Abstract
This manuscript details the application of Isothermal Titration Calorimetry (ITC) to characterize the kinetics of 3CLpro, the main protease from the Severe Acute Respiratory Syndrome CoronaVirus-2 (SARS-CoV-2), and its inhibition by Ensitrelvir, a known non-covalent inhibitor. 3CLpro is essential for producing the proteins necessary for viral infection, which led to the COVID-19 pandemic. The ITC-based assay provided rapid and reliable measurements of 3CLpro activity, allowing for the direct derivation of the kinetic enzymatic constants KM and kcat by monitoring the thermal power required to maintain a constant temperature as the substrate is consumed. The manuscript highlights several advantages of the proposed ITC-based assay over traditional methods used to study 3CLpro, such as Förster Resonance Energy Transfer (FRET) and Liquid Chromatography-Mass Spectrometry (LC–MS) and overcomes the need for non-biological substrates or discontinuous post-reaction steps. The ease of application of the ITC method allowed for the determination of the temperature dependence of the catalytic constants, enabling the estimation of the reaction activation energy. Additionally, the assay was used to determine the inhibition mode and kinetic parameters for 3CLpro inhibition by Ensitrelvir. This molecule was revealed to act as a slow- and tight-binding inhibitor that forms an initial E•I complex (KI = 9.9 ± 0.7 nM) quickly transitioning to a tighter E•I* assembly (KI* = 1.1 ± 0.2 nM). This versatile calorimetric method is proposed for general use in the discovery and development of drugs targeting 3CLpro.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-81990-y.
Subject terms: Enzyme mechanisms, Proteases
Introduction
Severe Acute Respiratory Syndrome CoronaVirus-2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), has been responsible for over seven million deaths worldwide, posing a significant threat to the global economy and healthcare system1. During SARS-CoV-2 infection, the viral proteases 3CLpro and PLpro are crucial for catalyzing the hydrolysis of two polyproteins, pp1a and pp1ab, which are translated by host ribosomes upon recognition of the viral positive single-stranded RNA2,3. The enzymatic cleavage of pp1a and pp1ab releases a set of non-structural viral proteins essential for SARS-CoV-2 replication4,5.
SARS-CoV-2 3CLpro is a homodimeric cysteine PA-clan protease6, composed of two 33.8-kDa protomers, each consisting of three structural domains (Fig. 1): domain I (residues 8 – 101), domain II (residues 102 – 184), and domain III (residues 201 – 306). The active site cleft, containing the His41—Cys145 (H41-C145) catalytic dyad, is located between domains I and II and comprises multiple subsites (S4, S3, S2, S1, and S1’). During catalysis, these subsites are occupied by specific sequences of substrate amino acid residues (P4, P3, P2, P1, and P1’, respectively). The substrate recognition motif, highly conserved among several coronavirus 3CLpro, prefers the Leu-Gln-Ser (LQS) sequence in the P2-P1-P1’ position7.
Fig. 1.
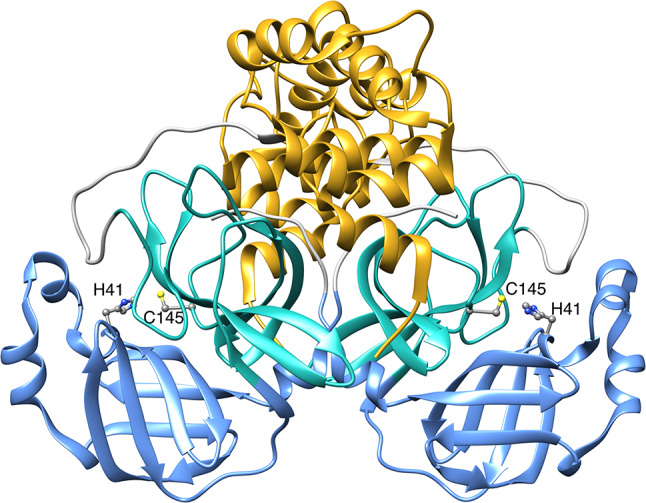
Ribbon representation of SARS-CoV-2 3CLpro (PDB code 8CDC18). Domains I, II, and III are colored in cornflower blue, light sea green and goldenrod, respectively. The side chains of the H41—C145 catalytic dyad are shown as ball-and-stick and colored according to the CPK code. Figure made using Chimera19.
3CLpro exhibits minimal catalytic activity in its monomeric state, with dimerization deemed essential for full protease activity8,9. Crystallographic evidence indicates that the dimerization process involves the first seven N-terminal residues of each protomer (N-fingers), which contribute to dimer stabilization and active site architecture, particularly the S1 subsite, through interactions with domain II of the adjacent protomer and domains II and III of the parent protomer10. Conversely, solution studies using native mass spectroscopy suggest that N-terminal processing is not critical for dimerization that, instead, appears to be triggered by an induced fit resulting from covalent linkage between the active site cysteine thiol S atom and the carbonyl C of the substrate during catalysis8.
Given its crucial role in the viral life cycle and the absence of closely related homologs in the human genome, drug discovery targeting 3CLpro has been a pivotal research area during the SARS-CoV-2 pandemic. Several 3CLpro inhibitors have been identified11–14, which can be categorized into two major families: peptidomimetics and non-peptide small molecules. Peptidomimetics are designed based on natural substrate scaffolds and typically share with them the same binding subsites. These inhibitors usually exploit an electrophilic warhead group (i.e., aldehydes, ketones, Michael acceptors, and nitriles) near the P1 moiety to covalently bind the nucleophilic thiol group of C145, thereby inactivating 3CLpro. Non-peptide small molecules inhibitors of 3CLpro encompass a diverse range of compounds, including flavonoids, terpenoids, quinoline analogs, pyridinyl esters, Ebselen analogs, benzotriazole-based compounds, pyrimidine analogs, acrylamide and related compounds, isatin analogs, triazine compounds, and metal-containing analogs. Additionally, certain macrocyclic inhibitors of other PA-clan proteases have also been found to inhibit SARS CoV-2 3CLpro15–17.
Despite the critical role of 3CLpro in the spread of COVID-19, detailed kinetic data on its activity and inhibition remain limited and often highly variable. This inconsistency arises from differences in methodologies, the form of 3CLpro used in assays, and the substrates analyzed20,21. The two most common methods for characterizing 3CLpro kinetics are Förster Resonance Energy Transfer (FRET)20–26 and Liquid Chromatography-Mass Spectrometry (LC–MS)27,28. The kinetic parameters KM and kcat obtained by these techniques span several orders of magnitude. For instance, KM values reported using FRET range from 17 to 60 µM for SARS-CoV 3CLpro21,28,29 and 28 to 230 µM for SARS-CoV-2 3CLpro21,30,31. In contrast, results based on LC–MS show KM values in the range of 0.2 to 2.6 mM for SARS-CoV 3CLpro21,32 and 0.9 mM for 3CLpro from SARS-CoV-221. Similarly, kcat values obtained by FRET range from 0.2 to 2 s-1 for SARS-CoV 3CLpro21,28,29 and 0.05 to 0.23 s-1 for SARS-CoV-2 3CLpro21,30,31, while LC–MS yields kcat values of 0.54 to 6.4 s-1 for SARS-CoV 3CLpro21,32 and 2.2 s-1 for SARS-CoV-2 3CLpro21. Thus, the KM values estimated by LC–MS (in the mM range) are generally much higher than those reported for FRET-based methods (in the sub-millimolar range), while the kcat values span at least two orders of magnitude independently of the technique used.
