Abstract
Rice (Oryza sativa L.) is a staple food for more than half of the world’s population, but its yields are increasingly threatened by environmental problems, including soil compaction. This problem limits root growth which limits water and nutrient foraging capacity thus reduces productivity due to, restricted diffusion of ethylene, a key plant hormone playing an important role in exacerbating these effects. Elevated ethylene levels in compacted soils can further inhibit root development. However, rice varieties that are less sensitive to ethylene may have an advantage as they exhibit better root growth and resource utilization under such conditions. In this study, 220 diverse rice accessions were analyzed to uncover the genetic factors that influence root length reduction (RLR) in response to ethylene. Genome-wide association studies (GWAS) identified a significant QTL on chromosome 10, named qRLR10, associated with ethylene response. Within this region, 20 candidate genes were identified, with three F-box genes namely Os10g0124700, Os10g0126600 and Os10g0128200 showing a strong correlation with RLR variations. These genes are involved in protein degradation, root development and hormone signaling, indicating their possible role in regulating ethylene sensitivity. The results suggest that rice varieties with lower ethylene sensitivity may have better root growth in compacted soils, making them ideal targets for breeding programs aimed at improving resilience to harsh environmental conditions. These results underscore the critical role of ethylene in rice root development and provide valuable insights for future rice improvement strategies aimed at mitigating the effects of soil compaction.
Keywords: rice, root, ethylene sensitivity, GWAS, F-box
1. Introduction
Rice (Oryza sativa L.) is one of the most important staple crops that feeds more than half of the world’s population. Thailand, a major player in global rice production, ranks as the sixth-largest rice producer, following China, India, Indonesia, Bangladesh, and Vietnam (FAO, 2017). Farming practices in Thailand and other countries have significantly evolved over the years, with mechanization becoming increasingly central. The use of heavy machinery, such as power tillers, tractors, and harvesters, has become widespread in agricultural operations, including soil preparation, sowing, and harvesting (FAO, 2017). While these advancements have boosted agricultural productivity, they have also led to unintended consequences, particularly soil compaction, which poses a significant challenge to crop growth and yield (Hassan et al., 2007). Soil compaction, a condition where soil particles are pressed together, reducing macropore spaces, is a major constraint in modern agriculture. It negatively impacts crop production by restricting root growth, limiting soil water and nutrient availability, and decreasing soil porosity (Arocena, 2000; Khan et al., 2012). Additionally, soil compaction hinders gas exchange between plant roots and the surrounding environment, further exacerbating plant stress. In this context, plants naturally produce various volatile phytohormones, such as ethylene, which can diffuse through air spaces between soil aggregates in noncompacted soil. However, soil compaction restricts ethylene diffusion in soil micropores, leading to inhibited root elongation and reduced overall root growth, which negatively impacts the plant’s ability to access water and nutrients (Pandey et al., 2021; Arocena, 2000). Elevated ethylene levels in compacted soils can trigger stress responses that further restrict root development, thereby compromising plant growth and yield (Lin et al., 2010).
Ethylene, a gaseous phytohormone synthesized from methionine in the presence of oxygen, is integral to plant stress responses. It is produced in most plant tissues and serves as a key regulator of various physiological processes, particularly under stress conditions (Lin et al., 2010). Ethylene Response Factors (ERFs) are central to ethylene’s role in stress response, acting as regulatory hubs that interact with other stress-related signals, such as abscisic acid and (ABA) and jasmonate (Müller and Munné-Bosch, 2015). These interactions are critical in modulating plant responses to diverse abiotic stresses, including salinity, drought, low temperature, heat, and variations in light availability. The ethylene signaling pathway is a complex network involving several key components, including receptors, EIN2, and transcription factors like EIN3 (Ethylene insensitive 3)/EIL1 (Ethylene insensitive-Like1). These components activate ERFs, which, in turn, regulate the expression of genes involved in stress and downstream hormone signaling pathway. Notably, rice mutant lines with alterations in EIN3/EIL1 maintain normal primary root growth even under compacted soil conditions, similar to wild-type plants in non-compacted soils (Pandey et al., 2021). This resilience is likely due to the disrupted ethylene signaling in these mutants, which prevents the typical ethylene-induced inhibition of root elongation in response to soil compaction.
The regulation of root growth and development in rice is a complex process influenced by multiple phytohormones, including auxin, ABA, cytokinin, and ethylene. These hormones are involved in various aspects of root growth and development, from biosynthesis to signal transduction, and play a crucial role in how plants respond and acclimate to environmental stresses (Dello Ioio et al., 2007; De Smet and Jürgens, 2007; McSteen, 2010). Ethylene, in particular, interacts with auxin by modulating its biosynthesis and transport, highlighting its role as a mediator in the regulation of root development (Le et al., 2001). Given the critical role of ethylene in plant stress response, manipulating its pathway offers a promising strategy for improving crop production, especially under adverse conditions. For instance, in maize, mutations in the ACC synthase gene, a key enzyme in ethylene biosynthesis, have been shown to delay leaf senescence and maintain photosynthesis during drought, leading to increased grain yield under stress (Habben et al., 2014). However, breeding for ethylene-responsive traits in crops remains challenging, particularly in non-GMO contexts. Nevertheless, utilizing diverse plant collections and innovative screening protocols can provide valuable genetic resources for breeding programs aimed at enhancing tolerance to unfavorable environments. In rice, developing ethylene-insensitive varieties could be a potential strategy to improve growth and production, particularly in compacted soils where water and nutrient extraction are critical for survival. This approach is especially relevant for countries like Thailand, where traditional breeding methods are preferred over genetic modification. By leveraging the genetic diversity found in local or landrace rice accessions, which often contain adaptive traits to harsh environments, breeders can identify and incorporate valuable alleles into commercial cultivars.
In this study, we aimed to investigate the root length response of 220 rice accessions under controlled and ethylene-treated conditions. By employing a genome-wide association study (GWAS), we sought to identify the genomic regions and key genes regulating root length and ethylene sensitivity in rice. The findings from this study are expected to contribute to the development of new rice varieties with enhanced root penetration ability and improved adaptation to soil compaction and other abiotic stresses.
