Abstract
Adult Schistosoma produces a large number of eggs that play essential roles in host pathology and disease dissemination. Consequently, understanding the mechanisms of sexual maturation and egg production may open a new avenue for controlling schistosomiasis. Here, we describe that Bantam miRNA and miR-1989 regulate Wnt signaling pathway by targeting Frizzled-5/7/9, which is involved in ovarian development and oviposition. Additionally, Frizzled-7 could cooperate with SjRho to maintain normal ovarian development and egg productions and SjRho may interact with Hsp60 to potentially support Frizzled-7 trafficking and signaling. Further in vivo inhibition of SjRho in mice model infected with Schistosoma results in a remarkable decrease in worm burden and egg productions. Our findings not only broaden the functions of Bantam miRNA and miR-1989 as well as Wnt signaling pathway, but also imply that interruption of Bantam/miR-1989-Frizzled-5/7/9-SjRho axis may serve as effective targets against schistosomiasis.
Subject terms: Parasite biology, Parasite development
Schistosoma sex-biased Bantam and miR-1989 microRNAs shed light on the roles of ovarian development and egg production in females by targeting Wnt signaling pathway.
Introduction
Schistosomiasis, a neglected tropical disease mainly caused by three major human disease-causing species, Schistosoma mansoni, S. japonicum, and S. haematobium, and affects more than 230 million people globally1. Adult schistosomes reside for years in the vertebrate host’s vascular system, where they lay eggs. Almost half of the eggs are trapped in the host tissue, inducing a primary inflammatory response. Since parasites themselves cause limited pathology2, understanding the biology of sexual maturation and egg production may help to develop new strategies for controlling schistosomiasis.
Unlike other trematodes, schistosomes have sexual dimorphism, and female sexual maturation depends on pairing with male worms. Continuous male-female pairing is essential for female development and reproductive maturation. Studies have shown that the interaction between male and female worms leads to the differentiation of germ cells in females into vitellocytes3–7. Unmated female schistosomes showed stunted growth and produced no eggs. Several studies have investigated the sexual differences on molecular and proteomics basis of schistosome development and sexual maturation8. They found that male schistosomes provide developmental signals cues for female sexual maturation8–10. For examples, Chen et al. demonstrated that a nonribosomal peptide produced by male schistosomes is responsible for inducing female sexual development and egg laying during pairing in S. mansoni9. However, the mechanism of female development and egg production still need to be further elucidated. Several signaling transduction pathways have essential functions in cell control involving non-linear integrated networks that interact primarily by switching the activity status of proteins. A study showed that MAPK signaling components are well conserved in different species of schistosomes11. The MAPK and TGF-β signaling pathways have been shown to regulate female reproductive development8,12.
miRNAs are a type of small non-coding RNAs that function as post-transcriptional regulators of gene expression13. We found certain miRNAs such as Bantam, miR-31, miR-2c and miR-1989, were highly expressed in females and significantly enriched in the ovary8. In S. mansoni, some miRNAs, including miR-31, miR-755, Bantam, and others, were enriched in female worms14. However, the regulatory roles of these female-enriched miRNAs in sexual maturation and egg production remain unknown. In addition, some of these miRNAs are shown to be invertebrate-specific, which could serve as specific drug targets against schistosomiasis.
Bantam miRNA was first reported in Drosophila melanogaster with the functions of cellular proliferation and apoptosis15,16. Next, Bantam has also been shown to interact with the oncogenic epithelial growth factor receptor (EGFR) during neoplastic tumor formation in Drosophila model17. Several Bantam targets, such as Suppressor of cytokine signaling at 3E, Head involution defective (hid), Meiotic P26, and others, have been identified in D. melanogaster16–21, suggesting that Bantam plays multiples roles in many biological processes. Corresponding to Bantam, miR-1989 is only found in the neotrochozoa clade22 with high expression in female schistosomes; however, its functions remain unknown. Here, we uncovered the regulatory roles of Bantam/miR-1989 in ovarian development and egg production in Schistosoma by interacting with Wnt signaling pathway and further determined the critical molecules involved in this signaling pathway for regulating these processes.
Results
Bantam/miR-1989 inhibition resulted in significant morphological defects of ovarian architecture and decreased egg production
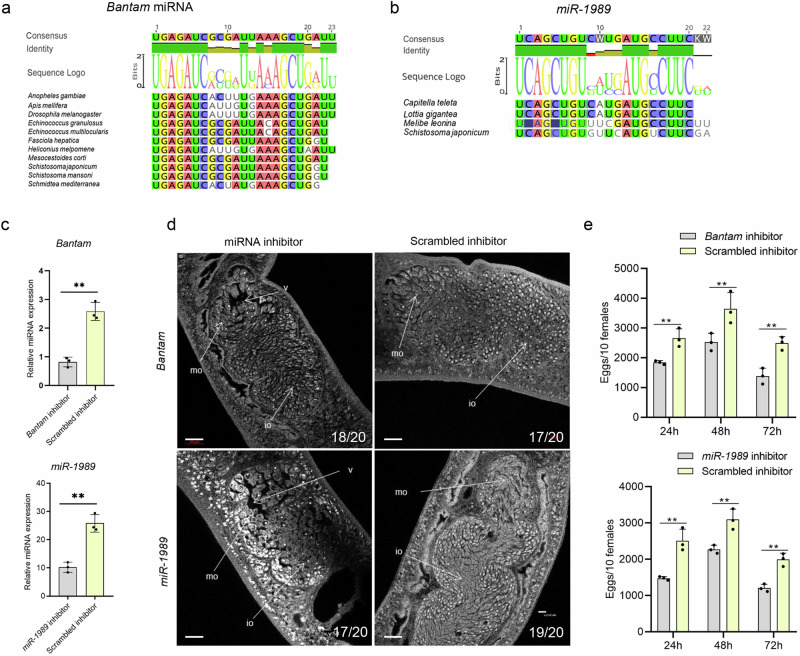
Previously, we found that Bantam and miR-1989 were significantly enriched in female ovary of S. japonicum. Sequence alignment of these miRNAs showed the conserved Bantam, especially in insects (Fig. 1a) and miR-1989 is a novel family in Annelids and Mollusca (Fig. 1b). To determine the regulatory functions of Bantam and miR-1989 in S. japonicum, we inhibited these miRNAs by delivering Bantam and miR-1989 inhibitor using electroporation in cultured parasites that collected from mice infected with S. japonicum cercariae at 26 days of post infection (dpi). At 4 days of post inhibition, we harvested the treated parasites and then analyzed the expressions of Bantam and miR-1989 by RT-qPCR. The results indicated these miRNAs have been significantly suppressed in the treated parasites (Fig. 1c and Supplementary Data 2). Then, we examined the morphological ovary of parasites treated with control inhibitor and Bantam/miR-1989 inhibitor using confocal microscopy combined with carmine red staining. The results displayed that inhibition of these miRNAs led to significant defects of ovarian architecture while these changes were not observed in the worms treated with control inhibitor (Fig. 1d). Additionally, we monitored time course of egg production in each well of cultured parasites that electroporated with Bantam and miR-1989 miRNA inhibitor as well as control inhibitor by microscopy. The results indicated that egg production was significantly reduced in parasites treated with Bantam or miR-1989 inhibitor as compared with that of controls (Fig. 1e and Supplementary Data 2). Overall, these results indicated that Bantam and miR-1989 miRNA have been successfully suppressed, leading to the defects of ovarian architecture and decreased egg production, in the treated parasites. Consequently, we used these suppressed parasites for the following RNA-seq analysis.
Fig. 1. Bantam and miR-1989 evolution and their inhibition resulted in significant morphological defects of S. japonicum ovary and decreased oviposition.
a Bantam miRNA alignment showed conserved sequences across different species. b miR-1989 alignment showed conserved sequences among different species. c RT-qPCR analyses of the expression of Bantam and miR-1989 treated with miRNA inhibitors. At 4 days of post-electroporation, parasites were collected and total RNAs were isolated for RT-qPCR analysis. Data illustrate representative results showing the mean and standard error from triplicate experiments, n = 3 independent experiments. d Effect of Bantam and miR-1989 suppression on female ovary morphology. S. japonicum worms were stained with carmine red and whole-mount preparations were imaged by confocal microscopy. Io immature oocyte, mo mature oocyte, v vacuole. The number of parasites similar to representative images is indicated in the lower right of each panel. n = 20 parasites, two biological replicates. Scale bar = 20 µm. e Effect of Bantam/miR-1989 suppression on egg production in female schistosomes. Ten females were electroporated as one shot and then cultured in one well of 12 well cell plate. Data illustrate representative results of the experiment contain 30 worms from three replicate shots and then cultured in 3 wells, three biological replicates. The results showing the mean and standard error, n = 3 independent experiments. ** P ≤ 0.01.