These discrepancies reveal less-than-optimal reliability for both types of assays. Additionally, each method presents significant disadvantages: LC–MS is a discontinuous method requiring several time-consuming sample manipulation steps, potentially leading to non-negligible experimental errors. Furthermore, FRET-based methods are susceptible to the inner filter effect, where fluorescent light is absorbed by quenching groups on neighboring substrates or cleaved reaction products, resulting in only a fraction of the fluorescence reaching the detector system of the fluorimeter33,34. FRET can also be affected by interactions between the substrate fluorophore and the enzyme, as well as by the absorbance or fluorescence of the inhibitor itself. These drawbacks highlight the need for alternative approaches that can rapidly and effectively evaluate the catalytic and inhibitory efficiency of SARS-CoV-2 3CLpro and related PA-clan proteases. Such improvements are crucial for optimizing drug screening and optimization processes, as well as for gaining deeper insights into the fundamental mechanisms of enzyme inhibition.
Isothermal Titration Calorimetry (ITC) is a technique used to characterize enzyme kinetics by monitoring the heat generated upon rapid mixing of small-volume injections of a substrate (or enzyme) solution into a sample cell containing an enzyme (or substrate) solution, either in the absence or presence of varying concentrations of an inhibitor35–41. This approach is highly versatile, as most chemical reactions involve heat production or consumption. Unlike other techniques that infer catalysis rates indirectly from substrate or product concentrations, ITC provides real-time detection of heat flow, offering a direct measurement of enzyme activity and its modulation by inhibitors. ITC does not require the development of customized assays using fluorophores or chromophores as substrates or products as needed for FRET, nor does it require post-reaction separation of products and substrates, as with LC–MS. Additionally, compared to reported measurements where enzyme, substrate, and inhibitor solutions are combined with different incubation times before the measurement, ITC measures heat flow rapidly, minimizing dead time. Despite its considerable potential, no study to date has utilized ITC to characterize the kinetics of catalysis and inhibition of 3CLpro.
Here we provide a comprehensive characterization of the activity of 3CLpro from SARS-CoV-2 using ITC. This method delivers kinetic parameters in less than an hour, with high reliability. Additionally, we describe the application of ITC to study the inhibition of 3CLpro by Ensitrelvir, a well-established 3CLpro inhibitor. Ensitrelvir is the first oral non-covalent and non-peptide inhibitor from the triazine compound family (Fig. 2), developed by Shionogi42. It was approved for emergency use in Japan in November 2022 and is marketed under the brand name Xocova.
Fig. 2.
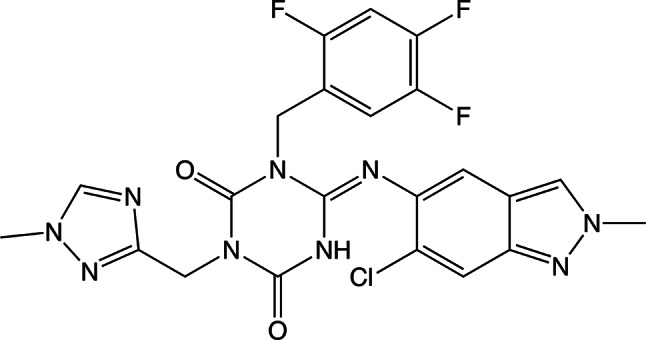
Chemical structure of Ensitrelvir.
Results
General description of the methodology
The most used approach to mathematically describe a classical enzyme-catalyzed reaction (Fig. 3) is the Michaelis–Menten model (Eq. 1). This model describes the reaction rate, expressed as the time-dependent decrease of substrate concentration [S], as a function of [S], the enzyme concentration [E], and the parameters KM and kcat.
| 1 |
Fig. 3.
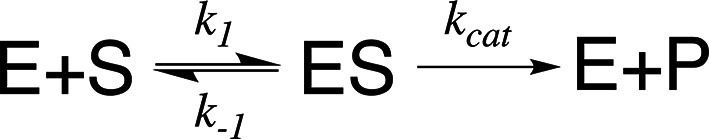
Classical enzymatic reaction.
In Eq. 1, KM (commonly known as Michaelis constant) is the pseudo-equilibrium constant [(k-1 + kcat)/k1] under steady-state conditions, and it represents the substrate concentration required to achieve half-maximal reaction rate. kcat is the catalytic rate constant (also known as turnover number) describing the limiting number of substrate molecules converted per second by the enzyme. The term kcat •[E], also defined as Vmax, is the maximum reaction rate theoretically achieved in the presence of an infinite amount of substrate. Complete characterization of any enzyme-catalyzed reaction involves determining KM and kcat. To achieve this objective, the reaction can be studied using ITC35–41, as briefly described here.
The isothermal calorimeter (Fig. 4) consists of an adiabatic shield encompassing two cells: a reference cell (usually filled with deionized water) and a sample cell. A computer-controlled syringe is mounted on the sample cell, where it dispenses its content using a rotating, paddle-shaped needle that ensures complete mixing of the solutions after each injection. During the ITC experiment, a thermoelectric device continuously measures the temperature difference between the sample and reference cells and, using a cell feedback network, it maintains this difference (ΔT) at zero by adding or removing heat from the sample cell.
Fig. 4.
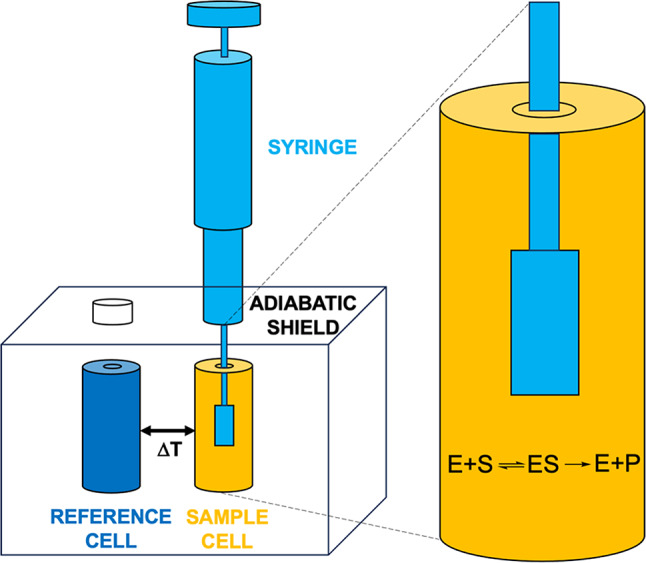
Schematic representation of the isothermal titration calorimeter: reference and sample cells are indicated in dark blue and orange, respectively, and the titration syringe is colored in light blue.
The amount of heat (Q) added or removed by the system over time (t) is defined as the thermal power (TP), often also referred to as heat flow (Eq. 2):
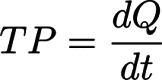 |
2 |
In an enzyme-catalyzed reaction, the heat associated with the conversion of n moles of substrate to product at constant pressure is described by Eq. 3:
| 3 |
Here, ΔHapp represents the total apparent molar enthalpy for the reaction, [S] is the molar concentration of converted substrate, and Vcell is the volume of the sample cell where the reaction occurs. The reaction rate, defined as the change in substrate concentration over time, can be related to the thermal power by Eq. 4:
| 4 |
Therefore, to calculate the reaction rates as a function of the substrate concentration, which then can be fitted using the Michaelis–Menten equation (Eq. 1) to derive the kinetic parameters KM and kcat, two key components are required: (i) knowledge of the total apparent molar enthalpy ΔHapp and (ii) measurements of dQ/dt at various substrate concentrations.