2. Materials and methods
2.1. Plant materials and phenotypic screening experiment
A rice diversity panel consisting of 220 rice accessions, including Thai landraces, Thai improved rice varieties, and rice varieties from other countries, were used to represent the diverse geographical regions and rice ecosystems such as rainfed lowland, lowland, upland, deepwater and irrigated system which are found in Thailand ( Supplementary Table S1 ). All rice accessions were evaluated ethylene sensitivity using an ethylene chamber to observe root characteristics in response to ethylene. A total of 10-20 seeds per accession were used to screen the ethylene sensitivity. Seeds were first sterilized with 10% sodium hypochlorite and rinsed with distilled water. The seeds were pre-germinated by soaking with distilled water at 30°C in the dark for 2 days then placed on stainless net, each accession was separated with a well. The samples were then put in an air-tight plastic box (27.5 x 39.5 x 25 cm) with a fitted net. The water (200 ml) was added into the box to provide enough moisture and humidity to allow the seedlings to grow in a rice growth room (25°C). The experiments were set into two conditions, ethylene treatment and control. The experimental design was a complete randomized design (CRD) with three replications. For ethylene treatment, the ethylene gas was injected into the plastic box by a calibrated syringe to get 20 ppm concentration. The root length phenotype was observed at 2 days after ethylene treatment following the modified protocols (Ma et al., 2013 and Pandey et al., 2021). For ethephon treatment, the rice seeds were sterilized with 10% sodium hypochlorite for 10 minutes, then rinsed with deionized (DI) water until clean. Next, they were transferred to plates with 20 mL of DI water and incubated in the dark at 30°C for 2 days. After germination, seedlings were placed on stainless steel mesh channels in two chambers: one with 3 L of DI water (control) and one with 100 µM ethephon in 3 L of DI water (treatment). Seedlings were treated for 48 hours, and root lengths were measured using ImageJ software to calculate root length reduction. The percentage of root length reduction is calculated by subtracting the root length in ethylene from the root length in control and multiplied by 100, then divided by root length in control. R version 4.4.1 (R Core Team, 2024) was used to perform data distribution and statistical analysis with R packages in Rstudio (Wickham, 2016; RStudio Team, 2020).
2.2. Population study and genome wide association study analysis
A total of 2,340,775 SNPs with a minor allele frequency (MAF) greater than 5%, missing data less than 10%, and pruned with a 1-kb window were used for population studies and GWAS ( Supplementary Figure S1 ). These SNPs were derived from a whole-genome resequencing project conducted by the National Center for Genetic Engineering and Biotechnology, Thailand (Theerawitaya et al., 2022) using Nipponbare IRGSP 1.0 as the reference rice genome. A SNP density heatmap was generated using the CMplot package in RStudio (Yang et al., 2022). Population structure and individual relatedness were determined through principal component analysis (PCA) and kinship analysis using the GAPIT software (Wang and Zhang, 2021). GWAS analysis was performed using the FarmCPU model in GAPIT, with significant SNP associations defined by a uniform threshold of -log10(p-value) > 6.
2.3. Linkage disequilibrium decay, haplotype analysis and candidate gene identification
Genome-wide LD decay was estimated by PopLDdecay (Zhang et al., 2019) and the LD structure and haplotypes within the QTL region were inferred using LDBlockshow (Zhang et al., 2019). The identification of genes located within the QTL region was based on the annotations from RAP-DB (https://rapdb.dna.affrc.go.jp) (Kawahara et al., 2013; Sakai et al., 2013). The effects of SNP variation in the genes were identified using the Variant Effect Predictor (VEP - https://asia.ensembl.org/info/docs/tools/vep/index.html). The gene-based haplotyped were analyzed using GeneHapR (Zhang et al., 2023) based on the SNPs with a moderate or high effect present in the genes. Candidate genes were identified by conducting an analysis of variance (ANOVA) to test for significant differences between the major haplotypes (each consisting of at least 7 samples) within the QTL.
3. Results
3.1. Phenotypic variation in root length reduction percentage under ethylene gas exposure in a panel of 220 rice accessions
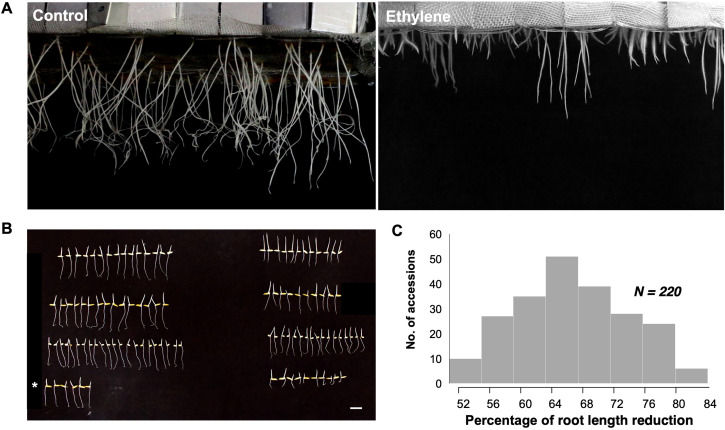
Root responsiveness to ethylene was assessed by calculating the percentage of reduction in root length (%RLR) for each rice accession by comparing the root lengths under ethylene treatment (20 ppm) with control conditions ( Figure 1A ). KDML105 served as a sensitive control to validate ethylene response and ensure reproducibility ( Figure 1B ). The %RLR in this diversity panel ranged from 52.06% to 82.98%, with an average of 67.76% ( Figure 1A ). Rice accessions with a %RLR of 60.67% or lower were classified as low ethylene sensitivity, while those with a %RLR of 74.9% or higher were considered highly sensitive. Naew Ubon 2 exhibited the lowest %RLR (52.06%), and Phitsanulok 80 had the highest %RLR (82.98%). The %RLR distribution across the panel displayed a slightly skew towards low values, forming a continuous curve ( Figure 1C ). The RLR phenotype in rice accessions with low and high sensitivity was confirmed using an ethephon solution treatment ( Supplementary Figure S2 ).
Figure 1.
(A) Root length reduction under control and ethylene treatments, showing contrasting phenotypes of root length among different accessions. (B) Comparison of root length between the two treatments for selected accessions, with KDML105 as the reference check (denoted by *). Left: control; right: ethylene treatment. Scale bar: 2 cm. (C) Distribution of data showing the percentage of root length reduction across accessions.