Bantam and miR-1989 inhibition resulted in significantly different expressions of genes
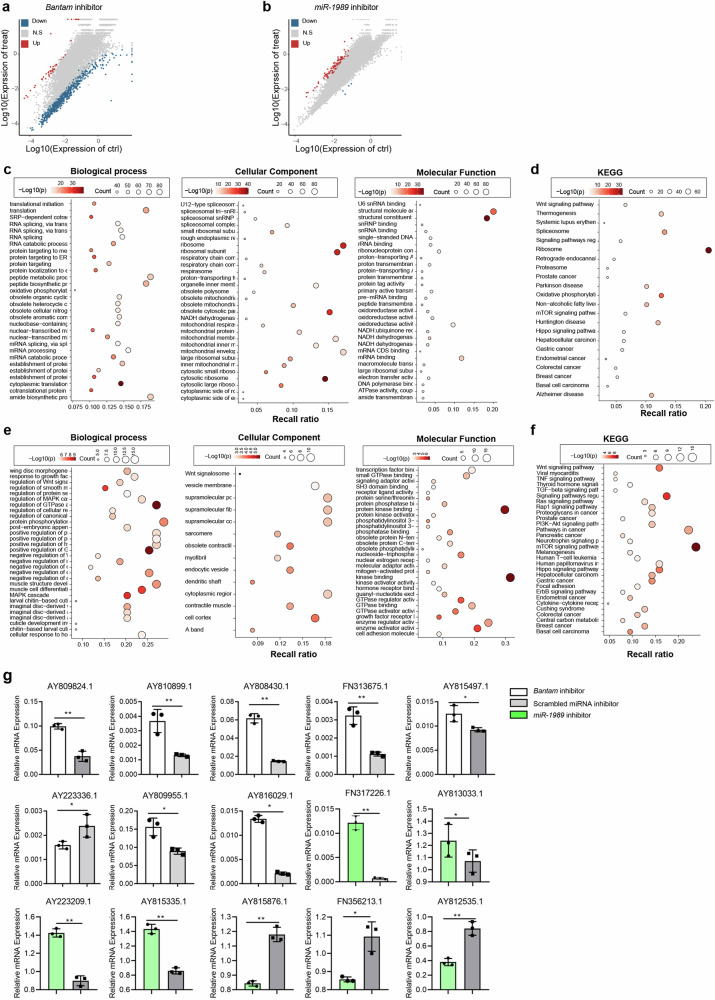
The treated and control parasites were subject to RNA-seq analyses. Upon the sequencing of 12 libraries with three biological replicates for each treatment or control, the average number of obtained clean reads for Bantam inhibition and its control were approximately 45.47, 44.57, 41.54 million for inhibition and 43.67, 44.56, 42.87 million for control; the average number of obtained clean reads from miR-1989 inhibition and its control were approximately 44.13, 41.96, 43.59 million and 44.06, 42.28, 41.97 million, respectively. Bioinformatic analysis indicated that Bantam inhibition led to 780 differently expressed genes (Fold Change ≥ 2 and adjusted P value ≤ 0.001) in the treated parasites as compared to that of control (Fig. 2a). Gene Ontology (GO) enrichment analysis of differently expressed genes (DEGs) for biological processes showed that most of these genes are associated with biological regulations, cellular process and regulation of cellular process (Fig. 2c). Likewise, GO term of the cellular component analysis revealed that DEGs are associated with cytoplasm and intracellular anatomical structure (Fig. 2c) and GO term analysis of molecular function analysis showed that most of the DEGs are associated with protein binding, ion binding and catalytic activity (Fig. 2c and Supplementary Table 7). Similarly, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEGs in Bantam inhibition showed that most genes are associated with metabolic pathways, axon guidance, and mTOR signaling pathways (Fig. 2d and Supplementary Table 8). Similarly, miR-1989 inhibition resulted in 2963 upregulated and 88 downregulated genes in treated parasites as compared to that of control worms (Fig. 2b). GO term of the biological process of DEGs in miR-1989 inhibited worms was shown to be mostly associated with biological regulation, regulation of biological process and regulation of cellular process (Fig. 2e). Similarly, cellular components analysis revealed that miR-1989 inhibition induced DEGs are linked to the cell periphery, membrane and plasma membrane (Fig. 2e). Similarly, GO term analysis of molecular function analysis showed that most of the DEGs are associated with protein binding, ion binding and cation binding (Fig. 2e and Supplementary Table 9). KEGG pathway enrichment analysis of DEGs in miR-1989 inhibited worms showed that most genes are associated with axon guidance, regulation of actin cytoskeleton, focal adhesion and RAP1 signaling pathway (Fig. 2f and Supplementary Table 10).
Fig. 2. RNA-seq analyses of differently expressed genes (DEGs) in S. japonicum treated with Bantam and miR-1989 inhibitor.
a Scatter-plot distribution of DEGs in worms treated with Bantam inhibitor. The X and Y axes represent the logarithmic value of gene expression in control and Bantam inhibitor-treated worms. Red represents upregulated and blue represents downregulated genes, and grey represents non-differentially expressed genes. b Scatter-plot distribution of DEGs in worms treated with miR-1989 inhibitor. The X and Y axes represent the logarithmic value of gene expression in control and bantam inhibitor-treated worms. Red represents upregulated and blue represents downregulated genes, and grey represents non-differentially expressed genes. c GO enrichment analysis of the DEGs for biological process, cellular components and molecular functions in parasites treated with Bantam inhibitor. d KEGG pathway enrichment analysis of the DEGs in worms treated with Bantam inhibitor. e GO enrichment analysis of the DEGs in worms treated with miR-1989 inhibitor. f KEGG pathway enrichment analysis of the DEGs in worms treated with miR-1989 inhibitor. g RT-qPCR validation of RNA seq results in the parasites treated with Bantam and miR-1989 inhibitor. Data illustrate representative results showing the mean and standard error derived from triplicate experiments. Statistical analysis compared miRNA inhibitor treatment vs scrambled miRNA inhibitor treatment using Students t-test, n = 3 independent experiments. * P ≤ 0.05, ** P ≤ 0.01.
To corroborate the DEGs identified by RNA-seq results, we selected some differently expressed genes to verify their expressions in independently treated parasites using RT-qPCR. We found that the changes of these selected genes were consistent with the RNA-seq results (Fig. 2g and Supplementary Data 2).
Bantam and miR-1989 regulate several critical signal pathways
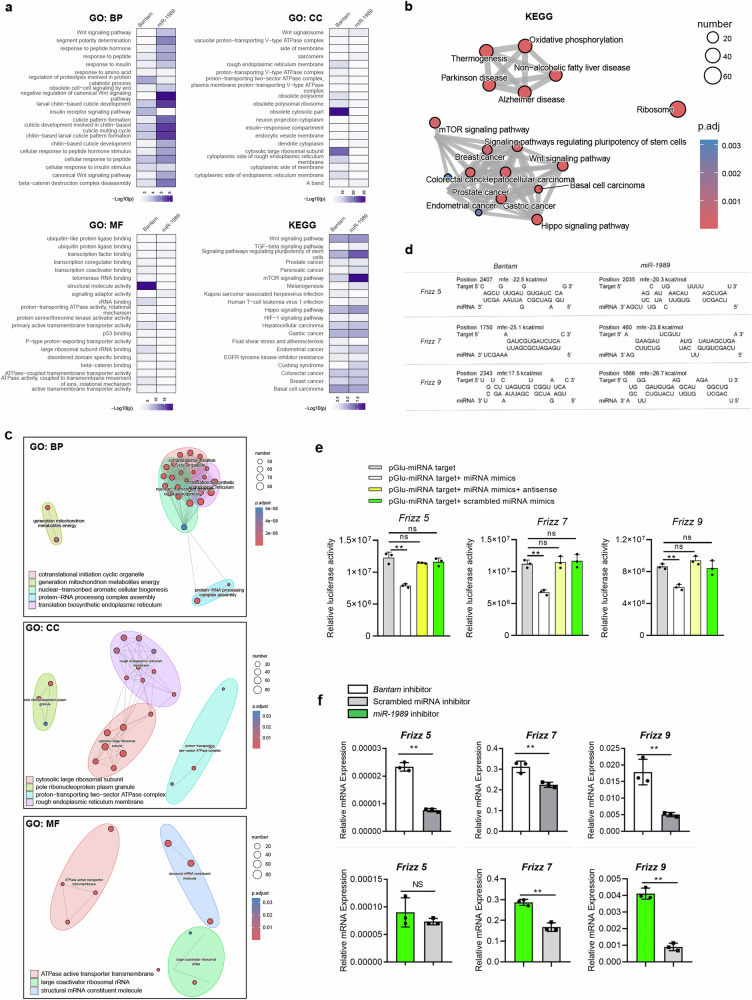
By delivering miRNA inhibitors in the parasites, the target genes of miRNAs were supposed to be increased in the treated parasites. Bioinformatic analyses of DEGs in the treated parasites were related to developmental process, biological regulation and anatomical structure development (Fig. 3a). Further analysis highlighted that these DEGs are related to signal pathways such as Wnt, Hippo and mTOR signals as well as enzyme regulator activity such as GTPase regulator activity (Fig. 3b, c), suggesting that these signal pathways may be regulated by Bantam or miR-1989.