ΔHapp is typically determined using the direct single-injection setup. In this method, a small amount of substrate, at a final concentration smaller than the expected value of KM, is injected into a solution containing the enzyme in the nM–μM range. The thermal power generated by the reaction is monitored over time until the substrate is completely consumed and the signal returns to the pre-injection baseline. The enzyme and substrate concentrations are chosen so that complete conversion occurs within minutes. ΔHapp is thus calculated by integrating the area under the curve, according to Eq. 5, where [S]total is the total concentration of substrate present in the sample cell at the start of the experiment:
| 5 |
The determination of dQ/dt values at various substrate concentrations typically employs the multiple-injection method. This involves sequential small injections of concentrated substrate solution into a diluted (in the pM–nM range) enzyme solution. Each injection increases the substrate concentration, causing a shift in the baseline that reflects a change in thermal power within the sample cell. These injections are timed to allow the thermal power to stabilize at the new baseline level but must be short enough to prevent significant substrate conversion (less than 5%), ensuring measurements are conducted under steady-state conditions. The dQ/dt value at each substrate concentration can thus be determined by measuring the difference between the original baseline and the new baseline after each injection. Using Eq. 4, the resulting reaction rates as a function of substrate concentration can then be computed and fitted to Eq. 1 to derive the kinetic parameters KM and kcat.
A drawback of the multiple-injection method is the typically low total thermal power generated, especially if the reaction has a small ΔHapp. This can make the assay sensitive to baseline drifts and instrumental noise, potentially affecting the reliability of KM and kcat estimates. Additionally, achieving high substrate concentrations in the syringe solution may be challenging for sparingly soluble compounds, while the large heat of substrate dilution could significantly interfere with heat of reaction measurements. In our investigation of 3CLpro enzymatic activity, we encountered these typical challenges of the multiple-injection method and thus explored the inverse single-injection method as an alternative approach, described below.
Derivation of the kinetic parameters of 3CLpro enzymatic catalysis
Using the inverse single-injection method, a small volume of 3CLpro solution (in the tens of µM range) is injected into the sample cell containing a substrate solution (ca. 1.5 mL) at a concentration significantly higher than the expected KM. This approach ensures substantial enzyme saturation while avoiding solubility issues. The rapid hydrolysis of the concentrated substrate upon enzyme injection leads to a marked decrease in the thermal power dQ/dt (Fig. 5A), indicative of an exothermic reaction. This decrease reaches its maximal effect and then gradually returns to the pre-injection baseline as the substrate is depleted (Fig. 5A). The heat of mixing, measured separately by injecting enzyme solution into buffer alone, is negligible. Integration of these data using Eq. 5 yields ΔHapp. The change in substrate concentration over any time interval can be calculated using Eq. 6:
| 6 |
Fig. 5.
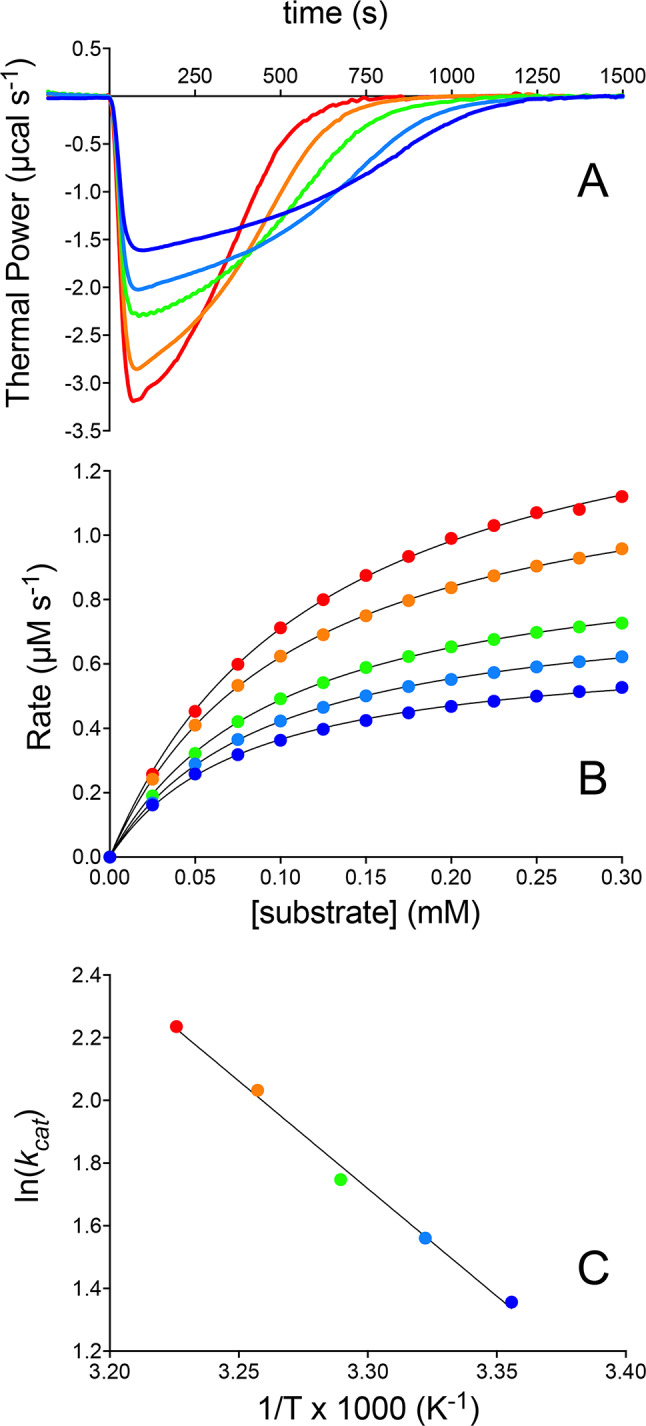
3CLpro kinetics at pH 7.5 characterized by ITC. (A) Thermal power recorded over time following injection of 150 nM 3CLpro into a sample cell containing 0.35 mM substrate at 298 K (dark blue line), 301 K (cyan line), 304 K (green line), 307 K (orange line), and 310 K (red line). (B) Corresponding reaction rates calculated using Eq. 4 and fits to the Michaelis–Menten equation shown as colored lines. The reaction rates represent a subset of the experimentally measured data points (a few hundred in total), selected every 0.025 mM for clarity. (C) Arrhenius plot depicting the logarithm of the kcat values against the inverse temperature, 1/T, used to determine the activation energy. In panels (B,C), the color-code corresponds to that in (A).
In this equation, t1 and t2 denote two consecutive time points (typically separated by 2 s). Reaction rates as a function of substrate concentration are then derived using Eq. 4 and fitted to the Michaelis–Menten equation (Eq. 1) to determine KM and kcat.
Using this methodology, the kinetics of 3CLpro were studied at five different temperatures ranging from 298 to 310 K (Fig. 5). At 298 K, the obtained values were ΔHapp = -2.1 kcal mol-1, KM = 81 ± 2 µM and kcat = 3.9 ± 0.1 s-1. These values are fully consistent with those previously reported for SARS-CoV-2 3CLpro using a similar calorimetric method, but a different data treatment43.
The measured values of ΔHapp are invariant with respect to temperature (Table 1) and consistent with values reported for peptide bond hydrolysis under similar buffer and temperature conditions44. KM increases slightly with temperature, from 81 to 124 µM, indicating a modest reduction in enzyme affinity for the substrate at higher temperatures, while kcat significantly increases from 3.9 to 9.3 s-1 in the explored temperature range. The temperature dependence of kcat allowed for the determination of the activation energy (Ea = 13.1 ± 1.4 kcal mol-1) for the 3CLpro hydrolytic reaction, derived from the Arrhenius equation (Eq. 7) and the corresponding Eyring plot (Fig. 5C):
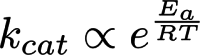 |
7 |
Table 1.