3.2. Population study and linkage disequilibrium analysis
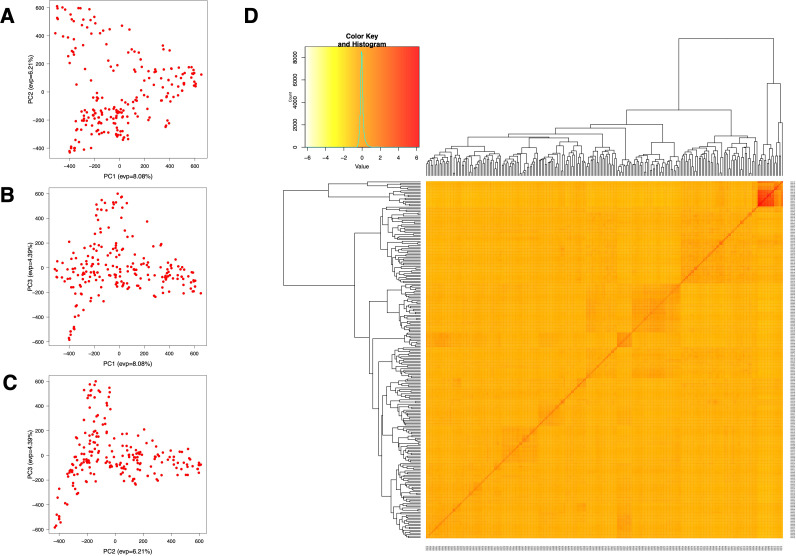
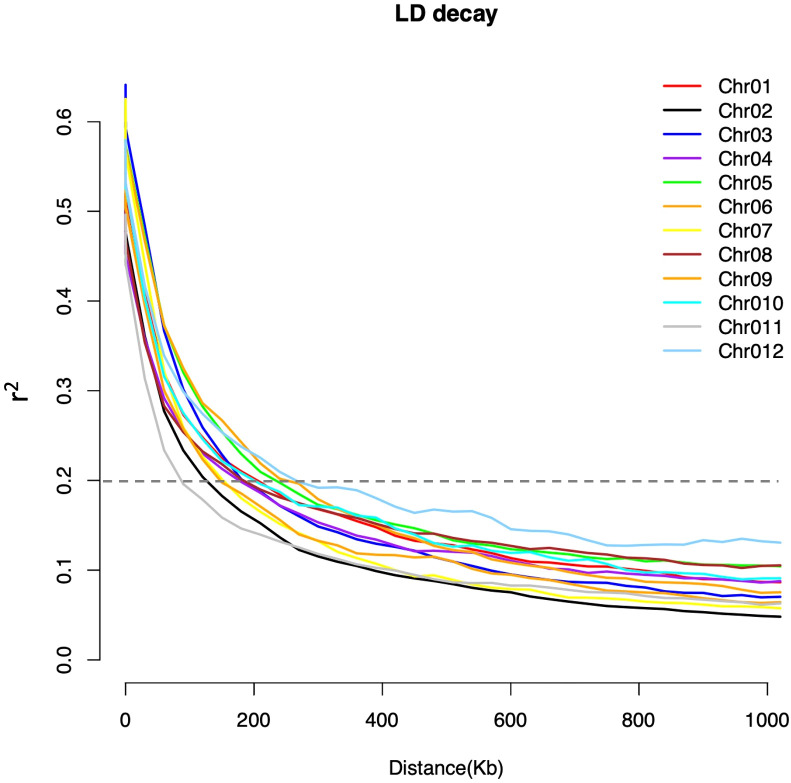
The population structure and cryptic relationship among the 220 rice accessions were analyzed using Principal Component Analysis (PCA) and a kinship matrix ( Figure 2 ). All rice accessions in this study belong to the indica ecotype ( Supplementary Table S1 ). The PCA and VanRaden kinship matrix results indicated the absence of distinct subpopulations within the diversity panel. The kinship coefficient is in the range of 0 to 0.5 which is often used for relatedness in GWAS. To estimate genome-wide linkage disequilibrium (LD) decay, we calculated the pairwise LD index (r2) and plotted the LD decay across the rice diversity panel. The average LD decay for the entire genome of the 220 rice accessions was approximately 190 kb (r2 < 0.2). Chromosome-wise, the shortest LD decay distance was around 100 kb on chromosome 11, while the longest was approximately 280 kb on chromosome 12 ( Figure 3 ).
Figure 2.
Population structure for GWAS analysis of 220 rice accessions. (A–C) Variation captured by the first three principal components (PCs): (A) PC1 vs. PC2, (B) PC1 vs. PC3, and (C) PC2 vs. PC3. (D) Kinship matrix of the 220 individuals, presented as a heatmap.
Figure 3.
Chromosome-wide linkage disequilibrium (LD) decay estimated from SNP genotypes of 220 rice accessions. Each line represents the smoothed r2 values for all marker pairs on each chromosome as a function of the distance between marker pairs. The LD decay threshold (r2 = 0.2) is indicated by the dotted line.
3.3. Genome-wide association analysis and candidate gene identification
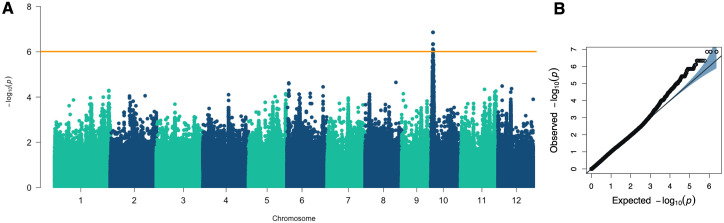
To identify genomic regions associated with root responsiveness to ethylene, we conducted a genome-wide association study (GWAS) on 220 rice accessions, utilizing 2,340,775 SNP as genotype data. All SNPs had a minor allele frequency (MAF) greater than 0.05. We employed the FarmCPU model, incorporating the first three principal components (PCs) and the kinship matrix, to detect quantitative trait loci (QTLs). As a result, a quantitative trait nucleotide (QTN) was detected with the highest significance (-log10 p-value of 6.86) on chromosome 10 at position 1,562,158 bp ( Figure 4 ). Given the average linkage disequilibrium (LD) decay on chromosome 10, approximately 200 kb (r2 = 0.2), we defined the region between 1.36 and 1.76 Mb as the QTL region (named qRLR10) associated with root responsiveness to ethylene ( Table 1 ).