Fig. 3. Analysis of the signal pathways associated with DEGs co-detected in Bantam miRNA inhibited and miR-1989 inhibited parasites and prediction and validation of Bantam and miR-1989 miRNA targets.
a The DEGs associated with same terms in Bantam miRNA inhibited and miR-1989 inhibited parasites. b Diagram showing co-detected signal pathways in Bantam miRNA inhibited and miR-1989 inhibited parasites. c Diagram showing co-detected biological processes in Bantam miRNA inhibited and miR-1989 inhibited parasites. d Predicted target sites for Bantam and miR-1989 in Frizzled-5/7/9 of S. japonicum using RNAhybrid. e Validation of Bantam miRNA targets by luciferase assay. HEK293T cells were transfected with the recombinant plasmids. At 24 h post-transfection, a control miRNA or miRNA mimics transfected into these cells. At 24–48 h post-transfection, luciferase activity was evaluated using a luciferase reporter assay system with normalization as protein assay. Each experiment shows representative results and the mean and standard errors, n = 3 independent experiments. ** P ≤ 0.01 and NS means non-significant (student’s t-test, miRNA mimics treatment vs. scrambled miRNA mimics treatment). f RT-qPCR analysis of the expression of target genes in parasites treated with miRNA inhibitors and scrambled miRNA inhibitor. Data illustrate representative results showing the mean and standard error, n = 3 independent experiments. Statistical analysis compared miRNA inhibitor treatment vs scrambled miRNA inhibitor treatment using Student’s t-test and ** P ≤ 0.01 and NS denotes non-significant.
To identify the regulatory roles of Bantam and miR-1989, the upregulated genes in the treated parasites were predicted for miRNA targets using RNAhybrid and miRanDa (Supplementary Table 6). Among them, we noted that Frizzled-5/7/9 involved in Hippo signal- and Wnt signaling pathways may be targeted by Bantam and miR-1989 (Supplementary Table 6). The RNAhybrid predicted target sites for Bantam and miR-1989 target for Frizzled-5/7/9 shown in Fig. 3d. To validate these predictions for miRNA target, we introduced antisense of Bantam and miR-1989 mimics into in vitro cultured parasites by electroporation. Then, we determined whether the levels of target mRNAs were changed by RT-qPCR. As shown in Fig. 3f, suppression of these miRNAs in parasites resulted in the increased expressions of Frizzled-5/7/9, suggesting that Bantam and miR-1989 could target Frizzled-5/7/9 in S. japonicum (Supplementary Data 2).
Next, we used luciferase assay to further confirm that the Frizzled-5/7/9 was regulated by Bantam miRNA in mammalian cells. Briefly, the predicted miRNA-binding sites were cloned into the 3′ UTR of a luciferase reporter. The recombinant plasmids and the corresponding miRNA mimics and antisense miRNA were transfected into HEK293T cells, and then luciferase activities were determined. As shown in Fig. 3e, transfection of Bantam miRNA mimics resulted in a reduction of luciferase activity compared to a scrambled miRNA control, indicating that Bantam miRNA can downregulate the expression of an mRNA incorporating target regions. Importantly, co-transfection with Bantam miRNA mimics and miRNA antisense in the experiments led to the de-repression of luciferase (Supplementary Data 2). These results suggest that Bantam miRNA can downregulate the expression of the luciferase mRNA by interacting with the target region.
SjFrizzled-7 plays a dominant role in maintenance of ovarian architecture and egg production
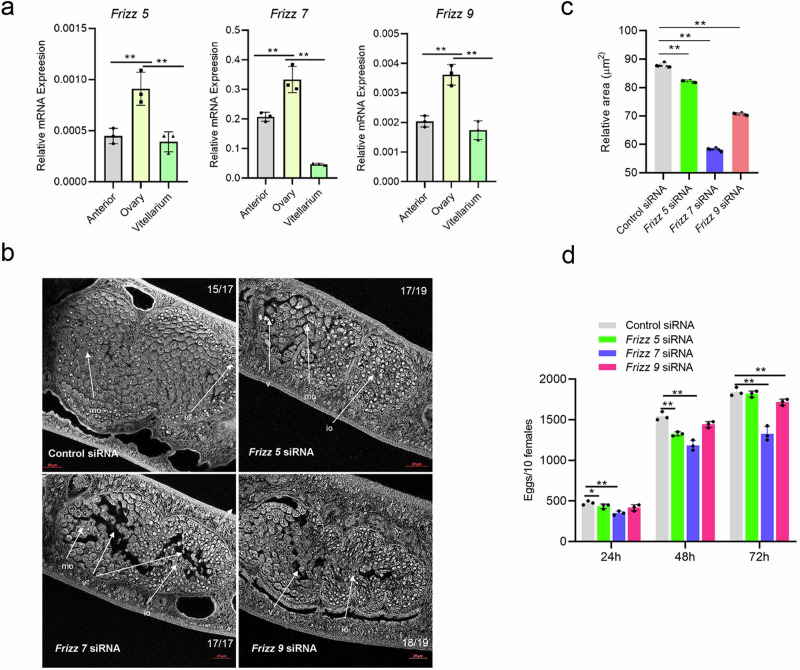
Previously, we found that Bantam and miR-1989 were significantly enriched in the ovaries of female schistosomes, here, we determined whether Frizzled-5/7/9 was highly expressed in ovary. We dissected females into three parts including anterior, ovary, and vitellarium. Total RNAs was isolated from each part and RT-qPCR was carried out. The results indicated that the Frizzled-5/7/9 were also significantly enriched in the ovaries (Fig. 4a and Supplementary Data 2). To determine the functions of Frizzled-5/7/9, we designed small interfering RNAs to knock down the transcript levels of these genes. Briefly, three siRNA duplexes based on each cDNA sequence were designed for RNAi (Supplementary Table 4) and each siRNA was electroporated into in vitro cultured parasites. RT-qPCR analysis was conducted at 4 days of post-treatment to evaluate the silencing effect for each siRNA (Supplementary Fig. 1). We noted that the best siRNAs maximally led to a 48.75% reduction of the transcript level for Frizzled-5, a 70.4% reduction of the transcript level for Frizzled-7 and a 40.8% reduction of the transcript level for Frizzled-9 (Supplementary Fig. 2). Then, we applied the best silencing siRNA to determine morphological changes in ovaries upon the suppression of Frizzled-5/7/9. At 4 days of post treatment, we found that the majority of treated parasites had notable morphological defects of ovaries including the vacuoles, crack-like appearances and damaged oocytes by confocal microscopy (Fig. 4b). Image J quantitative analysis of ovarian architecture by measuring and calculating the ratio of total area of ovary to the area occupied by oocytes, we found that Frizzled- 5/7/9 inhibition by RNAi showed a significant reduction of parenchymal cells and oocytes in the female schistosomes particularly for Frizzled-5/7/9 (Fig. 4c and Supplementary Data 2). Additionally, we also monitored egg production at different point after suppression, we noted that inhibitions of Frizzled-5/7/9 resulted in to some extend decreased egg production (Fig. 4d and Supplementary Data 2) and Frizzled-7 inhibition showed consistently and significantly lessoned egg number. Altogether, these results implied that Frizzled-7 might play a dominantly role in the regulation of ovarian architecture and egg production.
Fig. 4. Suppression of Frizzled-5/7/9 led to female worm ovary morphological alteration.
a RT-qPCR analyses of the transcript levels of Frizzled-5/7/9 in three different regions of the female S. japonicum. Data illustrate representative results showing the mean and standard error, n = 3 independent experiments. Statistical analysis was performed comparing ovary vs anterior or vitellarium using Students t-test and ** denotes P ≤ 0.01. b Inhibition of Frizzled-5/7/9 led to morphological defects of ovaries in females. Female worms were electroporated either with siRNA or control siRNA (3 μg) and then cultured for four days as described in Materials and Methods. S. japonicum worms stained with carmine red and whole-mount preparations of worms imaged by confocal microscopy. Io immature oocyte, mo mature oocyte, v vacuole. The number of parasites similar to representative images is indicated in the upper or lower right of each panel. n = 17–19 parasites, two biological replicates. Scale bar = 20 µm. c Analysis of the ratio of total area of ovary to the area occupied by oocytes as determined by Image J. Data illustrate representative results showing the mean and standard error, n = 6 individual parasites. Statistical analysis compared ovary areas vs areas of appearance of oocytes in ovary using Student’s t test. * P ≤ 0.05 and ** P ≤ 0.01. d Suppression of Frizzled-5/7/9 resulted in decreased egg production. Data illustrate representative results showing the mean and standard error, n = 3 independent wells for egg counts. ** P ≤ 0.01 (student’s t-test, Frizzled-5/7/9 siRNA vs control siRNA).
