Abstract
Fetal growth restriction (FGR) is a common complication of pregnancy, which seriously endangers fetal health and still lacks effective therapeutic targets. Clostridium difficile (C. difficile) is associated with fetal birth weight, and its membrane vesicles (MVs) are pathogenic vectors. However, the role of C. difficile and its MVs in FGR remains unclear. Here we found that supplementation with C. difficile altered the characteristics of gut microbiota and reduced the birth weight in mice. Interestingly, C. difficile MVs entered placenta, inhibited trophoblast motility, and induced fetal weight loss in mice. Mechanistically, C. difficile MVs activated the PPAR pathway via enhancing the transcriptional activity of PPARγ promoter, consequently inhibiting trophoblast motility. Moreover, PPARγ expression was significantly elevated in FGR placenta, and negatively correlated with fetal birth weight. Together, our findings reveal the significance of C. difficile and its MVs in FGR, providing new insights into the mechanisms of FGR development.
Subject terms: Microbiology, Cellular microbiology
Introduction
Fetal growth restriction (FGR), also known as intrauterine growth restriction, is a common complication of pregnancy associated with neonatal mortality and morbidity and long-term fetal health1–5. At present, there is no recognized effective treatment for FGR except for close prenatal monitoring and optimizing the timing of pregnancy termination to improve fetal prognosis3,6,7. However, until now, the specific pathogenesis of FGR is not completely clear.
The gut microbiota plays a vital and complex role in the maintenance of maternal and infant health, so it is gaining increasing attention8–11. Our previous studies also found that the dysbiosis of gut microbiota is an essential cause of placental disease preeclampsia, in which the abundance of Clostridium is significantly up-regulated8. Placental dysfunction is the main factor of FGR, suggesting that the dysregulation of Clostridium may be related to FGR12. In addition, the abundance of Clostridium is reported to be significantly elevated in pregnant women with FGR13. C. difficile is an important component of the Clostridium and associated with fetal weight14–17. Nevertheless, it is undefined whether C. difficile is involved in FGR.
Bacterial membrane vesicles (MVs) are vital mediums for bacteria to influence the host18,19. Our previous studies showed that Akkermansia relieve preeclampsia-like symptoms through outer MVs in mice20. C. difficile also produces MVs, thereby inducing inflammatory responses in intestinal epithelial cells21. Yet, the role of C. difficile MVs in FGR is still unknown. Given this, we hypothesized that C. difficile and its MVs might contribute to the occurrence of FGR.
In this study, we found that C. difficile and its MVs induced low birth weight in mice. Further studies have revealed that C. difficile MVs involved in FGR through inhibiting trophoblast motility via activating the PPARγ/RXRΑ /ANGPTL4 axis. Our findings provide new insights into the regulation of the gut microbiota in placenta-derived diseases.
Results
C. difficile is involved in FGR
Based on previous reports, we used dexamethasone (DEX) to induce FGR models in mice22. Since pregnant women with FGR did not present diarrhea symptoms caused by C. difficile toxins, we chose non-toxin-producing C. difficile for our study. The role of C. difficile in FGR was studied by exogenous supplementation of C. difficile by intragastric administration in mice. The treatment process of mice is shown in Supplementary Fig. 1A. Compared with the control group, the birth weight and length of mice in C. difficile, DEX, and C. difficile + DEX groups decreased significantly (P < 0.001, Supplementary Fig. 1B–D). Interestingly, C. difficile also affects the characteristics of gut microbiota. After C. difficile supplementation, no statistical differences in α-diversity represented by Simpson, ACE, PD-tree, and Good’s coverage indices, were found among all groups (Supplementary Fig. 1E–H). However, by calculating the weighted UniFrac distance between individual samples to estimate the β-diversity, both PCoA and NMDS plots revealed that after 14 days of C. difficile supplementation, the gut microbiota structure of C. difficile group had a significant deviation and formed a distinct cluster. Meanwhile, there was no significant change in the gut microbiota of the control group during the 14 days (Supplementary Fig. 1I, J). In addition, LEfSe analysis was used to identify differentially abundant taxa between the groups. Supplementary Fig. 1K shows that Verrucomicrobiales, Ileibacterium, and Clostridium-sensu-stricto-1 were highly abundant in the C. difficile group after 14 days of C. difficile supplementation.