Kinetic parameters of wild type 3CLpro measured at different temperatures.
| ΔHapp (kcal mol-1) | KM (µM) | kcat (s-1) | kcat/KM (M-1 s-1) | |
|---|---|---|---|---|
| 298 K | -2.1 ± 0.1 | 81 ± 2 | 3.9 ± 0.1 | (48 ± 2) × 103 |
| 301 K | -2.2 ± 0.1 | 92 ± 1 | 4.8 ± 0.1 | (52 ± 2) × 103 |
| 304 K | -2.2 ± 0.1 | 100 ± 1 | 5.7 ± 0.1 | (57 ± 2) × 103 |
| 307 K | -2.0 ± 0.1 | 109 ± 1 | 7.6 ± 0.1 | (70 ± 2) × 103 |
| 310 K | -2.0 ± 0.1 | 124 ± 3 | 9.3 ± 0.1 | (75 ± 3) × 103 |
Derivation of the inhibition mode of 3CLpro by Ensitrelvir using ITC
The inverse single-injection method is also adaptable for studying enzyme inhibition kinetics by introducing inhibitors into the substrate solution at saturating concentration in the sample cell prior to enzyme addition. We applied this approach to investigate the inhibition kinetics of 3CLpro by the non-covalent inhibitor Ensitrelvir (Fig. 7). Raw data (Fig. 7A) were collected from experiments with varying concentrations of Ensitrelvir (in the 12.5 – 250 nM range), encompassing the reported IC50 value (ca. 50 nM45). The results showed that the thermal power initially decreased upon enzyme injection to a level dependent on the inhibitor concentration, eventually returning slowly towards the pre-injection baseline without fully reaching it unless the inhibitor concentration significantly exceeded the enzyme concentration.
Fig. 7.
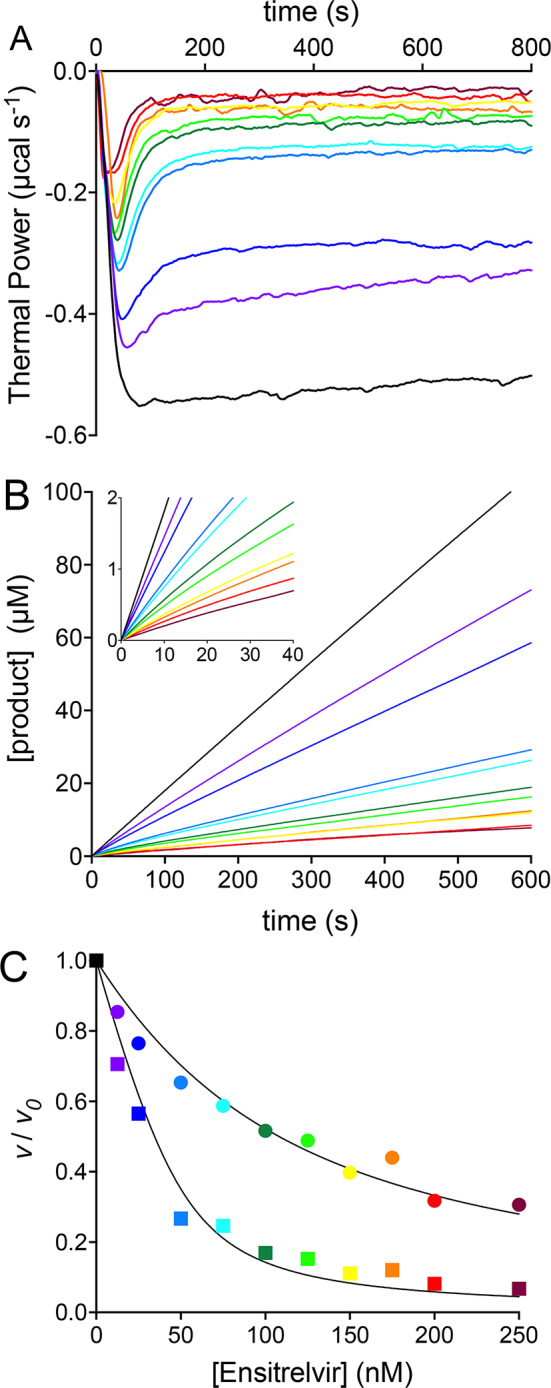
3CLpro inhibition by Ensitrelvir at pH 7.5 and 298 K characterized by ITC. (A) Thermal power recorded over time following the injection of 50 nM 3CLpro into the sample cell containing 0.60 mM substrate, in the absence of inhibitor (black) and supplemented with Ensitrelvir at concentrations of 12.5 nM (grape), 25 nM (blue), 50 nM (cyan), 75 nM (turquoise), 100 nM (dark green), 125 nM (light green), 150 nM (yellow), 175 nM (orange), 200 nM (red), 250 nM (maroon). (B) Progress curves derived from the raw data shown in (A); the inset shows the 0 – 40 s portion of each time course depicting the transition between the initial and the steady state phases. (C) vi / v0 (circles) and vs / v0 (squares) vs. Ensitrelvir concentration plots and corresponding fits to Eq. 8. In panels (B,C), the color-code corresponds to that in (A).
This behavior is characteristic of a reversible inhibitor that binds to the enzyme with association and dissociation kinetic constants (k3 and k-3) governing the equilibrium between the enzyme, the inhibitor, and the enzyme-inhibitor (E•I) complex (Fig. 6). These constants are of the same order of magnitude as those (k1 and k-1) governing the equilibrium between the enzyme, the substrate, and the enzyme–substrate (E•S) complex (Fig. 3). The observed pattern also implies the presence of a subsequent slow process, governed by k4 and k-4 constants that are smaller than k1 and k-1. This latter step is commonly interpreted as a rearrangement of E•I to a tighter E•I* complex46. The dissociation constants of the E•I and E•I* complexes are denoted here as KI and KI*
Fig. 6.
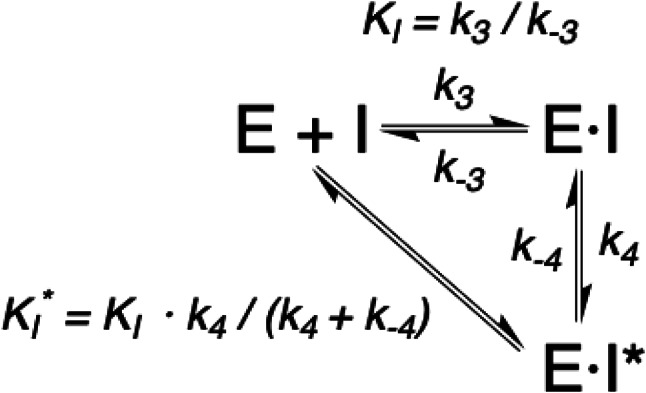
Enzymatic inhibition reaction.
To analyze this behavior, we extracted progress curves depicting the increase of product concentration as a function of time at increasing Ensitrelvir concentrations (Fig. 7B). The use of ITC and the inverse single injection method to obtain progress curves has been previously reported47–49. The progress curve recorded in the absence of inhibitor lies on a straight line with a slope corresponding to the 3CLpro reaction rate (v0). All progress curves in the presence of Ensitrelvir show an initial linear phase followed by a transition that eventually evolves toward a second linear phase with a smaller slope. The slope of the linear portions at early and late stages of each time course yielded the initial (vi) and the steady-state (vs) rate values, respectively. The steady-state rate is reached after the equilibrium involving E, I, E•I and E•I* has been fully established. Consistent with the inhibition mechanism reported in Fig. 6, both vi and vs progressively decreased in a concentration-dependent manner, corroborating the model. The sets of vi and vs values were used separately to quantitatively derive the apparent inhibition constants KIapp and K*Iapp, respectively for these two steps by using Eq. 846 (Fig. 7C):
| 8 |
Here, [E]T and [I]T are the total concentrations of enzyme and inhibitor, while Kapp refers to either Kiapp or to K*iapp depending on the use of vi or vs in place of v, respectively. Kiapp and K*iapp for the two equilibria were derived as 83 ± 6 nM and 9.5 ± 1.7 nM, respectively. Assuming a competitive inhibition mechanism, as indicated by the X-ray structures of the 3CLpro-Ensitrelvir complex42,50,51, the inhibition constants (KI and K*I) can be derived using Eq. 952:
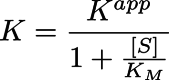 |
9 |
The two distinct values thus obtained for KI and K*I were 9.9 ± 0.7 nM and 1.1 ± 0.2 nM.