Figure 4.
GWAS results for the percentage of root length reduction. (A) Manhattan plot from the FarmCPU model displaying the association signals across the genome. (B) Quantile-Quantile (Q-Q) plot comparing the distribution of observed versus expected p-values from the GWAS analysis.
Table 1.
The QTLs for percentage of root length reduction based on significant SNPs.
| QTL | Chromosome | QTL region (Mb) | Lead SNP | P-value | MAF |
|---|---|---|---|---|---|
| qRLR10 | 10 | 1.36 – 1.76 | 1562158 | 1.39 × 10-07 | 0.44 |
We then identified candidate genes within the qRLR10 region by analyzing genes annotated within the LD block containing the lead SNP. A total of 20 genes with annotated functions were identified ( Table 2 ). Among these, several genes are related to root development and plant hormone responses, such as Os10g0122600 (nodulin domain-containing protein, OsSNDP1), which is associates with root hair elongation (Huang et al., 2013). In addition, 12 genes encode F-box domain-containing proteins, including Os10g0123200, Os10g0124500 (OsFbox507, OsFBX343), Os10g0124700 (OsFbox508, OsFBX344), Os10g0125300, Os10g0126000 (OsFbox510, OsFBX346), Os10g0126500 (OsFbox511, OsFBX347), Os10g0126600 (OsFbox512, OsFBX348), Os10g0126700 (OsFbox513, OsFBX346), Os10g0126800 (OsFbox514, OsFBX350), Os10g0127000 (OsFbox515, OsFBX351), Os10g0127900 (OsFbox516, OsFBX352), and Os10g0128200 (OsFbox517, OsFBX353). Furthermore, genes encoding CCR4-associated factor 1 (CAF1) proteins, Os10g0123900 (OsCAF1-14) and Os10g0124200 (OsCAF1-15), which are important enzymes for catalysis of mRNA de-adenylation in eukaryotes, were also identified. CAF1 proteins have been reported to response to abiotic stresses in plants. A search of Rice Expression Profile Database (http://ricexpro.dna.affrc.go.jp/) showed that 16 gene expression profiles are in database, and they have different expression patterns, referring their different biological functions. Particularly, Os10g0122000, Os10g0122600, Os10g0124700, Os10g0125700, and Os10g0126000 are expressed in various rice organs throughout rice development, including high expression in root. According to root gene expression profile, Os10g0122000 is highly expressed in maturation zone, Os10g0124700 is highly expressed in division zone, Os10g0122600, Os10g0125700, and Os10g0126000 are expressed in elongation zone, and maturation I.
Table 2.
The 20 associated genes within the QTL region containing nonsynonymous SNPs causing missense and nonsense effect.
| QTL | Chr. | Start | Stop | Locus ID | Description |
|---|---|---|---|---|---|
| qRLR10 | 10 | 1405692 | 1407206 | Os10g0122000 | UDP-glucuronosyl/UDP-glucosyltransferase family protein. (Os10t0122000-00) |
| 1431142 | 1432407 | Os10g0122300 | Transferase domain containing protein. (Os10t0122300-00) | ||
| 1447351 | 1451372 | Os10g0122600 | Sec14 and nodulin domain-containing protein, Root hair elongation (Os10t0122600-01) | ||
| 1479530 | 1481991 | Os10g0123200 | Cyclin-like F-box domain containing protein. (Os10t0123200-01) | ||
| 1525512 | 1526367 | Os10g0123900 | Similar to CAF1 family ribonuclease containing protein. (Os10t0123900-00) | ||
| 1530563 | 1532176 | Os10g0124000 | Ribosomal protein L7A/RS6 family domain containing protein. (Os10t0124000-01) | ||
| 1539462 | 1540349 | Os10g0124200 | Similar to CAF1 family ribonuclease containing protein. (Os10t0124200-00) | ||
| 1556778 | 1559803 | Os10g0124500 | Cyclin-like F-box domain containing protein. (Os10t0124500-01) | ||
| 1562608 | 1565335 | Os10g0124700 | Cyclin-like F-box domain containing protein. (Os10t0124700-01) | ||
| 1622222 | 1635958 | Os10g0125300 | F-box domain, cyclin-like domain containing protein. (Os10t0125300-01);F-box protein, Tapetum cell development, Pollen formation, Control of anther development (Os10t0125300-02) | ||
| 1642565 | 1646770 | Os10g0125700 | NB-ARC domain containing protein. (Os10t0125700-00) | ||
| 1656688 | 1659249 | Os10g0126000 | Similar to F-box domain containing protein, expressed. (Os10t0126000-01) | ||
| 1681092 | 1684225 | Os10g0126500 | Cyclin-like F-box domain containing protein. (Os10t0126500-01) | ||
| 1688331 | 1691224 | Os10g0126600 | Cyclin-like F-box domain containing protein. (Os10t0126600-01) | ||
| 1694920 | 1697854 | Os10g0126700 | Similar to F-box domain containing protein, expressed. (Os10t0126700-01) | ||
| 1700332 | 1702953 | Os10g0126800 | Cyclin-like F-box domain containing protein. (Os10t0126800-01) | ||
| 1711433 | 1713322 | Os10g0127000 | Similar to F-box domain containing protein, expressed. (Os10t0127000-01) | ||
| 1726495 | 1727783 | Os10g0127350 | Glycosyl transferase, family 14 domain containing protein. (Os10t0127350-00) | ||
| 1743579 | 1752159 | Os10g0127900 | Similar to F-box domain containing protein, expressed. (Os10t0127900-01; Regulation of glucose-induced delay of seed germination (Os10t0127900-03) | ||
| 1755462 | 1757886 | Os10g0128200 | Similar to F-box domain containing protein, expressed. (Os10t0128200-01) |
3.4. Haplotype analysis
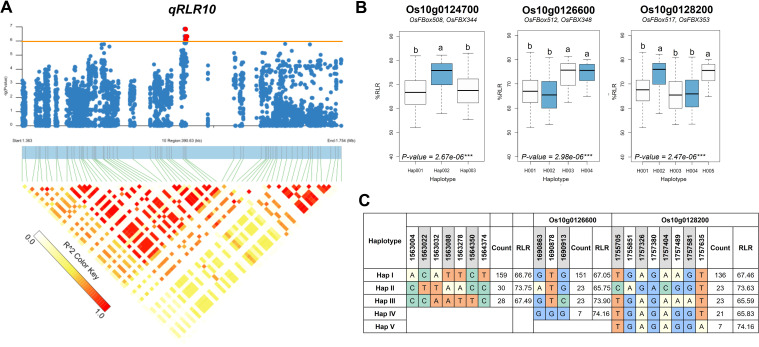