Taken together, these data provided evidence that C. difficile affected the occurrence of FGR.
C. difficile MVs induce FGR in vivo
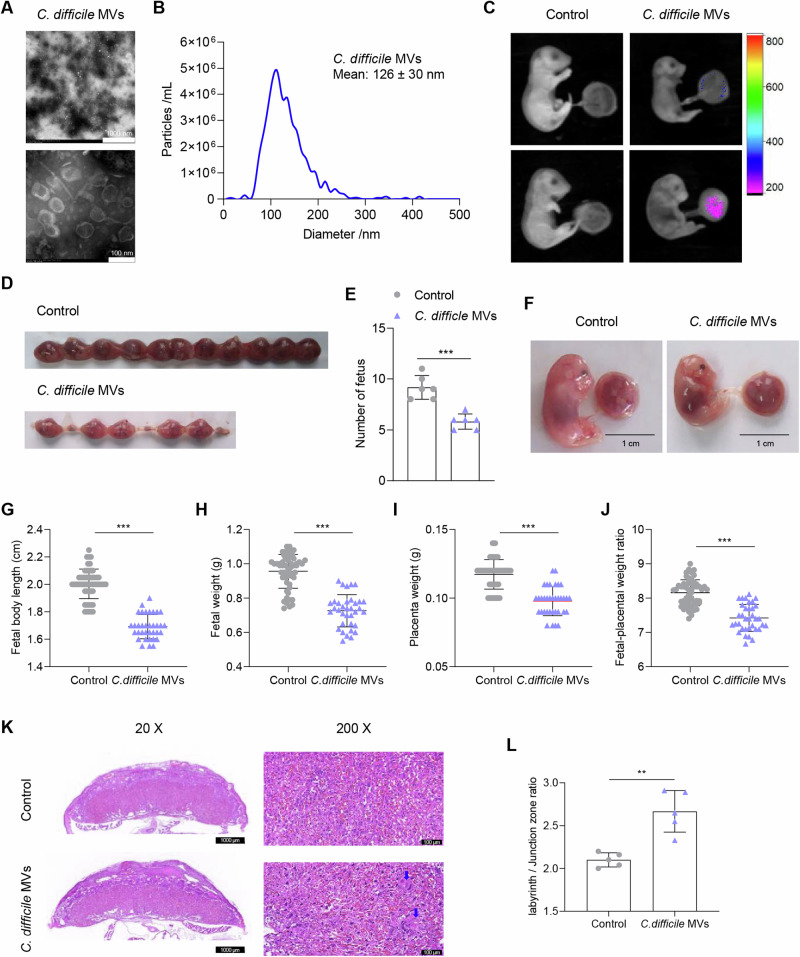
C. difficile MVs were isolated using ultracentrifugation. Transmission electron microscopy showed that C. difficile MVs had a spherical structure (Fig. 1A). The mean diameter of C. difficile MVs was 126 ± 30 nm, as measured using Nanoparticle tracking analysis (NTA) (Fig. 1B). To explore whether C. difficile MVs could enter the placenta, C. difficile MVs were labeled with Cy7 and administered to mice by intragastric administration on E17.5. Interestingly, Cy7 fluorescence was detected in the placenta at 12 h after intragastric administration (Fig. 1C). These data indicated that C. difficile MVs may enter the placenta and affect its function.
Fig. 1. C. difficile MVs are involved in the occurrence of FGR.
A TEM of isolated C. difficile MVs. B Size distribution of C. difficile MVs analyzed by NTA. C The distribution of Cy7-labeled C. difficile MVs in the mouse fetuses and placenta was examined by fluorescence imaging after oral administration of PBS and Cy7-C. difficile MVs for 12 h. D Representative photograph of the number of fetuses in both groups. E The number of fetuses in the C. difficile MVs group was significantly reduced. F Representative photograph of the fetuses and placentas from both groups. Fetal body length (G), fetal weight (H), placental weight (I), and fetal-placental weight ratio (J) were significantly decreased in the C. difficile MVs group compared with the control group. K The placentas from each group were collected on E17.5 and stained with HE (scale bar is 1000, 100 and 50 um respectively). L The labyrinth/junction zone ratio was significantly higher in the C. difficile MVs group. Data are represented as mean ± SD including at least three replicates. Data was analyzed using t-test. *P < 0.05; **P < 0.01; ***P < 0.001.