Discussion
This study represents the first successful application of isothermal titration calorimetry for characterizing the inhibition of the enzymatic activity of SARS-CoV-2 3CLpro by Ensitrelvir, an anti-SARS-CoV-2 drug for COVID-19 selectively acting as a non-covalent inhibitor of this viral protease. ITC has proven to be an effective, rapid, and reliable method for obtaining kinetic and inhibition parameters. Using ITC, we can generate Michaelis–Menten profiles in under an hour. Our experimentation with different setups, including direct multiple-injections of substrate into enzyme solution and inverse single-injection of enzyme into substrate solution coupled with the use of progress curves, identified the latter as the optimal method for accurate kinetic and inhibition parameters determination.
The inverse single-injection approach using ITC offers several advantages over other previously employed assays like FRET or LC–MS for 3CLpro kinetics and inhibition characterization. Unlike these methods, ITC continuously measures enzyme catalytic rates without requiring complex substrate or product concentration measurements in batch experiments. This approach allows for robust determination of Michaelis–Menten kinetics across a large number (hundreds) of substrate concentrations, avoiding the need for specific probes or post-reaction sample manipulations. The versatility of ITC is demonstrated by its ability to quickly provide the catalytic parameters KM and kcat. The obtained value of KM for peptide hydrolysis catalyzed by SARS-CoV-2 3CLpro at 298 K (81 ± 2 µM) aligns more closely with FRET-derived values (28–230 µM21,30) rather than those obtained from LC–MS (0.9 mM21). On the other hand, the value of kcat (3.9 ± 0.1 s-1) is more consistent with LC–MS values (2.2 s-1)21 rather than those from FRET (0.05–0.23 s-121,30,31). Furthermore, the calorimetric assay provided the first experimentally determined activation energy (Ea = 13.1 ± 1.4 kcal mol-1) for 3CLpro catalytic hydrolysis. This value is consistent with theoretical and experimental data for other cysteine proteases53–55, highlighting the robustness and novel insights enabled by ITC in enzyme kinetics studies.
However, some potential issues and their resolutions must be discussed regarding the calorimetric assay used in this study. One possible concern is substrate or product inhibition. This was ruled out by analyzing the shape of the Michaelis–Menten curves and testing various substrate concentrations (0.15 – 0.35 mM), which showed no significant effect on KM and kcat (Fig. 1-SI). Another critical consideration is the presence of a residual concentration of 3CLpro monomer in equilibrium with the dimer, as dimerization is essential for full enzymatic activity8,9. Reports of the dimer dissociation constant (KD) for SARS-CoV 3CLpro range from 0.25 to 1 nM20, while values of KD for the SARS-CoV-2 3CLpro dimer vary more widely, ranging from 7 ± 1 μM (determined using small-angle X-ray scattering)56, to ~ 2.5 μM (from sedimentation velocities obtained by analytical ultracentrifugation)57, down to 0.14 ± 0.03 μM (from mass spectrometry data)58. In our approach, the enzyme concentration in the syringe is 15 µM, significantly higher than the above mentioned dissociation constants and much greater than concentrations used in FRET assays (0.1 – 2.0 µM21,30). It has been reported that the dissociation occurs over days at room temperature for SARS-CoV 3CLpro59 and is further disfavored in the presence of substrate10. Therefore, the inverse single-injection method used here likely maintains the enzyme in its active dimeric form as dilution from 15 µM in the syringe to 50 nM (or larger) in the sample cell occurs rapidly and in the presence of substrate. This was confirmed by the linear dependence of Vmax as a function of a wide range of enzyme concentrations (0.050 – 1.2 µM, see Fig. 2-SI). This might explain the reason for the value of kcat determined using ITC, (3.9 ± 0.1 s-1) being significantly higher than those determined by FRET and LC–MS methods, (0.05—2.2 s-1)21,30,31. Indeed, in the latter assays the reaction is initiated by the addition of the substrate into a solution containing the enzyme already equilibrated at the final concentration (in the absence of substrate), which might be so diluted that the enzyme could already be partially dissociated into the scarcely active monomeric form. The FRET-based assays also employed a wide range of enzyme-inhibitor incubation times (1.5 – 180 min) during which dissociation could occur at different degrees thus resulting in the high variability of the reported kcat values. The E290A/R298A SARS-CoV-2 3CLpro double mutant, a prevalently monomeric form of the enzyme9, was additionally tested using ITC and was indeed shown to be inactive (Fig. 3-SI), further indicating that the proposed experimental setup maintains 3CLpro in a dimeric and fully active form.
The methodology adopted in this study to investigate the mode of action of Ensitrelvir as a non-covalent inhibitor of SARS-CoV-2 3CLpro contrasts with the approach reported by Stille et al.43 to evaluate covalent inhibitors of the same enzyme using ITC. In our approach, the substrate concentration (0.60 mM) is deliberately set much higher than the estimated KM value to maintain substrate saturation throughout the calorimetric measurement. At the same time, the enzyme concentration is kept low (0.05 µM) to reduce the hydrolysis rate, enabling the recording of ITC traces without substantial decrease in the reaction rate due to substrate depletion. As a result, any observed decrease in heat flow over time reflects only the interaction between the enzyme and the inhibitor. On the other hand, Stille et al.43 employed a lower substrate concentration (ca. 0.3 mM) and a significantly higher enzyme concentration (0.400 µM), leading to significant substrate depletion during the assay. Consequently, the reaction rates were affected by both substrate depletion and enzyme-inhibitor interactions, conditions that required complex data treatment involving convoluted algorithms to integrate differential equations and fit the results to obtain inhibition constants.
The methodology described in this study offers significant advantages, particularly in distinguishing between fast and slow inhibitors, as well as between tight-binding, non-tight-binding, and covalent modes of action. By isolating the enzyme-inhibitor interaction from the complicating effects of substrate depletion, this method enables a detailed characterization of inhibition properties. Our ITC-based assay revealed that Ensitrelvir is a potent inhibitor of SARS-CoV-2 3CLpro that acts as a tight- and slow-binding inhibitor, forming an initial E•I complex with a dissociation constant KI = 9.9 ± 0.7 nM that evolves into a tighter E•I* complex with K*I = 1.1 ± 0.2 nM within seconds. The key outcome of this study is therefore the identification of an initial step in the enzyme-inhibitor interaction, for which an estimate of its equilibrium constant can be inferred. This step is followed by a slower and tighter binding equilibrium, from which the enzyme-inhibitor affinity is also determined.
The previously reported affinity of Ensitrelvir for 3CLpro in the absence of substrate resulted in the determination of a single dissociation constant KD = 8.43 nM45 using ITC binding experiments, consistently with the KI determined in this study for the E•I complex in the presence of substrate. The detection of an additional tighter equilibrium, with a dissociation constant of one order of magnitude smaller, suggests that the presence of substrate causes the enzyme to adopt a conformation that has higher affinity for the inhibitor in the E•I* complex.
ITC is the first method to directly estimate the 3CLpro inhibition constants by Ensitrelvir, without the need to convert the IC50 values commonly derived from FRET45,51,60 and LC-MS42 assays via the Cheng-Prusoff equation (which implies the knowledge of the inhibition mode of the compound under study). The only reported value of KI = 9 ± 0.7 nM for this inhibitor was indeed calculated from the IC50 value assuming a competitive mechanism51, consistently with the X-ray crystal structure reported for the enzyme-inhibitor complex45,60. In addition, while the enzyme-inhibitor incubation times used for FRET or LC–MS might determine the formation of a stable E•I complex prior to its exposure to the substrate, the described calorimetric assay provides a true competition experiment for the enzyme to either substrate or inhibitor.