To prioritize potential candidate genes, we performed a gene-based haplotype analysis on each of the 20 genes to identify those with haplotypes associated with variations in root length reduction (RLR) within the diversity panel. The analysis focused on SNPs with moderate to high effects, such as nonsynonymous and nonsense SNPs, within each gene. Among the 20 genes analyzed, three—Os10g0124700, Os10g0126600, and Os10g0128200—were found to contain haplotypes significantly associated with the phenotype differences. For Os10g0124700, three distinct major haplotypes were identified. Haplotype H002 (n = 30) was significantly associated with a higher %RLR (73.75), whereas haplotypes H001 (n = 159) and H003 (n = 28) were associated with lower %RLR values (66.76 and 67.49, respectively: Figure 5 ). In the case of Os10g0126600, four major haplotypes were found. Haplotypes H001 (n = 151) and H002 (n = 23) were associated with lower %RLR values (67.05 and 65.75, respectively), while the haplotypes H003 (n = 23) and H004 (n = 7) were associated with higher %RLR values (73.90 and 74.16, respectively). For Os10g0128200, five major haplotypes were identified. Among these, the haplotypes H002 (n = 23) and H005 (n = 7) were associated with higher %RLR values (73.63 and 74.16, respectively), whereas haplotypes H001 (n = 136), H003 (n = 23) and H004 (n = 21) were associated with lower %RLR values (67.46, 65.59 and 65.83, respectively).
Figure 5.
Associated regions and haplotype analysis of qRLR10. (A) Regional Manhattan plots and LD heatmap across the 390.6-kb region surrounding the significant SNP. Red dots above the red line indicate significant SNPs. Triangles in the LD heatmap represent haploblocks with varying R2 values. (B) Boxplots displaying the distribution of root length reduction (RLR) across haplotype groups of Os10g0124700, Os10g0126600, and Os10g0128200. The middle line in each boxplot represents the median. Different letters above each haplotype group show significant difference at p ≤ 0.05. (C) Haplotype analysis of the three genes in the region.
4. Discussion
Root plays a vital role in rice growth and performance under both normal and stress conditions, serving as the primary interface between the plant and its environment. The growth, development, and adaptive responses of rice are regulated by a complex network of internal and external factors, including phytohormones such as ethylene, auxin and cytokinin. The hormones can act synergistically or antagonistically to modulate plant growth, particularly under stress conditions (He et al., 2005; Ueda et al., 2020; Yamauchi et al., 2020). Ethylene, often referred to as the stress hormones, is a key mediator in plant adaptive responses, interacting with other hormones to regulate growth under adverse conditions (Huang et al., 2020; Ma et al., 2013; Yang et al., 2015). Roots are particularly sensitive to changes in ethylene concentration, which can lead to reduced root length and increased radial cell expansion (Vaseva et al., 2018). Additionally, ethylene has been shown to influence root hair growth and development (Feng et al., 2017) and plays a role in the plant’s response to soil compaction by reducing root length and increasing root thickness in rice, a process mediated by abscisic acid (ABA) and auxin (Huang et al., 2022). Transgenic rice plants overproducing ethylene exhibit lower 1000-grain weight and faster leaf senescence compared to those with reduced ethylene production, indicating the significant impact of ethylene on overall plant development and yield (Ma et al., 2013). In the ethylene signaling pathway, the ethylene gas triggers the translocation of CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) from the endoplasmic reticulum to the nucleus. This process involves the phosphorylation of ETHYLENE-INSENSITIVE2 (EIN2) by CTR1, which inhibits EIN2 from signaling in the absence of ethylene (Ju et al., 2012; Jun et al., 2004). In Arabidopsis, plants with enhanced nuclear-localized CTR1 have shown to improve tolerance to drought and salinity stress (Park et al., 2023). These findings suggest that manipulating ethylene sensitivity could be a valuable strategy for improving crop resilience under unfavorable environmental conditions, such as those exacerbated by climate change, while maintaining productivity.
In rice, ethylene’s role in root response to soil compaction has been further elucidated through studies involving rice mutants with varying levels of ethylene sensitivity. For instance, ethylene-insensitive mutants exhibited significantly longer root than those with high ethylene sensitivity under soil compaction, highlighting the genetic basis of ethylene responsiveness (Pandey et al., 2021). In this study, 220 rice accessions were collected from many areas in Thailand in order to have a diverse genetic source to study GWAS. According to Kinship and PCA analysis, our germplasm showed the low levels of relatedness within the rice indica panel (Aesomnuk et al., 2021). Thus, this diversity panel can be used in GWAS analysis. Importantly, the higher genetic diversity of germplasm, the more genetic variation in target trait, leading to successfully identify the crucial gene underlying the target trait (Quero et al., 2018). This will help plant breeder to enhance breeding programs to genetically improve target traits (Mourad et al., 2020). Our genome-wide association study (GWAS) using 220 indica rice accessions identified a QTL, qRLR10, associated with difference in root length reduction (RLR) in response to ethylene. Within this QTL region, 20 candidate genes were identified. The subsequent gene-based haplotype analysis revealed that specific haplotypes within three of these genes—Os10g0124700, Os10g0126600, and Os10g0128200—are significantly associated with variations in RLR. This association underscores the potential functional importance of these genes in the regulation of ethylene responsiveness.