SjFrizzled-7 interacts with SjRho and other interactors
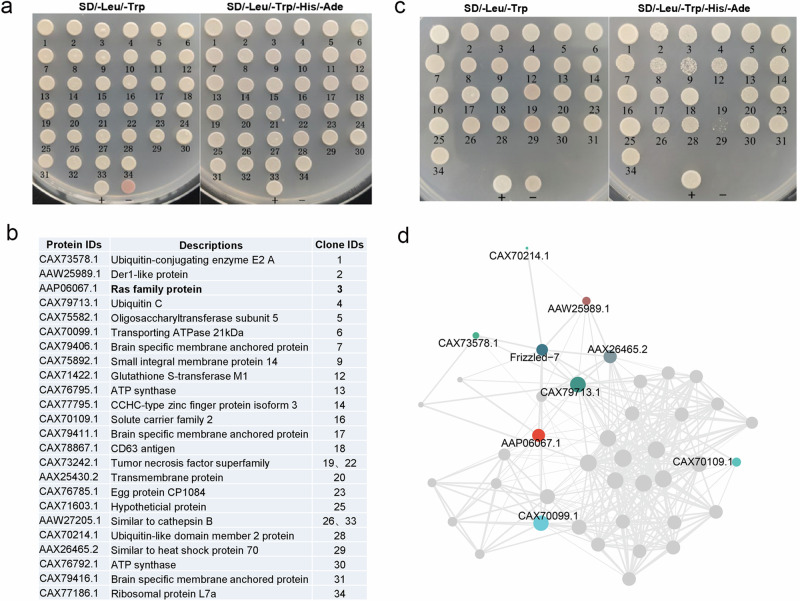
To explore the regulatory mechanism of Frizzled-7, we used a Yeast Two Hybrid (YTH) system to screen the interactors of Frizzled-7. Briefly, the recombinant plasmid expressing Frizzled-7 was constructed as a bait plasmid. Then, the recombinant plasmid was used to screen the YTH library of S. japonicum to identify the interacting partners of Frizzled-7. Upon evaluation of no self-activation activity from the Frizzled-7 bait fusion (Supplementary Fig. 3), a total of 34 clones were obtained (Fig. 5a) and 32/34 clones were successfully obtained for their sequences. Blastx analysis of these sequences indicated that six clones showed no significant homology to the NCBI database and the remaining 26 clones showed high identity to 24 S. japonicum proteins (Fig. 5b). To further validate the interaction between Frizzled-7 and the identified interactors, we further isolated the prey and bait plasmids and then co-transformed them into the NMY32 yeast cells independently and then inoculated in selective plates. Finally, 23 yeast clones were shown to survive in selective plates (Trp2/Leu2/Ade2/His2), indicating that Frizzled-7 interacts with these partners in yeast cells (Fig. 5c). The interactions between bait and prey plasmid were also validated by β-galactosidase assay (Supplementary Fig. 4). Using String database and Cytoscape by imputing Frizzled-7 and the identified interactors, the network of Frizzled-7 and its interactors was constructed (Fig. 5d).
Fig. 5. Identification of the interacting partners of Frizzled-7 by Yeast two hybridization.
a Screening of the yeast two-hybrid library using pBT3SUC-SjFrizz7 as the bait plasmid. NMY32 yeast cells containing the pBT3SUC-SjFizzled-7 bait plasmid transformed with plasmids from the Y2HSJ library and cultured on SDTLH + 5 mM 3AT plates. Colonies that grew on the selective plate were further inoculated onto SD-TL and SD-TLHA + 5 mM 3AT plates. b List of the interacting interactors identified by Y2H. c Re-transformation of SjFrizzled-7 bait plasmid and identified prey plasmids into yeast cells for confirming protein interactions. All diploid yeast strains for which the density was normalized before plating grew equally well under selection for both bait and prey plasmids (SD-Leu-Trp). d Diagram showing the protein-protein interaction network of Frizzled-7.
Suppression of SjRho leads to morphological alternation of ovary in S. japonicum
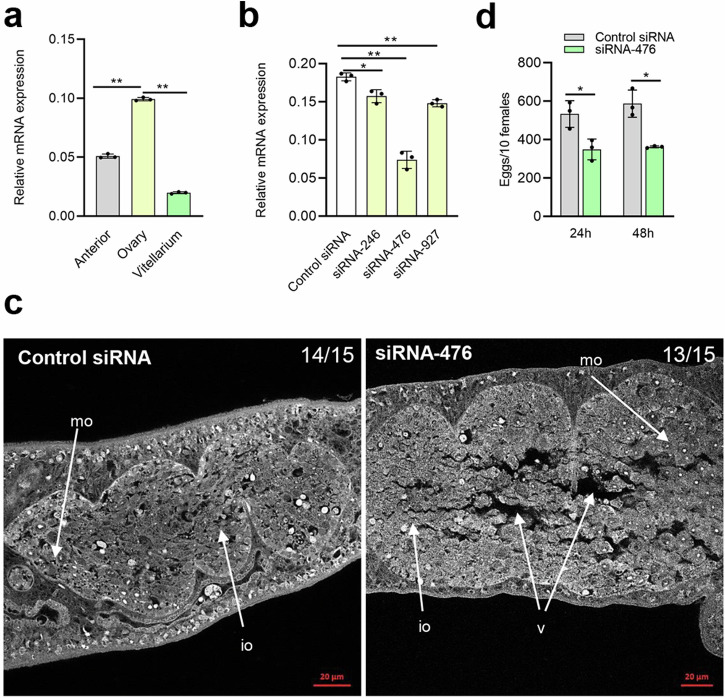
Given that Frizzled-7 is highly expressed in ovaries and SjRho is shown to interact with Frizzled-7, we also determined whether SjRho is highly expressed in ovaries of females. As expected, SjRho shown highest expression in ovarian part (Fig. 6a and Supplementary Data 2). Then, we designed and synthesized three siRNAs based on the cDNA sequence of SjRho. Upon the screening of the best siRNA for silencing SjRho (Fig. 6b and Supplementary Data 2). Next, we selected siRNA-476 to determine its inhibitory effect on worm motility, oviposition and ovary architecture. After 4 days of monitoring, we observed that worm viability decreased mainly at 3 days and 4 days of post-treatment (Supplementary Fig. 5). Additionally, we observed significantly morphological alterations in ovarian architecture in the treated females with siRNA-476 that include the dilated oocyte and crack-like appearances while these changes were not observed in control siRNA-treated worms (Fig. 6c). Additionally, we found that egg production was significantly reduced during at 24 h and 48 h of post treatment (Fig. 6d and Supplementary Data 2). These results indicated that SjRho is also involved in ovarian development and egg production.
Fig. 6. Identification of SjRho functions in ovarian development and egg production.
a Characterization of SjRho expression in different part of females. Data illustrate representative results showing the mean and standard error, n = 3 independent experiments. Statistical analysis compared ovary vs anterior and ovary vs vitellarium by Students t test. b Screening of the best siRNA for silencing SjRho. Data illustrate representative results showing the mean and standard error, n = 3 independent experiments. Statistical analysis was performed comparing control siRNA vs siRNA-246/476/927 using Students t-test and * P ≤ 0.05, ** P ≤ 0.01. c Inhibition of SjRho resulted in morphological defects of ovaries in female schistosomes. Female worms were electroporated either with siRNA (3 μg) or control siRNA (3 μg) and then incubated for 4 days as described in Materials and Methods. Worms were stained with carmine red and whole-mount preparations of worms imaged by confocal microscopy. Io immature oocyte, mo mature oocyte, v vacuole. The number of parasites similar to representative images is indicated in the upper right of each panel. n = 15 parasites, two biological replicates. Scale bar = 20 µm. d Suppression of SjRho induced significantly decreased egg production. Data illustrate representative results showing the mean and standard error, n = 3 independent experiments. Statistical analysis was performed comparing control siRNA vs siRNA-476 using Students t-test and * P ≤ 0.05.