To determine whether C. difficile acts in vivo through MVs, pregnant mice were treated with C. difficile MVs. We observed that compared with the control group, the number of fetuses in C. difficile MVs group decreased significantly (P < 0.001, Fig. 1D, E). In addition, placental weight, fetal weight, and fetal body length were significantly lower in C. difficile MVs group than in the control group (P < 0.001, Fig. 1F–I). Furthermore, the fetal-placental weight ratio in C. difficile MVs group was significantly lower than that in the control group, suggesting decreased placental utilization efficiency and inadequate placental function (P < 0.001, Fig. 1J). Placental structure was further analyzed by HE staining to evaluate the effect of C. difficile MVs on placental function. As shown in Fig. 1K, L, exogenous supplementation of C. difficile MVs resulted in increased infarct, interstitial collagen deposition, and dysfunction of spiral artery recast in mouse placenta. The labyrinth/junctional zone ratio of C. difficile MVs group was also higher than that of the control group. Overall, these data indicated that C. difficile MVs impaired placental structure and function, thereby inducing FGR in mice.
C. difficile MVs inhibit the motility of HTR-8/SVneo cells
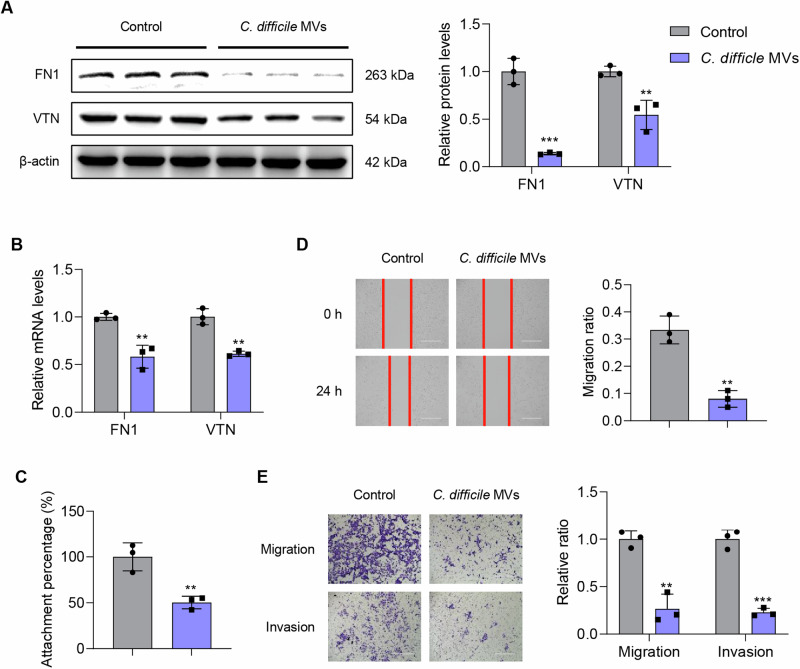
To further explain the role of C. difficile MVs in placental function, we performed a series of cell function and molecular biological experiments to investigate the effects of C. difficile MVs on trophoblast motility, including cell adhesion, migration and invasion functions. The decrease of trophoblast motility leads to spiral artery recasting disorder, placental insufficiency, and promotes FGR23–25. Fibronectin1 (FN1) and vitronectin (VTN), both extracellular matrix proteins, are major controllers of the extracellular environment and directly affect cell motility26,27. In HTR-8/SVneo cells, the protein expression levels of FN1 and VTN in C. difficile MVs group were significantly lower than those of control group (P < 0.01, Fig. 2A). Meanwhile, we observed the same trend at the mRNA level (P < 0.01, Fig. 2B). The results of the adhesion assay showed that C. difficile MVs reduce the adhesion ability of HTR-8/SVneo cells (P < 0.01, Fig. 2C). The results of the wound healing assay showed that C. difficile MVs stimulation caused a decrease in the migration ability of HTR-8/SVneo cells compared with the control cells (P < 0.01, Fig. 2D). The results of the Transwell assay also showed that C. difficile MVs treatment inhibited the migration and invasion of HTR-8/SVneo cells (P < 0.01, Fig. 2E). Meanwhile, the treatment of C. difficile MVs had no significant effect on apoptosis of HTR-8/SVneo cells (Supplementary Fig. 2). These findings indicated that C. difficile MVs inhibited the adhesion, migration and invasion of HTR-8/SVneo cells.