The gene Os10g0124700 (OsFbox508, OsFBX344), which encodes an F-box protein, showed the highest expression in rice roots before the flowering stage according to the strand-specific RNA-Seq database in RAP-DB (Wang et al., 2015). F-box proteins are components of the SCF (SKP1-CUL1-F-box protein) complex, which functions as an E3 ubiquitin ligase, tagging target proteins for degradation. This pathway is crucial for regulating various physiological processes, including growth, development, and stress responses (Gagne et al., 2002; Lechner et al., 2006; Dreher and Callis, 2007; Zhang et al., 2008; Guo et al., 2018; Xu et al., 2021). F-box proteins are likely involved in the fine-tuning of ethylene responses by regulation the stability of key component signaling pathway. For example, in Arabidopsis, the F-box protein EBF1/EBF2 targets the ethylene-responsive transcription factor EIN3 for degradation, thus modulating the ethylene response. Given the association of Os10g0124700 with variations in RLR, it is plausible that this gene plays a similar role in rice. Os10g0126600 (OsFbox512, OsFBX348) is another F-box domain-containing protein, which has been linked to drought resistance in Shanlan upland rice through its involvement with long non-coding RNA (lncRNA) (Wang et al., 2015). F-box proteins are known to integrate with signaling pathways, including those mediated by phytohormones like ethylene, auxin, and ABA (Koops et al., 2011; Sasaki et al., 2003; Wang et al., 2009; Xu et al., 2021; Yan et al., 2011). The association of Os10g126600 with RLR suggests its involvement in the regulation of root architecture in response to ethylene, potentially through interactions with other hormones and stress-related pathways. In drought conditions, ethylene production is often elevated, and the ability of roots to adapt to this stress is critical for plant survival. The role of Os10g0126600 in drought resistance suggests that it may be crucial for modulating root growth under conditions where ethylene levels are altered, helping the plant to balance growth and survival (Yang et al., 2022). This gene’s involvement in RLR further supports its importance in the adaptive response of rice roots to environmental stresses, likely through the modulation of hormone signaling networks. Os10g0128200 (OsFbox517, OsFBX353) has also been identified as a gene associated with RLR, though its specific role in ethylene signaling and root development is less characterized compared to the other two genes. However, its significant association with root length reduction suggest that it may be involved in key regulatory process that control root growth in response to ethylene.
Although not all identified candidate genes showed significant differences in RLR among haplotypes, they may still play roles in root growth and ethylene response. Evidence suggests crosstalk among plant hormones, with ethylene inhibiting rice root growth in compacted soil via ABA and auxin mechanisms (Huang et al., 2022). For example, OsSNDP1 (Os10g0122600), involved in root hair elongation, might contribute to root responses under compacted soil conditions (Huang et al., 2013; Kong et al., 2024). OsFbx352 (Os10g0127900) influences ABA biosynthesis and catabolism, affecting root architecture under stress (Song et al., 2012). OsCAF1-15 (Os10g0124200) is involved in stress hormone responses, suggesting a role in ethylene signaling under compaction (Liang et al., 2009). Additional functional studies on these genes, as well as research on root growth in compacted soil conditions, will be necessary to clearly understand the roles of these genes in ethylene signaling and root development. Understanding the specific functions of these genes could provide valuable insights into the genetic basis of stress resilience in rice, with potential applications in breeding programs aimed at improving crop performance under adverse environmental conditions.
5. Conclusion
In this study, we identified a QTL on chromosome 10, qRLR10, associated with root length reduction (RLR) in response to ethylene using GWAS in 220 indica rice accessions. Within this QTL region, a cluster of genes encoding F-box proteins were identified as candidate genes, with three of these F-box genes considered particularly promising. These findings highlight the crucial role of ethylene in root development and stress adaptation, and the identification of these candidate genes lays the groundwork for future research and crop improvement strategies.
Acknowledgments
The authors are grateful to Assist. Prof. Dr. Wachiraya Imsabai from Postharvest Technology Center, Department of Horticulture, Faculty of Agriculture at Kasetsart University, Kamphaeng Sean campus for supporting the ethylene gas in this study.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the Office of the Ministry of Higher Education, Science, Research and Innovation; and the Thailand Science Research and Innovation through the Kasetsart University Reinventing University Program 2021. This work was also partially financially supported by the Thailand Rice Science Research Hub of Knowledge (NRCT Grant Number: N34E670027), and the National Science, Research and Innovation Fund, Thailand Science Research and innovation (TSRI) (Grant No.: FFB670076/0337). SK was supported by Thailand Graduate Institute of Science and Technology (TGIST) Scholarships (Grant No. SCA-CO-2563-12042-TH), NSTDA, Thailand, and Department of Agronomy, Faculty of Agriculture at Kamphaeng Saen, Kasetsart University, Thailand.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.