Conclusions
We propose the generalized use of the calorimetric assay developed in this study for investigating the kinetics of catalysis and inhibition of SARS-CoV-2 3CLpro. ITC can be used to validate novel hits from a 3CLpro inhibitor high-throughput screen and to identify potential pan-assay interference compounds (PAINS) that can yield false positives in widely used high-throughput fluorescence-based platforms61. Moreover, ITC is invaluable not only for determining inhibition modes and relevant kinetic parameters but also for providing critical thermodynamic signatures, such as binding enthalpy and entropy, for protein–ligand interactions. These thermodynamic insights can significantly enhance drug design efforts toward improved 3CLpro inhibitors. Specifically, focusing on optimizing enthalpic versus entropic contributions to binding for hit compounds has been shown to lead to better prioritization and optimization of ligands during hit selection and hit-to-lead processes. This approach can ultimately result in the development of more effective therapeutic agents61,62.
Materials and methods
Enzyme, substrate and inhibitor sources
Native 3CLpro (Mr monomer = 33.8 kDa, pI = 5.95) was expressed using the plasmid vector pGTM_COV2_NSP5_004_SUMO (available from AddGene, ID: 190,062), and BL21(DE3) Escherichia coli cells, and purified following a previously described protocol18. The plasmid for the E290A/R298A 3CLpro double mutant was obtained from GenScript (Rijswijk, Netherlands) starting from the above construct as a template. The E290A/R298A 3CLpro double mutant was purified using the same procedure as for the native enzyme. 3CLpro and its double mutant (purity > 98% as checked by SDS-PAGE) were stored as 150 µM and 900 µM stock aliquots, respectively (protein concentration is referred to the monomer throughout the manuscript) at -80 °C in 20 mM Tris–HCl buffer, 50 mM NaCl, 1 mM EDTA, at pH 7.5. The peptide substrate WKTSAVLQ/SGFRKMEW (Mr = 1.95 kDa, pI = 9.99) was designed with the 3CLpro cleavage site Q/S, and was purchased from GenScript (Rijswijk, Netherlands). It includes two non-native tryptophan (W) residues at the N and C termini to ensure a measurable absorption at 280 nm. Solutions of substrate were freshly prepared before every experiment (see below for details). Protein and peptide quantification was carried out, prior to each experiment, by measuring the absorbance at 280 nm and considering a molar extinction coefficient (ε280) of 33,000 M-1 cm-1 and 11,000 M-1 cm-1 for 3CLpro and the substrate respectively, estimated using ProtParam63. Ensitrelvir fumarate (molar mass = 647.95 g mol-1) was purchased from Cabru S.A.S. (Arcore, Italy), dissolved at 10 mM in pure DMSO, and stored as 10 µL aliquots at -80 °C.
Calorimetric studies on the enzymatic hydrolysis by 3CLpro
The determination of the 3CLpro kinetic parameters at fixed enzyme concentration was carried out using a high-sensitivity VP-ITC micro-calorimeter (MicroCal LLC, Northampton, MA, USA). For each experiment, the reference cell was filled with deionized water and the temperature of the reference and sample cells was set and stabilized at five temperatures in the range 298—310 K. Stirring speed was 300 rpm and the thermal power was monitored every 2 s using high instrumental feedback. Solutions of 3CLpro for the assay were prepared by diluting a stock solution down to 15 µM in 600 µL of 20 mM Tris–HCl, 50 mM NaCl, 1 mM EDTA, 1% DMSO, at pH 7.5 (buffer A) and loaded into the injection syringe. The measured enzymatic activity was not affected over several days at 4 °C in the absence of DTT. The substrate was prepared by dissolving the purchased powder in 2 mL of the same buffer A, obtaining a final concentration of 0.35 mM, and loaded in the sample cell. The inverse single-injection experiment was carried out by injecting 15 µL of the 15 µM 3CLpro solution (final enzyme concentration in the sample cell = 0.15 µM) from the syringe into the 0.35 mM substrate solution. The thermal power (TP, µcal s-1) was recorded over 2000 s, ensuring the instrument baseline shift caused by the heat flow generated from the enzymatic hydrolysis of the substrate to return to its original pre-injection level. The raw calorimetric data were processed using the Method 1 available in the MicroCal Origin software to derive the total apparent molar enthalpy ΔHapp of 3CLpro-catalyzed substrate hydrolysis (according to Eq. 5). The reaction rates as a function of substrate concentration were obtained according to Eqs. 4 and 6, discarding the data obtained immediately after the injection of the substrate and before the calorimeter equilibrated with the ongoing reaction. The obtained reaction rates were fit using the canonical Michaelis–Menten equation (Eq. 1) to derive the kinetic parameters KM and kcat. The determination of the 3CLpro kinetic parameters at increasing monomer concentration (in the range 0.050 – 1.2 µM) was carried out at 298 K following the same procedure, using injection volumes in the 60 – 5 µL range. The same experimental setup was used for the E290A/R298A 3CLpro double mutant by injecting 60 µL of a 900 µM protein (final concentration in the sample cell = 36 µM) into the sample cell containing 0.35 mM substrate. The experiments were performed as multiple technical replicates, each carried out with a minimum of three repetitions. The obtained values deviated by less than 5% across all measurements. The reported errors reflect the precision of the parameters obtained from a representative fit.
Calorimetric studies on the enzymatic inhibition of 3CLpro by Ensitrelvir
The determination of the inhibition parameters of Ensitrelvir on 3CLpro was carried out using an inverse single-injection assay slightly modified with respect to that previously described. In this case, the 600-µL enzyme solution was prepared by diluting the stock 3CLpro down to 5.0 µM using buffer A, while the 2-mL substrate solution was prepared at 0.60 mM in the same buffer. A first experiment was carried out by injecting 15 µL of the 3CLpro solution (final enzyme concentration in the sample cell = 0.050 µM) into the substrate-containing sample cell. The use of smaller concentrations of enzyme and larger concentrations of substrate than those used for the Michaelis–Menten experiments ensured that, throughout the reaction time, the system fulfilled saturating and steady state conditions, thus ensuring a constant initial velocity. The thermal power was recorded over ca. 1000 s, a time that guaranteed the signal generated from the enzymatic hydrolysis of the substrate to be at a constant value, in turn signifying that the reaction rate is constant according to Eq. 4. A set of experiments was also carried out at increasing concentrations of Ensitrelvir in the 12.5 – 250 nM range, the latter being added by dilution of the 10 mM stock in buffer A to the substrate solution. This experimental setup therefore consisted in the injection of 15 µL of 3CLpro in the sample cell (final enzyme concentration = 0.05 µM) that contained both the substrate and the inhibitor, thus providing a true competition experiment for the enzyme to either the substrate or the inhibitor. The recorded thermal power was integrated over time starting from the time at which it reached its minimum, the latter invariably occurring well after the nominal response time of the VP-ITC (10–20 s); the resulting total heat (expressed as µcal) was converted to the corresponding concentration of product formed during the reaction by considering ∆Happ = -2.0 kcal mol-1. Taking into account the enzyme concentration, the sample cell volume, and the molar enthalpy for Ensitrelvir binding to 3CLpro in the absence of substrate—previously determined to be -15.4 kcal/mol64—it can be estimated that the heat of inhibitor-enzyme binding is negligible compared to the overall heat measured in the assays, contributing approximately 1:2000. The obtained [P] formation was then plotted as a function of reaction time to yield progress curves. The initial (vi) and the steady-state (vs) rates in the presence of Ensitrelvir were derived by a linear fit of the first and last ten data points at early and late stages of each time course, respectively. The constant reaction rate in the absence of Ensitrelvir (v0) was derived as the slope of the corresponding linear progress curve. The obtained vi/v0 and vs/v0 values were used to derive the dissociation constants of the E•I and E•I* complexes according to Eqs. 8 and 9. Additional experiments, performed in the presence of fixed concentrations of Ensitrelvir (0.10 µM) and substrate (500 µM) and by injecting the enzyme solution (final concentration of 0.050 µM) at different injection rates (2.34 μL s-1, 1.17 μL s-1, and 0.59 μL s-1, corresponding to 5 s, 10 s, and 20 s of injection time), were carried out, with no significant variation of the derived parameters, thus ruling out a possible dependance of the injection rate (Fig. 4-SI). Finally, to consider possible effects of the instrumental response time on the accurate determination of the reaction rates, we estimated the inhibition constants by selectively removing the reaction rates determined at progressively increasing Ensitrelvir concentrations. The analysis of the results showed that the selective removal of the reaction rates does not significantly affect the result.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
LM and SC acknowledge financial support from the University of Bologna and the Consorzio Interuniversitario di Risonanze Magnetiche di Metallo-Proteine (CIRMMP). This study was also supported by grant R35-GM141818 (to GTM) from the National Institutes of Health, National Institute of General Medical Sciences. RG-C was supported in part by a NIGMS Graduate Training Grant (T32-GM141865).