Identification of SjRho interacting protein by YTH
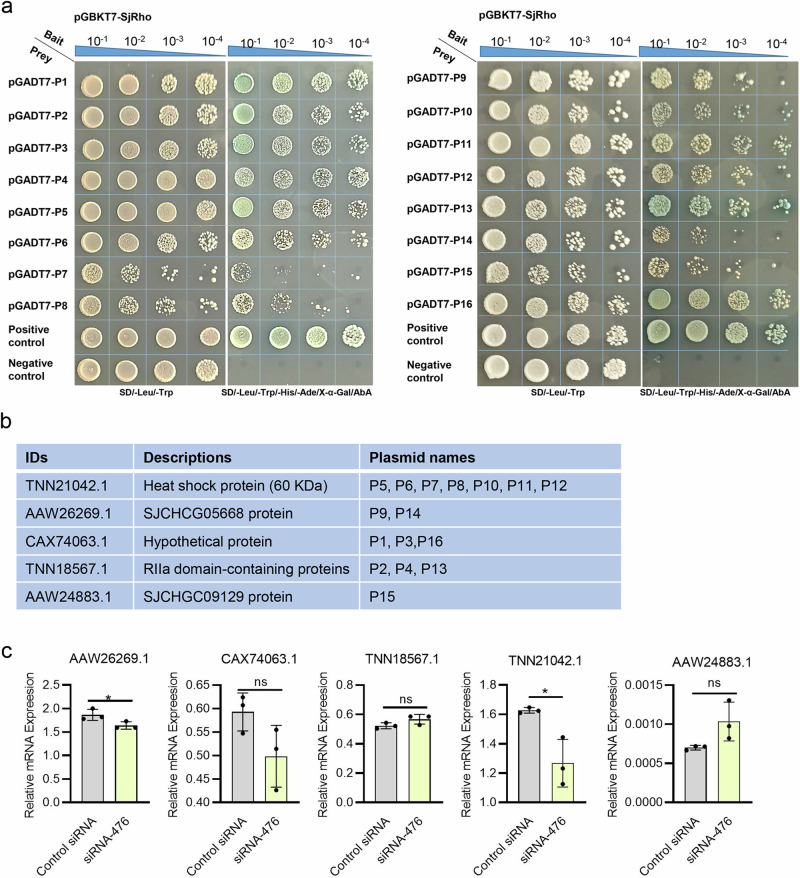
To further reveal regulatory role of SjRho, we used YTH to identify its interactor. Briefly, the full-length cDNA of SjRho was cloned and fused to the binding domain of the pGBKT7 vector as a bait plasmid. Upon evaluating self-activation and expression, the bait vector pGBKT7-SjRho was used to screen the Yeast Two Hybrid library of S. japonicum. We found that 17 prey clones could grow on the selective plates. After the isolation of these prey plasmids, 16/17 clones were successfully sequenced. Blastx analysis of the sequences of 16 clones indicated high identity to five S. japonicum proteins, including hypothetical protein, RIIa domain-containing proteins, heat shock protein (60 KDa), SJCHCG05668 protein and SJCHGC09129 protein (Fig. 7a, b).
Fig. 7. Yeast two-hybrid assay to screen the interacting partners of SjRho.
a Re-transformation of SjRho- and its prey plasmids into yeast cells to confirm protein interactions. All diploid yeast strains for which the density was normalized before plating grew equally well under selection for both bait and prey plasmids (SD-Leu-Trp). Tenfold dilutions for each diploid sample were plated onto each selection plate. Colonies that grew on the selective plate were further inoculated onto SD-TLHA + 5 mM 3AT plates. pNubG-Fe65 + pTSU2-APP was used as a positive control, and pGADT7-BD+pGADT7 was used as a negative control. b List of identified interactors of SjRho by Y2H technique. c Effect of SjRho silencing on the expression of its interacting partners evaluated by RT-qPCR. Data are shown as the mean ± standard error, n = 3 independent experiments. * P ≤ 0.05, ns means non-significant.
To further corroborate the interaction between SjRho and identified proteins, we independently prepared bait plasmids (pGBKT7-SjRho), isolated the prey plasmids, and then co-transformed them into NMY32 yeast cells. Then, the transformed cells were inoculated with selective plates with different dilutions. The results indicated that the yeast clones survived (SD/-Leu/-Trp; Fig. 7a) while some grew up on selective agar (SD/-Leu/-Trp/-His/-Ade), indicating that SjRho can interact with the putative interactors. In addition, we also determined the transcript levels of the interactors of SjRho in SjRho-inhibited parasites using RT-qPCR. The results indicated that SjRho inhibition led to altered expressions of some interactors such as heat shock protein, SJCHCG05668 and Hypothetical protein (Fig. 7c and Supplementary Data 2).
Suppression of SjRho resulted in decreased worm burden and egg production in animal model
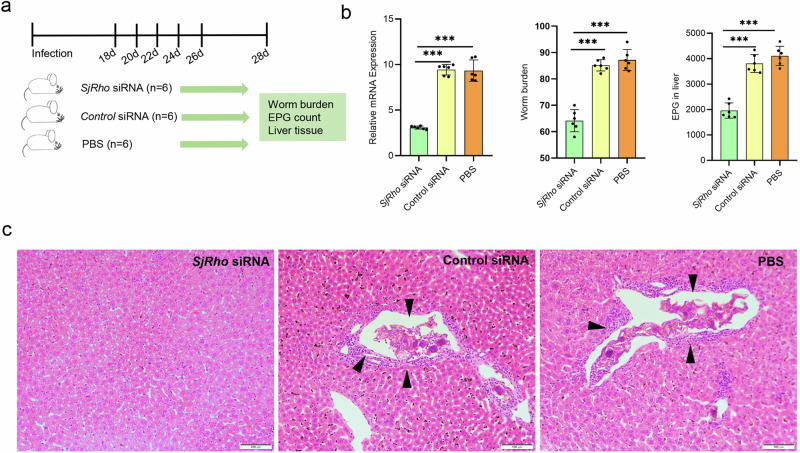
Given that Frizzled-7 and SjRho play important roles in maintenance of ovarian architecture and egg production in parasites, we further evaluated whether suppression of SjRho can influence worm burden and egg deposition in animal model infected with S. japonicum. Mice were infected with S. japonicum cercariae and then treated with siRNA targeting SjRho, irrelevant siRNA, and PBS control from 18–26 days of post-infection with 5 times at 2-day intervals (Fig. 8a). At 28 days of post infections, mice were sacrificed and worms were collected by perfusion. We firstly evaluated the effect of siRNA treatment on the expression of SjRho of worms collected from administrated mice by RT-qPCR. The results revealed that the expression of the SjRho was significantly decreased upon SjRho siRNA administration (Fig. 8b and Supplementary Data 2). Moreover, we noted that worm burden and egg deposition in the livers was also significantly reduced in mice administered SjRho siRNA (Fig. 8b and Supplementary Data 2). Egg-induced granuloma formation in the livers was lessened in mice administered SjRho siRNA compared to that from control mice (Fig. 8c). Overall, these results uncovered that SjRho play an important role in egg production as well as Schistosoma parasitism.
Fig. 8. Suppression of SjRho significantly decreased worm burden and liver egg deposition in mice infected with S. japonicum.
a Diagram showing the schedule for siRNA administration in mice. Each group contains 6 mice (n = 6). b RT-qPCR analysis of SjRho mRNA in worms isolated from the infected mice injected with SjRho siRNA; Effect of SjRho siRNA injection on the worm burden in mice at 28 days post-infection; Effect of SjRho siRNA suppression on egg deposition in the liver of mice. Data are shown as the mean ± standard error from 6 mice (n = 6), *** P ≤ 0.001. c Histopathological analysis of liver tissues from the mice treated with PBS, control siRNA and SjRho siRNA. Arrows indicate egg-induced granuloma formation. Scale bar = 100 µm.
Discussion
Understanding the mechanism of Schistosoma sexual development and egg production may effectively guide to develop novel strategies against schistosomiasis. miRNAs have been reported to regulate many biological processes in many organisms. Schistosoma miRNAs displayed diverse expression patterns in different sexes and at different developmental stages8,23–27. However, their functions remain poorly characterized. Here, we found the regulatory mechanism of Bantam and miR-1989 in ovarian development and egg production in Schistosoma.
