting in a reduction in target gene expression (Fig. 3A). We provide multiple lines of evidence ranging from promoter deletion and mutational analyses (Fig. 3B and C), along with use of a p65 DNA-binding-defective mutant (Fig. 3D), that DNA binding of p65 is not requisite for the p65-mediated inhibition of the KLF2 promoter. Furthermore, overexpression of coactivators, such as p300 and p/CAF (Fig. 3E), did not affect p65's inhibitory effects. Finally, the ability of TSA to rescue the TNF-mediated inhibition of KLF2 suggests that recruitment of corepressors, such as HDACs, may be involved.
The HDACs constitute a major class of repressor molecules, and a total of 11 have been identified to date (6). The first line of evidence implicating these factors is derived from our observation that the HDAC inhibitor TSA completely prevented TNF-α's ability to inhibit KLF2 expression in endothelial cells. This raised the possibility that p65 and HDACs may cooperate to inhibit the KLF2 promoter. Indeed, a precedent exists for such an interaction, derived from recent studies indicating that several members of the HDAC family (HDAC1, HDAC2, HDAC4 and HDAC5) (1, 5, 13), through interaction with p65, can inhibit target promoter expression. However, since the p65-mediated inhibition of the KLF2 promoter was not dependent on DNA binding and because HDACs do not directly bind DNA, we hypothesized that p65 and HDAC4 and -5 may cooperate to inhibit the function of a third, as yet unidentified, factor “X” that binds and regulates the KLF2 promoter (Fig. 3A).
A critical observation as to the identity of this factor “X” came from the observation that among six HDACs tested, only HDAC4 and -5 can inhibit the KLF2 promoter (Fig. 4B). This is a particularly noteworthy finding, because one of the distinguishing features of these two HDACs is that they are able to bind MEF factors (20, 21). Furthermore, the potential importance of the MEF-HDAC interaction was substantiated by the fact that a point mutant of HDAC4 that is unable to bind MEF2 factors was rendered completely incapable of inhibiting the KLF2 promoter (Fig. 4D). Indeed, our promoter and gel shift studies support an important role for MEF factors in regulating KLF2 expression (Fig. 5A to C). Furthermore, a combination of cotransfection studies and coimmunoprecipitation and ChIP assays suggest that following TNF-α treatment, p65 and HDAC4 cooperate to inhibit MEF2 function (Fig. 6 and 7). MEF2 factors are members of the MADS box (MCM1, Agamous, Deficiens, Serum response factor) family of transcription factors that bind to A/T-rich sequences (21). Although best known for their role in muscle development, an emerging literature implicates MEF2A and MEF2C as critical regulators of endothelial biology (10, 25, 31). For example, Wang et al. identified mutations in MEF2A in an inherited disorder with features of coronary artery disease (31). Furthermore, MEF2C has been implicated as a regulator of endothelial integrity and permeability (10). The basis for the favorable effects of MEF factors in endothelial cells is not understood. Our results raise the intriguing possibility that some of the favorable properties attributed to MEF protein in endothelial biology may be secondary to the induction of KLF2—particularly in light of the fact that KLF2 can induce key molecules, such as eNOS/thrombomudulin, and inhibit proadhesive factors, such as VCAM-1 (19, 29). Finally, given the importance of MEF factors in other cellular systems, these observations may have broader implications. For example, TNF-α is known to induce muscle wasting and atrophy (9, 14). While these effects are undoubtedly complex, our studies suggest that a cooperative inhibition of MEF factors by p65 and HDACs may be operative.
.
Acknowledgments
This work was supported by NIH grant HL-69477 (M.K.J.), HL-72952 (M.K.J.), HL-75427 (M.K.J.), HL-76754 (M.K.J.), American Heart Association Grant 0250030N (M.K.J.), American Diabetes Association Grant 1-02-JF-40 (M.K.J.), NIH grant F32 HL78183 (A.K.), and American Heart Association Postdoctoral Fellowship 0425789T (Z.L.).
REFERENCES
- 1.Ashburner, B. P., S. D. Westerheide, and A. S. Baldwin, Jr. 2001. The p65 (RelA) subunit of NF-κB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol. 21:7065-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beg, A. A., W. C. Sha, R. T. Bronson, S. Ghosh, and D. Baltimore. 1995. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature 376:167-170. [DOI] [PubMed] [Google Scholar]
- 3.Berk, B. C., J. J. Abe, W. Min, J. Suprapisitchat, and C. Yan. 2001. Endothelial atheroprotective and anti-inflammatory mechanisms. Ann. N. Y. Acad. Sci. 947:93-111. [DOI] [PubMed] [Google Scholar]
- 4.Bieker, J. J. 2001. Kruppel-like factors: three fingers in many pies. J. Biol. Chem. 276:34355-34358. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, K. J., S. Rocha, and N. D. Perkins. 2004. Active repression of antiapoptotic gene expression by RelA(p65) NF-kappa B. Mol. Cell 13:853-865. [DOI] [PubMed] [Google Scholar]
- 6.de Ruijter, A. J., A. H. van Gennip, H. N. Caron, S. Kemp, and A. B. van Kuilenburg. 2003. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feinberg, M. W., Z. Lin, S. Fisch, and M. K. Jain. 2004. An emerging role for kruppel-like factors in vascular biology. Trends Cardiovasc. Med. 14:241-246. [DOI] [PubMed] [Google Scholar]