Author contributions
L.M. and S.C. conceived the project; L.M., S.R., D.S. and R.G.-C. performed protein purification; L.M., S.R. and D.S. carried out the ITC experiments; L.M. and S.C. analysed the data with the contribution of S.R., D.S. , G.T.M. and C.C.; L.M. and S.C. co-wrote the paper with the collaboration of all the co-authors.
Data availability
The calorimetric raw data are available from the corresponding author upon request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Luca Mazzei, Email: luca.mazzei2@unibo.it.
Stefano Ciurli, Email: stefano.ciurli@unibo.it.
References
- 1.Forman, R. et al. Drawing light from the pandemic: Rethinking strategies for health policy and beyond. Health Policy126, 1–6. 10.1016/j.healthpol.2021.12.001 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brant, A. C., Tian, W., Majerciak, V., Yang, W. & Zheng, Z. M. SARS-CoV-2: from its discovery to genome structure, transcription, and replication. Cell Biosci11, 136. 10.1186/s13578-021-00643-z (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jahirul Islam, M., Nawal Islam, N., Siddik Alom, M., Kabir, M. & Halim, M. A. A review on structural, non-structural, and accessory proteins of SARS-CoV-2: Highlighting drug target sites. Immunobiology228, 152302. 10.1016/j.imbio.2022.152302 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suryawanshi, R. K., Koganti, R., Agelidis, A., Patil, C. D. & Shukla, D. Dysregulation of cell signaling by SARS-CoV-2. Trends Microbiol29, 224–237. 10.1016/j.tim.2020.12.007 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.To, K. K. et al. Lessons learned 1 year after SARS-CoV-2 emergence leading to COVID-19 pandemic. Emerg Microbes Infect10, 507–535. 10.1080/22221751.2021.1898291 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rawlings, N. D., Barrett, A. J. & Bateman, A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res40, D343-350. 10.1093/nar/gkr987 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao, Y. et al. Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2. Proc Natl Acad Sci USA119, e2117142119. 10.1073/pnas.2117142119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noske, G. D. et al. A crystallographic snapshot of SARS-CoV-2 main protease maturation process. J Mol Biol433, 167118. 10.1016/j.jmb.2021.167118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nashed, N. T., Aniana, A., Ghirlando, R., Chiliveri, S. C. & Louis, J. M. Modulation of the monomer-dimer equilibrium and catalytic activity of SARS-CoV-2 main protease by a transition-state analog inhibitor. Commun Biol5, 160. 10.1038/s42003-022-03084-7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noske, G. D. et al. An in-solution snapshot of SARS-COV-2 main protease maturation process and inhibition. Nat Commun14, 1545. 10.1038/s41467-023-37035-5 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, X. & Song, Y. Structure and function of SARS-CoV and SARS-CoV-2 main proteases and their inhibition: A comprehensive review. Eur J Med Chem260, 115772. 10.1016/j.ejmech.2023.115772 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, K. et al. Perspectives on SARS-CoV-2 main protease inhibitors. J Med Chem64, 16922–16955. 10.1021/acs.jmedchem.1c00409 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.She, Z. et al. M(pro)-targeted anti-SARS-CoV-2 inhibitor-based drugs. J Chem Res47, 17475198231184800. 10.1177/17475198231184799 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kronenberger, T., Laufer, S. A. & Pillaiyar, T. COVID-19 therapeutics: Small-molecule drug development targeting SARS-CoV-2 main protease. Drug Discov Today28, 103579. 10.1016/j.drudis.2023.103579 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bafna, K. et al. Hepatitis C virus drugs that inhibit SARS-CoV-2 papain-like protease synergize with remdesivir to suppress viral replication in cell culture. Cell Rep35, 109133. 10.1016/j.celrep.2021.109133 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bafna, K., Cioffi, C. L., Krug, R. M. & Montelione, G. T. Structural similarities between SARS-CoV2 3CL(pro) and other viral proteases suggest potential lead molecules for developing broad spectrum antivirals. Front Chem10, 948553. 10.3389/fchem.2022.948553 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lockbaum, G. J. et al. Pan-3C protease inhibitor rupintrivir binds SARS-CoV-2 main protease in a unique binding mode. Biochemistry60, 2925–2931. 10.1021/acs.biochem.1c00414 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazzei, L. et al. Protocol for production and purification of SARS-CoV-2 3CLpro. STAR Protocols4, 102326. 10.1016/j.xpro.2023.102326 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pettersen, E. F. et al. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem.25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Grum-Tokars, V., Ratia, K., Begaye, A., Baker, S. C. & Mesecar, A. D. Evaluating the 3C-like protease activity of SARS-Coronavirus: recommendations for standardized assays for drug discovery. Virus Res133, 63–73. 10.1016/j.virusres.2007.02.015 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, F., Fang, T., Guo, F., Zhao, Z. & Zhang, J. Comprehensive understanding of the kinetic behaviors of main protease from SARS-CoV-2 and SARS-CoV: New data and comparison to published parameters. Molecules.10.3390/molecules28124605 (2023). [DOI] [PMC free article] [PubMed]
- 22.Jin, Z. et al. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature582, 289–293. 10.1038/s41586-020-2223-y (2020). [DOI] [PubMed] [Google Scholar]
- 23.Coelho, C., Gallo, G., Campos, C. B., Hardy, L. & Wurtele, M. Biochemical screening for SARS-CoV-2 main protease inhibitors. PLoS ONE15, e0240079. 10.1371/journal.pone.0240079 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breidenbach, J. et al. Targeting the main protease of SARS-CoV-2: From the establishment of high throughput screening to the design of tailored inhibitors. Angew Chem Int Ed Engl60, 10423–10429. 10.1002/anie.202016961 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuzikov, M. et al. Identification of inhibitors of SARS-CoV-2 3CL-pro enzymatic activity using a small molecule in vitro repurposing screen. ACS Pharmacol Transl Sci4, 1096–1110. 10.1021/acsptsci.0c00216 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu, W. et al. Identification of SARS-CoV-2 3CL protease inhibitors by a quantitative high-throughput screening. ACS Pharmacol Transl Sci3, 1008–1016. 10.1021/acsptsci.0c00108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan, K. et al. The substrate specificity of SARS coronavirus 3C-like proteinase. Biochem Biophys Res Commun329, 934–940. 10.1016/j.bbrc.2005.02.061 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue, X. et al. Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J Mol Biol366, 965–975. 10.1016/j.jmb.2006.11.073 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuo, C. J., Chi, Y. H., Hsu, J. T. & Liang, P. H. Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate. Biochem Biophys Res Commun318, 862–867. 10.1016/j.bbrc.2004.04.098 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sacco, M. D. et al. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M(pro) and cathepsin L. Sci Adv.10.1126/sciadv.abe0751 (2020). [DOI] [PMC free article] [PubMed]