Bantam has been shown to prevent apoptosis in Drosophila16. We found that Bantam miRNA inhibition led to the altered genes related to mTOR-, Wnt- and Hippo signaling pathways as well as the Rap1 signaling pathway, suggesting that these signaling pathways might be directly or indirectly regulated by Bantam miRNA. The Hippo signaling pathway plays crucial roles in regulating organ size and controlling tissue homeostasis in many organisms28. Wnt signaling pathway can regulate cell proliferation and differentiation during development in mammalian cells29. Although a recent study documented that miR-1 from S. japonicum regulates schistosomiasis hepatic fibrosis by targeting host secreted Frizzled-related protein 1, the Frizzled-related protein 1 is from mice and many studies have characterized the Wnt pathway including Frizzled receptor proteins in mammalian model30–32. Unfortunately, these critical signaling pathways that widely studied in mammalian and other model organisms remain under characterized in Lophotrochozoa particularly for platyhelminthes, while a few Wnt receptors were cloned and preliminarily analysis of their expression patterns was preliminarily carried out in S. japonicum33,34. In the present study, by exploring the function of Bantam and miR-1989, we demonstrated that Schistosoma Frizzled proteins (Frizzled-5/7/9) were targeted by the female-enriched miRNAs including Bantam and miR-1989. Frizzled proteins are the receptors of Wnt acting as an extracellular signaling pathway for involvement in animal developmental processes in several model organisms, such as Drosophila, C. elegans, Zebrafish, and Xenopus embryos35. However, the functions of Frizzled-5/7/9 in Schistosoma remain unknown. By using small interfering RNAs, we noted that SjFrizzled-5/7/9 inhibition resulted in the decreased egg production and the altered ovarian architecture. These results not only uncovered that the homeostasis of Bantam/miR-1989-Frizzled-5/7/9 axis is essential for ovarian development and maintenance of egg production in Schistosoma (Fig. 9), but also added novel functions for Bantam/miR-1989 and Frizzled-5/7/9 in Lophotrochozoa.
Fig. 9. Proposed model for Bantam/miR-1989-mediated Wnt signal pathway for regulating ovary development and egg production in S. japonicum.
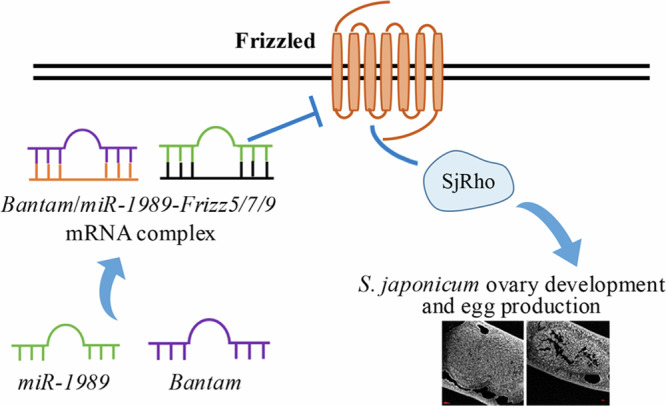
Bantam/miR-1989 targets Frizzled-5/7/9 and Frizzled-7 interacts with SjRho and other interactors. SjRho interacts with Hsp60 and others to regulate ovary development and egg production.
Since Frizzled-7 appeared to play relatively important roles in regulation of egg production and ovarian development, we further revealed that Frizzled-7 may coordinate with 23 proteins (its interactors) to implement the regulatory functions. In addition, our results also suggested that S. japonicum Frizzled-7 may have diverse functions that is involved in the regulations of metabolism, protein translation, and ubiquitination in Schistosoma. In particular, we found that Frizzled-7 may strongly interact with SjRho to potentially manage small GTPases that regulates egg production and ovarian development. Consider that Bantam/miR-1989 inhibition resulted in the DEGs related to the regulation of GTPase activity and Wnt signal pathway is targeted by Bantam/miR-1989, our findings displayed that Bantam/miR-1989 mediated Wnt signal pathway is critical for regulating ovarian development and egg production in Schistosoma that also expand current understanding of the functions of Wnt signal pathway in platyhelminthes.
Moreover, we also found that Hsp60 likely extensively and strongly interacts with SjRho in S. japonicum as identified by YTH. Hsps constitute a large family of chaperone proteins involved in protein folding and maturation36. In humans, Hsp60 was shown to regulate the expression of lipid metabolism in ovarian cancer37 and potential roles in gonadal development38–40. Consequently, we deduced that Hsp60 may serve as an intracellular scaffold to some extend support Frizzled-7 trafficking and signaling in Schistosoma. Overall, our results suggested that Hsp60 may hold specific functions for Schistosoma by involving in the Wnt signaling pathway and that targeting the species-specific molecules involved in this important signaling pathway may be considered for developing novel strategies against schistosomiasis.
Materials and methods
Ethics statement
All animal experiments were carried out following the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People’s Republic of China. Animal protocols were approved by the Animal Management Committee and the Animal Care and Use Committee of the Shanghai Science and Technology Commission of the Shanghai municipal government for the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, China (Permit number: SYXK 2016-0010). We have complied with all relevant ethical regulations for animal use.
Schistosome culture
The life cycle of S. japonicum (Anhui isolated, China) was maintained using BALB/c mice (male, six-week old) and the intermediate snail host Oncomelania hupensis obtained from the Center of National Institute of Parasitic Disease of Chinese Center for Disease Control and Prevention (Shanghai, China). Mice were challenged with 100–150 S. japonicum cercariae via abdominal skin penetration. Schistosomes were collected at 25–28 days post-infection (dpi) and thoroughly and gently washed three times with PBS (pH 7.4). Male and female worms were separated manually. Schistosomes were cultured in 12-well flat-bottom plates containing 2 mL of complete RPMI-1640 medium (ThermoFisher Scientific, USA) supplemented with 10% fetal bovine serum (Gibco, Gaithersburg, MD, USA) and 3% penicillin/streptomycin (10,000 U penicillin and 10 mg/streptomycin in 0.9% NaCl, Gibco) in a humidified 5% CO2 incubator at 37 °C.
miRNAs inhibition by electroporation
For studying the silencing effects of miRNAs on ovary morphology, female worms (25–28 days) were electroporated with miRNA inhibitors and scrambled miRNA inhibitors (3 μg per experiment, chemically synthesized in Shanghai GenePharma, China)8. Then worms were transferred into 12-well cell culture plates containing 2 mL of fresh media. At indicated times, worm mobility and survival were observed under an inverted microscope (Olympus, Japan). The parasites were collected at 96 h post electroporation for RT-qPCR and confocal microscopy as described below. All siRNA sequence details are given Supplementary Table 1.
Ovary architecture observation by confocal microscopy
At indicated time of post-electroporation, worms were fixed, stained, and mounted41. Briefly, worms were fixed in 10% formalin, 48% alcohol and 2% glacial acetic acid, stained with a carmine red, cleared in 0.5% hydrochloric alcohol solution, and mounted the whole worms. Images were obtained with a Nikon CLSI laser confocal microscope (Nikon, Japan), using a 488 nm He/Ne laser.
Bioinformatic analyses of RNA-seq data
Twelve transcriptome libraries (six transcriptome libraries for Bantam and control with biological triplicates and six transcriptome libraries for miR-1989 and control with biological triplicates) were sequenced using the BGISEQ-500 sequencing platform. Clean reads were obtained by removing low-quality reads, adaptors, and any reads containing poly-N from raw reads. The high-quality clean reads were mapped to the NCBInr database by HISAT2 v2.1.042. Then mapped reads were assembled using StringTie v1.0.4 with a reference-based approach43. RSEM v1.2.8 was used to estimate the expression level of each gene by the FPKM method11. DEseq2 was used to analyze the differential expression, Benjamini–Hochberg method and Storey-Tibshirani method were applied to calculate Q-values44. Differential expression analysis was carried out on DEseq2 R package, a widely-used tool to estimate variance-mean dependence in count data from high-throughput sequencing assays based negative binomial distribution model45. DEGs were defined by fold change ≥ 2 and adjusted p-value ≤ 0.001.The GO analysis of differentially expressed genes was performed using BLAST2GO and the KEGG pathway annotation was performed using the KEGG database (https://www.kegg.jp/). The raw sequencing data were uploaded to the National Genomics Data Center (https://ngdc.cncb.ac.cn/?lang=zh).
Validation of miRNA expression and RNA-seq results by RT-qPCR
Schistosomes were collected at 96 h after treatment. Total RNA was extracted and RT-qPCR8. A miScript II RT Kit (Qiagen, Germany) was used to reverse-transcribe RNA to cDNA. qPCR was performed using miScript SYBR Green PCR Kit (Qiagen) in a Mastercycler ep realplex (Eppendorf, Germany). The reactions were incubated in a 96-well plate at 95 °C for 15 min, followed by 40 cycles of 94 °C for 15 s, 55 °C for 30 s and 70 °C for 30 s. The miScript Primers for Bantam and miR-1989 were ordered from Qiagen (Qiagen’s proprietary). S. japonicum Nicotinamide adenine dinucleotide (NADH) dehydrogenase was used as an internal control. All reactions were run in at least triplicate. We used the 2−ΔCt method to calculate relative expression46. The other primers used for RT-qPCR are listed in Supplementary Table 2.