- 8.Gimbrone, M. A., Jr., J. N. Topper, T. Nagel, K. R. Anderson, and G. Garcia-Cardena. 2000. Endothelial dysfunction, hemodynamic forces, and atherogenesis. Ann. N. Y. Acad. Sci. 902:230-239,239-240. [DOI] [PubMed] [Google Scholar]
- 9.Guttridge, D. C., M. W. Mayo, L. V. Madrid, C. Y. Wang, and A. S. Baldwin, Jr. 2000. NF-kappaB-induced loss of MyoD messenger RNA: possible role in muscle decay and cachexia. Science 289:2363-2366. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi, M., S. W. Kim, K. Imanaka-Yoshida, T. Yoshida, E. D. Abel, B. Eliceiri, Y. Yang, R. J. Ulevitch, and J. D. Lee. 2004. Targeted deletion of BMK1/ERK5 in adult mice perturbs vascular integrity and leads to endothelial failure. J. Clin. Investig. 113:1138-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayden, M. S., and S. Ghosh. 2004. Signaling to NF-kappaB. Genes Dev. 18:2195-2224. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann, A., T. H. Leung, and D. Baltimore. 2003. Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. EMBO J. 22:5530-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong, C. Y., J. H. Park, K. H. Seo, J. M. Kim, S. Y. Im, J. W. Lee, H. S. Choi, and K. Lee. 2003. Expression of MIS in the testis is downregulated by tumor necrosis factor alpha through the negative regulation of SF-1 transactivation by NF-kappa B. Mol. Cell. Biol. 23:6000-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackman, R. W., and S. C. Kandarian. 2004. The molecular basis of skeletal muscle atrophy. Am. J. Physiol. Cell Physiol. 287:C834-C843. [DOI] [PubMed] [Google Scholar]
- 15.Katz, J. P., N. Perreault, B. G. Goldstein, C. S. Lee, P. A. Labosky, V. W. Yang, and K. H. Kaestner. 2002. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development 129:2619-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ke, S., A. B. Rabson, J. F. Germino, M. A. Gallo, and Y. Tian. 2001. Mechanism of suppression of cytochrome P-450 1A1 expression by tumor necrosis factor-alpha and lipopolysaccharide. J. Biol. Chem. 276:39638-39644. [DOI] [PubMed] [Google Scholar]
- 17.Kuo, C. T., M. L. Veselits, K. P. Barton, M. M. Lu, C. Clendenin, and J. M. Leiden. 1997. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 11:2996-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo, C. T., M. L. Veselits, and J. M. Leiden. 1997. LKLF: a transcriptional regulator of single-positive T cell quiescence and survival. Science 277:1986-1990. [DOI] [PubMed] [Google Scholar]
- 19.Lin, Z., A. Kumar, S. Senbanerjee, K. Staniszewski, K. Parmar, D. E. Vaughan, M. A. Gimbrone, Jr., V. Balasubramanian, G. Garcia-Cardena, and M. K. Jain. 2005. Kruppel-like factor 2 (KLF2) regulates endothelial thrombotic function. Circ. Res. 96:e48-e57. [DOI] [PubMed] [Google Scholar]
- 20.McKinsey, T. A., C. L. Zhang, J. Lu, and E. N. Olson. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408:106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2002. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27:40-47. [DOI] [PubMed] [Google Scholar]
- 22.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2002. Signaling chromatin to make muscle. Curr. Opin. Cell Biol. 14:763-772. [DOI] [PubMed] [Google Scholar]
- 23.Novitskiy, G., J. J. Potter, L. Rennie-Tankersley, and E. Mezey. 2004. Identification of a novel NF-kappaB-binding site with regulation of the murine alpha2(I) collagen promoter. J. Biol. Chem. 279:15639-15644. [DOI] [PubMed] [Google Scholar]
- 24.Nuez, B., D. Michalovich, A. Bygrave, R. Ploemacher, and F. Grosveld. 1995. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375:316-318. [DOI] [PubMed] [Google Scholar]
- 25.Olson, E. N. 2004. Undermining the endothelium by ablation of MAPK-MEF2 signaling. J. Clin. Investig. 113:1110-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perkins, A. C., A. H. Sharpe, and S. H. Orkin. 1995. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375:318-322. [DOI] [PubMed] [Google Scholar]
- 27.Saccani, S., I. Marazzi, A. A. Beg, and G. Natoli. 2004. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor kappaB response. J. Exp. Med. 200:107-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segre, J. A., C. Bauer, and E. Fuchs. 1999. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 22:356-360. [DOI] [PubMed] [Google Scholar]
- 29.SenBanerjee, S., Z. Lin, G. B. Atkins, D. M. Greif, R. M. Rao, A. Kumar, M. W. Feinberg, Z. Chen, D. I. Simon, F. W. Luscinskas, T. M. Michel, M. A. Gimbrone, Jr., G. Garcia-Cardena, and M. K. Jain. 2004. KLF2 is a novel transcriptional regulator of endothelial proinflammatory activation. J. Exp. Med. 199:1305-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, A. H., and X. J. Yang. 2001. Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol. Cell. Biol. 21:5992-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, L., C. Fan, S. E. Topol, E. J. Topol, and Q. Wang. 2003. Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science 302:1578-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Werner, F., M. K. Jain, M. W. Feinberg, N. E. Sibinga, A. Pellacani, P. Wiesel, M. T. Chin, J. N. Topper, M. A. Perrella, and M. E. Lee. 2000. Transforming growth factor-b1 inhibition of macrophage activation is mediated via Smad3. J. Biol. Chem. 275:36653-36658. [DOI] [PubMed] [Google Scholar]
- 33.Zeng, M., A. Kumar, G. Meng, Q. Gao, G. Dimri, D. Wazer, H. Band, and V. Band. 2002. Human papilloma virus 16 E6 oncoprotein inhibits retinoic X receptor-mediated transactivation by targeting human ADA3 coactivator. J. Biol. Chem. 277:45611-45618. [DOI] [PubMed] [Google Scholar]