- 31.Rut, W. et al. SARS-CoV-2 M(pro) inhibitors and activity-based probes for patient-sample imaging. Nat Chem Biol17, 222–228. 10.1038/s41589-020-00689-z (2021). [DOI] [PubMed] [Google Scholar]
- 32.Kao, R. Y. et al. Characterization of SARS-CoV main protease and identification of biologically active small molecule inhibitors using a continuous fluorescence-based assay. FEBS Lett576, 325–330. 10.1016/j.febslet.2004.09.026 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, Y. et al. Use of a fluorescence plate reader for measuring kinetic parameters with inner filter effect correction. Anal Biochem267, 331–335. 10.1006/abio.1998.3014 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Kasparek, A. & Smyk, B. A new approach to the old problem: Inner filter effect type I and II in fluorescence. Spectrochim Acta A Mol Biomol Spectrosc198, 297–303. 10.1016/j.saa.2018.03.027 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Todd, M. J. & Gomez, J. Enzyme kinetics determined using calorimetry: A general assay for enzyme activity?. Anal Biochem296, 179–187. 10.1006/abio.2001.5218 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Olsen, S. N. Applications of isothermal titration calorimetry to measure enzyme kinetics and activity in complex solutions. Thermochimica Acta448, 12–18. 10.1016/j.tca.2006.06.019 (2006). [Google Scholar]
- 37.Bianconi, M. L. Calorimetry of enzyme-catalyzed reactions. Biophys Chem126, 59–64. 10.1016/j.bpc.2006.05.017 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Demarse, N. A., Killian, M. C., Hansen, L. D. & Quinn, C. F. Determining enzyme kinetics via isothermal titration calorimetry. Methods Mol Biol978, 21–30. 10.1007/978-1-62703-293-3_2 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Transtrum, M. K., Hansen, L. D. & Quinn, C. Enzyme kinetics determined by single-injection isothermal titration calorimetry. Methods76, 194–200. 10.1016/j.ymeth.2014.12.003 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Mazzei, L., Ciurli, S. & Zambelli, B. Isothermal titration calorimetry to characterize enzymatic reactions. Methods Enzymol567, 215–236. 10.1016/bs.mie.2015.07.022 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Wang, Y., Wang, G., Moitessier, N. & Mittermaier, A. K. Enzyme kinetics by isothermal titration calorimetry: Allostery, inhibition, and dynamics. Front Mol Biosci7, 583826. 10.3389/fmolb.2020.583826 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unoh, Y. et al. Discovery of S-217622, a noncovalent oral SARS-CoV-2 3CL protease inhibitor clinical candidate for treating COVID-19. J Med Chem65, 6499–6512. 10.1021/acs.jmedchem.2c00117 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stille, J. K. et al. Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CL(pro) covalent inhibitors. Eur J Med Chem229, 114046. 10.1016/j.ejmech.2021.114046 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rawitscher, M., Wadsö, I. & Sturtevant, J. M. Heats of hydrolysis of peptide Bonds1. Journal of the American Chemical Society83, 3180–3184. 10.1021/ja01476a003 (1961). [Google Scholar]
- 45.Lin, M. et al. Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants. Communications Biology6, 694. 10.1038/s42003-023-05071-y (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Copeland, R. A. Evaluation of enzyme inhibitors in drug discovery. A guide for medicinal chemists and pharmacologists. Methods of biochemical analysis46, 1–265 (2005). [PubMed]
- 47.Mazzei, L., Cianci, M., Contaldo, U., Musiani, F. & Ciurli, S. Urease inhibition in the presence of N-(n-butyl) thiophosphoric triamide, a suicide substrate: structure and kinetics. Biochemistry56, 5391–5404. 10.1021/acs.biochem.7b00750 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Mazzei, L., Cianci, M., Benini, S. & Ciurli, S. The impact of pH on catalytically critical protein conformational changes: the case of the urease, a nickel enzyme. Chem. Eur. J.25, 12145–12158. 10.1002/chem.201902320 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Mazzei, L., Cianci, M., Contaldo, U. & Ciurli, S. Insights into urease inhibition by N-(n-butyl) phosphoric triamide through an integrated structural and kinetic approach. J. Agric. Food Chem.67, 2127–2138. 10.1021/acs.jafc.8b04791 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Duan, Y. et al. Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir. Nature622, 376–382. 10.1038/s41586-023-06609-0 (2023). [DOI] [PubMed] [Google Scholar]
- 51.Noske, G. D. et al. Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease. Journal of Biological Chemistry299, 103004. 10.1016/j.jbc.2023.103004 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Copeland, R. A. Enzymes: A practical introduction to structure, mechanism, and data analysis. (Wiley-VCH, 2000).
- 53.Fernandes, H. S., Sousa, S. F. & Cerqueira, N. New insights into the catalytic mechanism of the SARS-CoV-2 main protease: An ONIOM QM/MM approach. Mol Divers26, 1373–1381. 10.1007/s11030-021-10259-7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swiderek, K. & Moliner, V. Revealing the molecular mechanisms of proteolysis of SARS-CoV-2 M(pro) by QM/MM computational methods. Chem Sci11, 10626–10630. 10.1039/d0sc02823a (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elsasser, B. & Goettig, P. Mechanisms of proteolytic enzymes and their inhibition in QM/MM studies. Int J Mol Sci.10.3390/ijms22063232 (2021). [DOI] [PMC free article] [PubMed]
- 56.Silvestrini, L. et al. The dimer-monomer equilibrium of SARS-CoV-2 main protease is affected by small molecule inhibitors. Sci Rep11, 9283. 10.1038/s41598-021-88630-9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, L. et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science368, 409–412. 10.1126/science.abb3405 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.El-Baba, T. J. et al. Allosteric inhibition of the SARS-CoV-2 main protease: Insights from mass spectrometry based assays**. Angew. Chem. Int. Ed.59, 23544–23548. 10.1002/anie.202010316 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kang, X. et al. Foldon unfolding mediates the interconversion between Mpro-C monomer and 3D domain-swapped dimer. Proc National Acad Sci109, 14900–14905. 10.1073/pnas.1205241109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin, C. et al. Structural basis for the inhibition of coronaviral main proteases by ensitrelvir. Structure31, 1016–1024. 10.1016/j.str.2023.06.010 (2023). [DOI] [PubMed]
- 61.Claveria-Gimeno, R., Vega, S., Abian, O. & Velazquez-Campoy, A. A look at ligand binding thermodynamics in drug discovery. Expert Opin Drug Discov12, 363–377. 10.1080/17460441.2017.1297418 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Ladbury, J. E., Klebe, G. & Freire, E. Adding calorimetric data to decision making in lead discovery: A hot tip. Nat Rev Drug Discov9, 23–27. 10.1038/nrd3054 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Gasteiger, E. et al. in The Proteomics Protocols Handbook (ed John M. Walker) 571–607 (Humana Press, 2005).
- 64.Kovalevsky, A. et al. Contribution of the catalytic dyad of SARS-CoV-2 main protease to binding covalent and noncovalent inhibitors. Journal of Biological Chemistry299, 104886. 10.1016/j.jbc.2023.104886 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The calorimetric raw data are available from the corresponding author upon request.