Alignment analysis of Bantam and miR-1989
To examine the evolutionary relationships of Bantam and miR-1989, mature miRNA sequences were obtained from the miRbase database (Release 22.1) and MirGeneDB47. Then, we used multiple sequence alignment and subsequently drew the phylogenetic tree by Geneious Prime software. We further used WebLogo to assess the conservation of the S. japonicum miRNAs48.
miRNA target prediction and validation by luciferase assay and in vivo suppression
Based on the RNA-seq results, increased expressions of genes involved in key biological processes and signal pathways were subject to in silico prediction of miRNA targets by RNAhybrid49 and miRanda50. cDNA fragments corresponding to target regions of target genes were PCR-amplified from S. japonicum cDNA and the PCR products were cloned into pCMV-Glu vector (Targeting Systems, USA) using standard molecular cloning methods. The primers for constructing recombinant plasmids were listed in Supplementary Table 3. HEK293T cells were cultured in 24 well cell culture plates in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% FBS, and 1% antibiotics. Cells were transfected with 5 ng recombinant plasmids (Promega, Madison, WI, USA) using Lipofectamine 3000 (Invitrogen). At 24 h post-transfection, the cells were transfected with 50 nM Bantam miRNA mimic, Bantam miRNA inhibitor and irrelevant control miRNA modified by 2ʹ-O-methyl or 2ʹ-O-methyl and phosphorothioate. After 24 h incubation, transfected cells were collected and processed for the luciferase reporter assay (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The relative reporter activity of transfected cells was obtained by normalizing it to protein assay. Protein assays on lysates were carried out using the Pierce BCA protein assay kit and Compat-Able protein assay preparation reagent set (Thermo Fisher Scientific).
Parasites were treated with Bantam/miR-1989 inhibitor using electroporation as described above. The total RNA was isolated and the transcript levels of selected targets were analysed by RT-qPCR. The primers were provided in Supplementary Table 5.
siRNA-mediated inhibition of the miRNA target and its interactor
Worms were collected from infected mice on day 24–28 post-infection. Males and females were manually separated and cultured in 12-well flat-bottom plates containing 2 mL RPMI1640 culture media with FBS, and antibiotics. SjFrizzled-5/7/9 siRNA, SjRho siRNA and control siRNAs (3 µg per experiment, Shanghai GenePharma, China) were electroporated (125 V, 20 ms, 1 pulse in 200 μL RPMI 1640 media) into cultured schistosomes. Following electroporation, worms were transferred into 12-well cell culture plates containing 2 mL of fresh media. Worm motility, mortality and egg numbers were counted under an inverted microscope (Olympus, Japan). The parasites were collected 96 h post-electroporation. For microscopic analysis, worms were preserved, stained, mounted and subjected to morphological examination using confocal microscopy (ZEISS, Germany)8. The other half of the worms were used for RT-qPCR analysis. All siRNA sequence details are given in Supplementary Table 4.
Cloning of the bait vectors and evaluation of self-activation
SjFrizzled-7 was amplified by performing PCR using recombinant DNA and the following primers (forward, 5′-AAGGCCATTACGGCCATGCATCTGACTCTTAGGGTATTG-3′; reverse, 5′- CCGGCCGAGGCGGCCCCTAGTAAACCTAGACCCGAGTTAGTC -3′, with each containing a Sfi I restriction site). PCR products were cloned via the Sfi I site into pBT3SUC vectors (Dualsystems Biotech, Schlieren, Switzerland). The recombinant pBT3SUC-SjFrizzled-7 plasmids were further confirmed by sequencing. To evaluate the potential self-activation of the recombinant pBT3SUC- SjFrizzled-7 plasmid, the following different combinations were transformed into NMY32 yeast cells: positive control (pNubG-Fe65 + pTSU2-APP), negative control (pPR3N + pTSU2-APP), self-activation evaluation (pPR3N + pBT3SUC-SjFrizzled-7), and functional assessment (pOST1-NubI+pBT3SUC-SjFrizzled-7). The transformed yeast cells were cultured at 30 °C for four days on different synthetic defined (SD) media (Dualsystems Biotech), including SD-Trp-Leu, SD-Trp-Leu-His, and SD-Trp-Leu-His-Ade, and the number of colonies counted.
Screening of S. japonicum Yeast Two Hybridization (Y2HSJ) library and sequencing analyses
Y2HSJ screening was performed using the SjFrizzled-7 as bait against the library. Briefly, plasmids from the Y2HSJ library were transformed into NMY32 yeast cells containing the pBT3SUC-SjFrizzled-7 bait plasmid and then cultured on SD-Trp-Leu-His + 5 mM 3AT agar. The yeast colonies that grew on the selective agar were inoculated to SD-Trp-Leu agar and analyzed for lacZ reporter expression using a β-galactosidase assay (HTX Kit: Dualsystems Biotech). To identify the cDNA sequence of each colony, each positive colony was inoculated in SD-Trp-Leu liquid media for culture. Then, the plasmids from yeast cells were isolated using the Yeast Plasmid Extraction Kit (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). The isolated plasmids were further transformed into competent Escherichia coli cells for plasmid amplification and subsequently isolated using the Axygen mini plasmid preparation kit (Axygen, NY, USA). The plasmids were then submitted for sequencing using the Sanger DNA sequence method. Sequences were analyzed by BLAST.
Confirmation of putative interacting partners by re-transformation
NMY32 yeast cells containing the bait plasmid pBT3SUC- SjFrizzled-7 were prepared as competent cells to confirm interacting partners further. Then, 34 prey plasmids were re-transformed into the yeast competent cells to ensure the interaction with SjFrizzled-7, respectively. Upon transformation, 100 µl of transformation solution was first plated onto SD-Trp-Leu agar. The cultured cells were then further inoculated onto an SD-Trp-Leu plate and SD-Trp-Leu-His-Ade + 10 mM 3AT plate. Positive colonies from the plate (SD-Trp-Leu-His-Ade + 10 mM 3AT) were further assessed for β-galactosidase activity (HTX Kit, Dualsystems Biotech).
Screening of a Y2HSJ for analyzing SjRho interactors
To identify the interactors of SjRho proteins, the full-length cDNA of SjRho was cloned and fused to the binding domain of the pGBKT7 vector as bait. The cDNA library was prepared from S. japonicum, and Y2H screening was conducted according to the Yeastmaker Yeast Transformation System 2 (Takara) as per the manufacturer’s instruction51. All positive clones were confirmed by sequencing. For the Y2H assay, the coding sequences of all positive clones and SjRho were individually cloned into pGADT7 prey and pGBKT7 bait vectors, respectively, and co-transformed into Y2HGold yeast competent cells (Takara). The yeasts were then grown using the lithium acetate method52 and cultured at 30 °C. The generated yeast transformants were screened on a medium lacking Trp and Leu and putative transformants were subsequently transferred to a medium lacking selection media supplemented with SD/-Trp/-Leu/-His/-Ade with or without X-α-gal.
In vivo inhibition of SjRho in mice infected with S. japonicum
Six to seven-week-old male BALB/c mice (mean weight 25 ± 2 g) were purchased from the Shanghai SLAC Laboratory Animal Co., Ltd. Eighteen mice were randomly divided into three groups. Each mouse was challenged with 100 ± 5 normal S. japonicum cercariae via abdominal skin penetration. At 18 days post-infection, mice in each group were injected with one µg siRNA, scrambled siRNA diluted in 200 μL of water. Similarly, one group was injected with an equal volume of PBS via the tail vein. Four additional injections were performed at 20, 22, 24- and 26-days post-infection. At 28 days post-infection, the parasites were collected by liver perfusion, and worm burden and egg production in the liver were counted. The expression of SjRho in collected worms was determined by RT-qPCR as described above. Liver tissue was fixed and processed for Hematoxylin and Eosin staining as described below.
Histopathology of mouse liver infected with S. japonicum
To assess the pathology of livers following SjRho inhibition in vivo, mouse livers were collected, cut into small pieces (1.0 cm × 1.0 cm × 0.3 cm) and fixed in 10% formalin. Then the fixed tissue was dehydrated in graded ethanol, cleared with xylene and embedded in paraffin. Then tissue was sectioned (6 µm thick) and stained with Hematoxylin and Eosin.
Statistics and reproducibility
Differential expression analysis was carried out on DEseq2 R package. DEGs were defined by fold change ≥ 2 and adjusted p-value ≤ 0.001. The results were analyzed in GraphPad Prism 8. Statistical analyses were performed using Student’s t tests or one-way ANOVA. P ≤ 0.05 were considered significant.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This study was supported, in whole or in part, by the National Natural Science Foundation of China (31672550 to G.C.) and the Key Program for International S&T Cooperation Projects of China (2021YFE0191600 to G.C.). The funders had no role in the study design, data collection, analysis, decision to publish, or manuscript preparation.