Fig. 2. The effect of C. difficile MVs on the adhesion, migration and invasion of HTR-8/SVneo cells.
A The protein expression levels of β-actin, FN1 and VTN were detected by western blotting. B The mRNA expression levels of FN1 and VTN was detected by qRT-PCR. C Adhesion ability of the HTR-8/SVneo cells treated with C. difficile MVs 5 μg/ml was detected by adhesion experiment. D Migration ability of the HTR-8/SVneo cells treated with C. difficile MVs 5 μg/ml for 24 h was detected by wound-healing assay. E Transwell assay was used to analyze the migration and invasion capabilities of HTR-8/SVneo cells treated with C. difficile MVs 5 μg/ml for 24 h. Data are represented as mean ± SD including at least three replicates. Data was analyzed using t-test. *P < 0.05; **P < 0.01; ***P < 0.001.
C. difficile MVs alter transcriptome and motility of HTR-8/SVneo cells
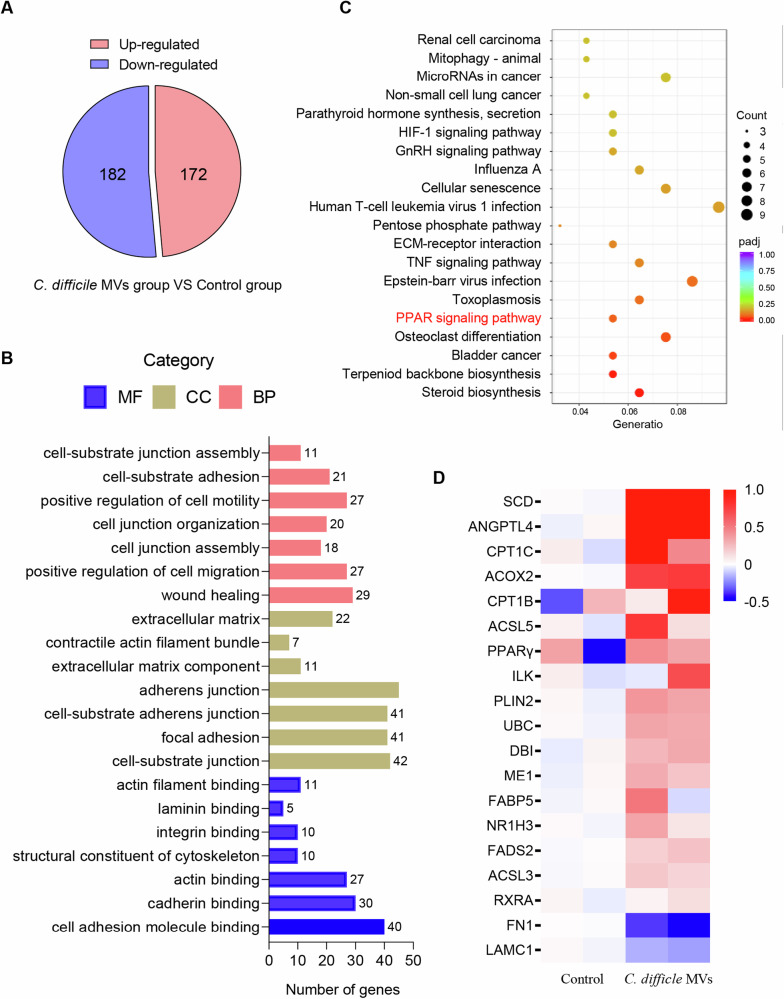
RNA sequencing was performed to detect differentially expressed genes between control and C. difficile MV-treated cells to determine the underlying mechanism by which C. difficile MVs modulate the motility of HTR-8/SVneo cells. C. difficile MVs treatment resulted in 172 up-regulated genes and 182 down-regulated genes in HTR-8/SVneo cells (Fig. 3A). GO analysis showed that the functions of these C. difficile MV-affected genes were significantly related to cell motility, migration, junction and adhesion, such as positive regulation of cell motility, wound healing, adherens junction, cell adhesion molecule binding, positive regulation of cell migration and cell-substrate junction (Fig. 3B). This suggests that C. difficile MVs can affect the adhesion migration and invasion abilities of HTR-8/SVneo cells. KEGG pathway analysis revealed that C. difficile MVs might significantly affect the PPAR signaling pathway (Fig. 3C). We showed the differential expression of genes involved in the PPAR signaling pathway, including PPARγ, RXRα, ANGPDL4, and FN1, as a heatmap (Fig. 3D). Collectively, our results indicated that C. difficile MVs may inhibit trophoblast motility through the PPAR signaling pathway.