Author contributions
SK: Formal analysis, Writing - original draft, Writing – review & editing. WA: Formal analysis, Writing – review & editing. PM: Formal analysis, Writing – review & editing. VR: Formal analysis, Writing – review & editing. JS: Conceptualization, Supervision, Writing – review & editing. BP: Formal analysis, Writing – review & editing. MB: Conceptualization, Supervision, Writing – review & editing. SW: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. SA: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1501533/full#supplementary-material
References
- Aesomnuk W., Ruengphayak S., Ruanjaichon V., Sreewongchai T., Malumpong C., Vanavichit A., et al. (2021). Estimation of the genetic diversity and population structure of. Agronomy 11 (5), 995. doi: 10.3390/agronomy11050995 [DOI] [Google Scholar]
- Arocena J. M. (2000). Cations in solution from forest soils subjected to forest floor removal and compaction treatments. For. Ecol. Manage. 133, 71–80. doi: 10.1016/S0378-1127(99)00299-6 [DOI] [Google Scholar]
- Dello Ioio R., Linhares F. S., Scacchi E., Casamitjana-Martinez E., Heidstra R., Costantino P., et al. (2007). Cytokinins determine arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17 (8), 678–682 doi: 10.1016/j.cub.2007.02.047 [DOI] [PubMed] [Google Scholar]
- De Smet I., Jürgens G. (2007). Patterning the axis in plants – auxin in control. Curr. Opin. Genet. Dev. 17, 337–343. doi: 10.1016/j.gde.2007.04.012 [DOI] [PubMed] [Google Scholar]
- Dreher K., Callis J. (2007). Ubiquitin, hormones and biotic stress in plants. Ann. Bot. 99, 787–822. doi: 10.1093/aob/mcl255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO . (2017). Rice technical manual for extension officers (Apia: Food and Agriculture Organization of the United Nations; ). [Google Scholar]
- Feng Y., Xu P., Li B., Li P., Wen X., An F., et al. (2017). Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 114, 13834–13839. doi: 10.1073/pnas.1711723115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J. M., Downes B. P., Shiu S. H., Durski A. M., Vierstra R. D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 99, 11519–11524. doi: 10.1073/pnas.162339999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Zhang Y., Tu Y., Wang Y., Cheng W., Yang Y. (2018). Overexpression of an EIN3-binding F-box protein2-like gene caused elongated fruit shape and delayed fruit development and ripening in tomato. Plant Sci. 272, 131–141. doi: 10.1016/j.plantsci.2018.04.016 [DOI] [PubMed] [Google Scholar]
- Habben J. E., Bao X., Bate N. J., Debruin J. L., Dolan D., Hasegawa D., et al. (2014). Transgenic alteration of ethylene biosynthesis increases grain yield in maize under field drought-stress conditions. Plant Biotechnol. J. 12, 685–693. doi: 10.1111/pbi.12172 [DOI] [PubMed] [Google Scholar]
- Hassan F. U., Ahmad M., Ahmad N., Abbasi M. K. (2007). Effects of subsoil compaction on yield and yield attributes of wheat in the sub-humid region of Pakistan. Soil Tillage Res. 96, 361–366. doi: 10.1016/j.still.2007.06.005 [DOI] [Google Scholar]
- He X. J., Mu R. L., Cao W. H., Zhang Z. G., Zhang J. S., Chen S. Y. (2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44, 903–916. doi: 10.1111/j.1365-313X.2005.02575.x [DOI] [PubMed] [Google Scholar]
- Huang L., Jiang Q., Wu J., An L., Zhou Z., Wong C. E., et al. (2020). Zinc finger protein 5 (ZFP5) associates with ethylene signaling to regulate the phosphate and potassium deficiency-induced root hair development in Arabidopsis. Plant Mol. Biol. 102, 143–158. doi: 10.1007/s11103-019-00937-4 [DOI] [PubMed] [Google Scholar]
- Huang G., Kilic A., Karady M., Zhang J., Mehra P., Hu H., et al. (2022). Ethylene inhibits rice root growth in compacted soil via ABA and auxin mediated mechanisms. PNAS 119 (30), e2201072119. doi: 10.1073/pnas.2201072119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Kim C. M., Xuan Y. H., Park S. J., Piao H. L., Je B., et al. (2013). OsSNDP1, a Sec14-nodulin domain-containing protein, plays a critical role in root hair elongation in rice. Plant Mol. Biol. 82, 39–50. doi: 10.1007/s11103-013-0033-4 [DOI] [PubMed] [Google Scholar]
- Ju C., Yoon G. M., Shemansky J. M., Lin D. Y., Ying Z. I., Chang J., et al. (2012). CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 109, 19486–19491. doi: 10.1073/pnas.1214848109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S. H., Han M. J., Lee S., Seo Y. S., Kim W. T., An G. (2004). OsEIN2 is a positive component in ethylene signaling in rice. Plant Cell Physiol. 45, 281–289. doi: 10.1093/pcp/pch033 [DOI] [PubMed] [Google Scholar]
- Kawahara Y., de la Bastide M., Hamilton J. P., Kanamori H., Mccombie W. R., Ouyang S., et al. (2013). Improvement of the Oryza sativa nipponbare reference genome using next generation sequence and optical map data. Rice 6, 3–10. doi: 10.1186/1939-8433-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. R., Abbasi M. K., Hussan A. U. L. (2012). Effect of induced soil compaction on changes in soil properties and wheat productivity under sandy loam and sandy clay loam soils : A greenhouse experiment. Commun. Soil Sci. Plant Anal. 43, 2550–2563. doi: 10.1080/00103624.2012.711877 [DOI] [Google Scholar]
- Kong X., Yu S., Xiong Y., Song X., Nevescanin-Moreno L., Wei X., et al. (2024). Root hairs facilitate rice root penetration into compacted layers. Curr. Biol. 34, 2039–2048.e3. doi: 10.1016/j.cub.2024.03.064 [DOI] [PubMed] [Google Scholar]
- Koops P., Pelser S., Ignatz M., Klose C., Marrocco-Selden K., Kretsch T. (2011). EDL3 is an F-box protein involved in the regulation of abscisic acid signalling in Arabidopsis thaliana. J. Exp. Bot. 62, 5547–5560. doi: 10.1093/jxb/err236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J., Vandenbussche F., van der Straeten D., Verbelen J.-P. (2001). In the early response of arabidopsis roots to ethylene, cell elongation is up- and down-regulated and uncoupled from differentiation1. Plant Physiol. 125, 519–522. doi: 10.1104/pp.125.2.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner E., Achard P., Vansiri A., Potuschak T., Genschik P. (2006). F-box proteins everywhere. Curr. Opin. Plant Biol. 9, 631–638. doi: 10.1016/j.pbi.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Liang W., Li C., Liu F., Jiang H., Li S., Sun J., et al. (2009). The Arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res. 19, 307–316. doi: 10.1038/cr.2008.317 [DOI] [PubMed] [Google Scholar]