Author contributions
G.C., P.D., and B.G. designed the study and wrote the manuscript. P.D., T.X., B.G., and X.L. performed most of the experiments and analyzed the data. S.L., C.F., and S.Y. helped to analyze data. G.C. obtained funding for the study.
Peer review
Peer review information
Communications Biology thanks Bastian Fromm and Philip LoVerde for their contribution to the peer review of this work. Primary handling editors Simona Chera and Kaliya Georgieva. A peer review file is available.
Data availability
The library data involved in this study has been deposited into the Short Read Archive (SRA) of the GenBank database under the BioProject accession no. PRJNA1166905. The plasmids were deposited into BRICS under the SP3196-3198. Supplementary Data 1 contains all Supplementary Tables in the manuscript. The source data for the graphs can be found in Supplementary Data 2. All other data are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Pengfei Du, Tianqi Xia, Xuxin Li.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07402-z.
References
- 1.Colley, D. G., Bustinduy, A. L., Secor, W. E. & King, C. H. Human schistosomiasis. Lancet383, 2253–2264 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang, J., Chen, R. & Collins, J. J. 3rd. Systematically improved in vitro culture conditions reveal new insights into the reproductive biology of the human parasite Schistosoma mansoni. PLoS Biol.17, e3000254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu, Z. et al. Schistosome sex matters: a deep view into gonad-specific and pairing-dependent transcriptomes reveals a complex gender interplay. Sci. Rep.6, 31150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LoVerde, P. T., Andrade, L. F. & Oliveira, G. Signal transduction regulates schistosome reproductive biology. Curr. Opin. Microbiol.12, 422–428 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang, J. et al. Dynamic transcriptomes identify biogenic amines and insect-like hormonal regulation for mediating reproduction in Schistosoma japonicum. Nat. Commun.8, 14693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, G. F. et al. Proteomic analysis of differentially expressed proteins between the male and female worm of Schistosoma japonicum after pairing. Proteomics5, 511–521 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Luo, R. et al. Identification of in vivo protein phosphorylation sites in human pathogen Schistosoma japonicum by a phosphoproteomic approach. J. Proteom.75, 868–877 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Zhu, L. et al. MicroRNAs are involved in the regulation of ovary development in the pathogenic blood fluke Schistosoma japonicum. PLoS Pathog.12, e1005423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, R. et al. A male-derived nonribosomal peptide pheromone controls female schistosome development. Cell185, 1506–1520 e1517 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou, Y. et al. The Schistosoma japonicum genome reveals features of host–parasite interplay. Nature460, 345–351 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrade, L. F. et al. Eukaryotic protein kinases (ePKs) of the helminth parasite Schistosoma mansoni. BMC Genomics12, 215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beckmann, S. et al. Schistosoma mansoni: signal transduction processes during the development of the reproductive organs. Parasitology137, 497–520 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Gebert, L. F. R. & MacRae, I. J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol.20, 21–37 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marco, A. et al. Sex-biased expression of microRNAs in Schistosoma mansoni. PLoS Negl. Trop. Dis.7, e2402 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson, B. J. & Cohen, S. M. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell126, 767–774 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Brennecke, J., Hipfner, D. R., Stark, A., Russell, R. B. & Cohen, S. M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell113, 25–36 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Herranz, H., Hong, X., Hung, N. T., Voorhoeve, P. M. & Cohen, S. M. Oncogenic cooperation between SOCS family proteins and EGFR identified using a Drosophila epithelial transformation model. Genes Dev.26, 1602–1611 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herranz, H. et al. The miRNA machinery targets Mei-P26 and regulates Myc protein levels in the Drosophila wing. EMBO J.29, 1688–1698 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becam, I., Rafel, N., Hong, X., Cohen, S. M. & Milan, M. Notch-mediated repression of bantam miRNA contributes to boundary formation in the Drosophila wing. Development138, 3781–3789 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Gerlach, S. U., Sander, M., Song, S. & Herranz, H. The miRNA bantam regulates growth and tumorigenesis by repressing the cell cycle regulator tribbles. Life Sci. Alliance2, e201900381 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herranz, H., Hong, X. & Cohen, S. M. Mutual repression by bantam miRNA and Capicua links the EGFR/MAPK and Hippo pathways in growth control. Curr. Biol.22, 651–657 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Wheeler, B. M. et al. The deep evolution of metazoan microRNAs. Evol. Dev.11, 50–68 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Cai, P. et al. A deep analysis of the small non-coding RNA population in Schistosoma japonicum eggs. PLoS ONE8, e64003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai, P. et al. Profiles of small non-coding RNAs in Schistosoma japonicum during development. PLoS Negl. Trop. Dis.5, e1256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, Z. et al. An “in-depth” description of the small non-coding RNA population of Schistosoma japonicum schistosomulum. PLoS Negl. Trop. Dis.4, e596 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao, L., Cai, P., Jiang, N., Wang, H. & Chen, Q. Identification and characterization of microRNAs and endogenous siRNAs in Schistosoma japonicum. BMC Genomics11, 55 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue, X. et al. Identification and characterization of novel microRNAs from Schistosoma japonicum. PLoS ONE3, e4034 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng, Z., Moroishi, T. & Guan, K. L. Mechanisms of Hippo pathway regulation. Genes Dev.30, 1–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komiya, Y. & Habas, R. Wnt signal transduction pathways. Organogenesis4, 68–75 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Etienne-Manneville, S. & Hall, A. Rho GTPases in cell biology. Nature420, 629–635 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Denk-Lobnig, M. & Martin, A. C. Modular regulation of Rho family GTPases in development. Small GTPases10, 122–129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He, X., Pu, G. B., Tang, R., Zhang, D. M. & Pan, W. Q. Activation of nuclear factor kappa B in the hepatic stellate cells of mice with schistosomiasis japonica (vol 9, e104323, 2014). PLoS ONE15 (2020). https://doi.org/ARTN e024366710.1371/journal.pone.0243667 [DOI] [PMC free article] [PubMed]
- 33.Wang, X. et al. Characterization and expression of a novel Frizzled 9 gene in Schistosoma japonicum. Gene Expr. Patterns11, 263–270 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Ye, Z. et al. Spatiotemporal expression pattern of Sjfz7 and its expression comparison with other frizzled family genes in developmental stages of Schistosoma japonicum. Gene Expr. Patterns32, 44–52 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Clevers, H. & Nusse, R. Wnt/β-catenin signaling and disease. Cell149, 1192–1205 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Wu, J. et al. Heat shock proteins and cancer. Trends Pharm. Sci.38, 226–256 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Li, N. et al. HSP60 regulates lipid metabolism in human ovarian cancer. Oxid. Med. Cell Longev.2021, 6610529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paranko, J., Seitz, J. & Meinhardt, A. Developmental expression of heat shock protein 60 (HSP60) in the rat testis and ovary. Differentiation60, 159–167 (1996). [DOI] [PubMed] [Google Scholar]
- 39.Zou, S. et al. Hsp60 expression and localization in different tissues and testis development of male cattle (cattle-yak and yak). Folia Morphol.10.5603/FM.a2020.0126 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Fang, D. A. et al. Developmental expression of HSP60 and HSP10 in the Coilia nasus Testis during upstream spawning migration. Genes8 (2017). 10.3390/genes8070189 [DOI] [PMC free article] [PubMed]
- 41.Zhang, W. N. et al. Worm morphology of Schistosoma japonicum using confocal laser scanning microscopy. J. Helminthol.86, 317–322 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol.33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, L., Feng, Z., Wang, X., Wang, X. & Zhang, X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics26, 136–138 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol.11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Fromm, B. et al. MirGeneDB 2.1: toward a complete sampling of all major animal phyla. Nucleic Acids Res.50, D204–D210 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res.14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rehmsmeier, M., Steffen, P., Hochsmann, M. & Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA10, 1507–1517 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Enright, A. et al. MicroRNA targets in Drosophila. Genome Biol.4, 1–27 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng, Z. Z. et al. Alternative splicingf regulation of doublesex gene by RNA-binding proteins in the silkworm Bombyx mori. RNA Biol.16, 809–820 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daniel Gietz, R. & Woods, R. A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. In Guide to Yeast Genetics and Molecular and Cell Biology - Part B Vol. 350 Methods in Enzymology (eds Guthrie, C. & Fink, G. R.) 87–96 (Academic Press, 2002). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The library data involved in this study has been deposited into the Short Read Archive (SRA) of the GenBank database under the BioProject accession no. PRJNA1166905. The plasmids were deposited into BRICS under the SP3196-3198. Supplementary Data 1 contains all Supplementary Tables in the manuscript. The source data for the graphs can be found in Supplementary Data 2. All other data are available from the corresponding author on reasonable request.