- Lin L. C., Hsu J. H., Wang L. C. (2010). Identification of novel inhibitors of 1-aminocyclopropane-1-carboxylic acid synthase by chemical screening in arabidopsis thaliana J. Biol. Chem. 285, 33445–33456. doi: 10.1074/jbc.M110.132498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B., He S. J., Duan K. X., Yin C. C., Chen H., Yang C., et al. (2013). Identification of rice ethylene-response mutants and characterization of MHZ7/OsEIN2 in distinct ethylene response and yield trait regulation. Mol. Plant 6, 1830–1848. doi: 10.1093/mp/sst087 [DOI] [PubMed] [Google Scholar]
- McSteen P. (2010). Auxin and monocot development. Cold Spring Harb. Perspect. Biol. 2, a001479. doi: 10.1101/cshperspect.a001479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourad A. M. I., Belamkar V., Baenziger P. S. (2020). Molecular genetic analysis of spring wheat core collection using genetic diversity, population structure, and linkage disequilibrium. BMC Genomics 21, 434. doi: 10.1186/s12864-020-06835-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Munné-Bosch S. (2015). Ethylene response factors: A key regulatory hub in hormone and stress signaling. Plant Physiol. 169, 32–41. doi: 10.1104/pp.15.00677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey B. K., Huang G., Bhosale R., Hartman S., Sturrock C. J., Jose L., et al. (2021). Plant roots sense soil compaction through restricted ethylene diffusion. Plant Sci. 371, 276–280. doi: 10.1126/science.abf3013 [DOI] [PubMed] [Google Scholar]
- Park H. L., Seo D. H., Lee H. Y., Bakshi A., Park C., Chien Y. C., et al. (2023). Ethylene-triggered subcellular trafficking of CTR1 enhances the response to ethylene gas. Nat. Commun. 14, 1–15. doi: 10.1038/s41467-023-35975-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quero G., Gutiérrez L., Monteverde E., Blanco P., Pérez de Vida F., Rosas J., et al. (2018). Genome-wide association study using historical breeding populations discovers genomic regions involved in high-quality rice. Plant Genome 11, 1–12. doi: 10.3835/plantgenome2017.08.0076 [DOI] [PubMed] [Google Scholar]
- R Core Team . (2024). R: A Language and Environment for Statistical Computing (Vienna, Austria: R Foundation for Statistical Computing; ). Available at: https://www.R-project.org/ (accessed September 1, 2024). [Google Scholar]
- RStudio Team (2020). RStudio: Integrated Development for R (Boston, MA: RStudio, PBC; ). Available at: http://www.rstudio.com/. [Google Scholar]
- Sakai H., Lee S. S., Tanaka T., Numa H., Kim J., Kawahara Y., et al. (2013). Rice annotation project database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 54. doi: 10.1093/pcp/pcs183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A., Itoh H., Gomi K., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., et al. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299 (5614), 1896–1898. doi: 10.1126/science.1081077 [DOI] [PubMed] [Google Scholar]
- Song S., Dai X., Zhang W. H. (2012). A rice F-box gene, OsFbx352, is involved in glucose-delayed in rice. J. Exp. Bot. 63, 695–709. doi: 10.1093/jxb/err313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theerawitaya C., Wanchana S., Ruanjaichon V., Tisaram R., Samphumphuang T., Sotesaritkul T., et al. (2022). Determination of traits responding to iron toxicity stress at different stages and genome-wide association analysis for iron toxicity tolerance in rice (Oryza sativa L.). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.994560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H., Ito T., Inoue R., Masuda Y., Nagashima Y., Kozuka T., et al. (2020). Genetic interaction among phytochrome, ethylene and abscisic acid signaling during dark-induced senescence in arabidopsis thaliana. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseva I. I., Qudeimat E., Potuschak T., Du Y., Genschik P., Vandenbussche F., et al. (2018). The plant hormone ethylene restricts Arabidopsis growth via the epidermis. Proc. Natl. Acad. Sci. U. S. A. 115, E4130–E4139. doi: 10.1073/pnas.1717649115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kong H., Hong M. (2009). F-box proteins regulate ethylene signaling and more. Genes Dev. 23, 391–396. doi: 10.1101/gad.1781609 [DOI] [PubMed] [Google Scholar]
- Wang H., Niu Q. W., Wu H. W., Liu J., Ye J., Yu N., et al. (2015). Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits. Plant J. 84, 404–416. doi: 10.1111/tpj.13018 [DOI] [PubMed] [Google Scholar]
- Wang J., Zhang Z. (2021). GAPIT version 3: boosting power and accuracy for genomic association and prediction. Genomics Proteomics Bioinf. 19 (4), 629-640. doi: 10.1016/j.gpb.2021.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2016). ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag New York; ). Available at: https://ggplot2.tidyverse.org (accessed September 1, 2024). [Google Scholar]
- Xu K., Wu N., Yao W., Li X., Zhou Y., Li H. (2021). The biological function and roles in phytohormone signaling of the F-box protein in plants. Agronomy 11, 1–17. doi: 10.3390/agronomy11112360 [DOI] [Google Scholar]
- Yamauchi T., Tanaka A., Tsutsumi N., Inukai Y., Nakazono M. (2020). A role for auxin in ethylene-dependent inducible. Plants 9, 610. doi: 10.3390/plants9050610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y.-S., Chen X.-Y., Yang K., Sun Z.-X., Fu Y.-P., Zhang Y.-M., et al. (2011). Overexpression of an F-box protein gene reduces abiotic stress tolerance and promotes root growth in rice. Mol. Plant 4, 190–197. doi: 10.1093/mp/ssq066 [DOI] [PubMed] [Google Scholar]
- Yang X., Liu C., Niu X., Wang L., Li L., Yuan Q., et al. (2022). Research on lncRNA related to drought resistance of Shanlan upland rice. BMC Genomics 23, 336. doi: 10.1186/s12864-022-08546-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Ma B., He S. J., Xiong Q., Duan K. X., Yin C. C., et al. (2015). MAOHUZI6/ETHYLENE INSENSITIVE3-LIKE1 and ETHYLENE INSENSITIVE3-LIKE2 regulate ethylene response of roots and coleoptiles and negatively affect salt tolerance in rice. Plant Physiol. 169, 148–165. doi: 10.1104/pp.15.00353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Dong S. S., Xu J. Y., He W. M., Yang T. L. (2019). PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 35, 1786–1788. doi: 10.1093/bioinformatics/bty875 [DOI] [PubMed] [Google Scholar]
- Zhang R., Jia G., Diao X. (2023). geneHapR: an R package for gene haplotypic statistics and visualization. BMC Bioinf. 24, 199. doi: 10.1186/s12859-023-05318-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xu W., Li Z., Deng X. W., Wu W., Xue Y. (2008). F-box protein DOR functions as a novel inhibitory factor for abscisic acid-induced stomatal closure under drought stress in arabidopsis. Plant Physiol. 148, 2121–2133. doi: 10.1104/pp.108.126912 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.