Fig. 3. The transcriptome of HTR-8/SVneo cells was altered by C. difficile MVs treatment.
A C. difficile MVs up-regulated 172 genes and down-regulated 182 genes in HTR-8/SVneo cells. B GO analysis showed that the adhesion and junction function of HTR-8/SVneo cells were affected by difficile MVs treatment. C KEGG analysis revealed significant enrichment of PPAR signaling pathway in C. difficile MV-treated HTR-8/SVneo cells. D Heat map of differentially expressed genes associated with PPAR signaling pathway in the C. difficile MVs and control groups.
C. difficile MVs activate PPAR signaling pathway in HTR-8/SVneo cells
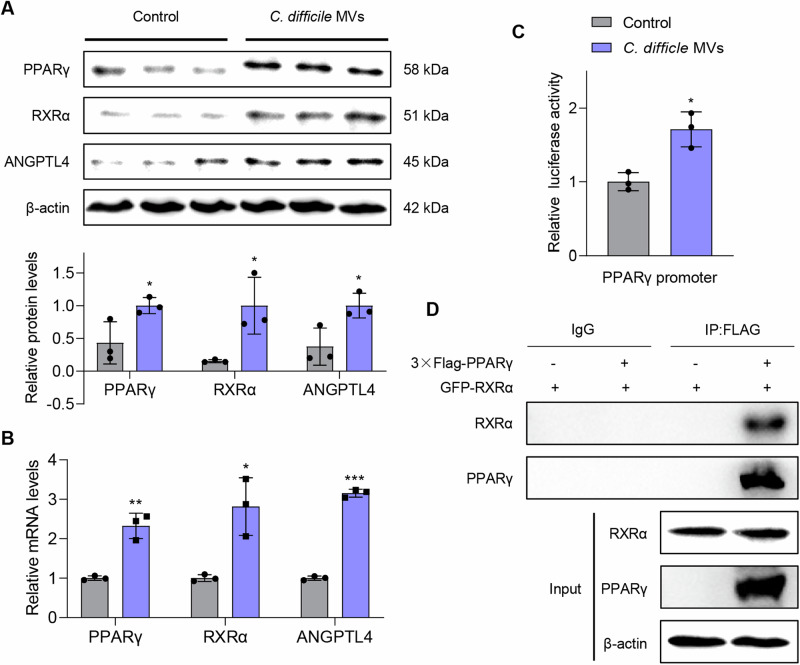
Further verification by Westen blot and qRT-PCR showed that C. difficile MVs promoted the protein and mRNA expression of PPARγ, ANGPTL4 and RXRα to activate PPAR signaling pathway (P < 0.05, Fig. 4A, B). To clarify the mechanism of C. difficile MVs activating PPAR pathway, we conducted double luciferase reporter gene experiments. The results showed that C. difficile MVs enhanced the transcriptional activity of PPARγ promoter (P < 0.05, Fig. 4C). Previous studies have shown that PPARγ forms dimers with RXRα to co-regulate the expression of ANGPTL4. In this study, we also found that PPARγ binds to RXRα protein (Fig. 4D). According to these results, C. difficile MVs promoted the transcriptional activity of PPARγ promoter and activated the PPAR pathway.
Fig. 4. C. difficile MVs enhanced transcriptional activity of PPARγ and activated PPAR pathway.
A C. difficile MVs treatment up-regulated the protein expression levels of PPARγ, ANGPTL4 and RXRΑ. B qRT-PCR was used to measure the mRNA expression levels of PPARγ, ANGPTL4 and RXRΑ in the two groups. C The transcriptional activity of PPARγ promoter was measured by dual luciferase reporter assay in HTR-8/SVneo cells treated with C. difficile MVs 5 μg/ml for 24 h. D The binding of PPARγ to RXRΑ protein was verified by Co-IP experiment. Data are represented as mean ± SD including at least three replicates. Data was analyzed using t-test. *P < 0.05; **P < 0.01; ***P < 0.001.
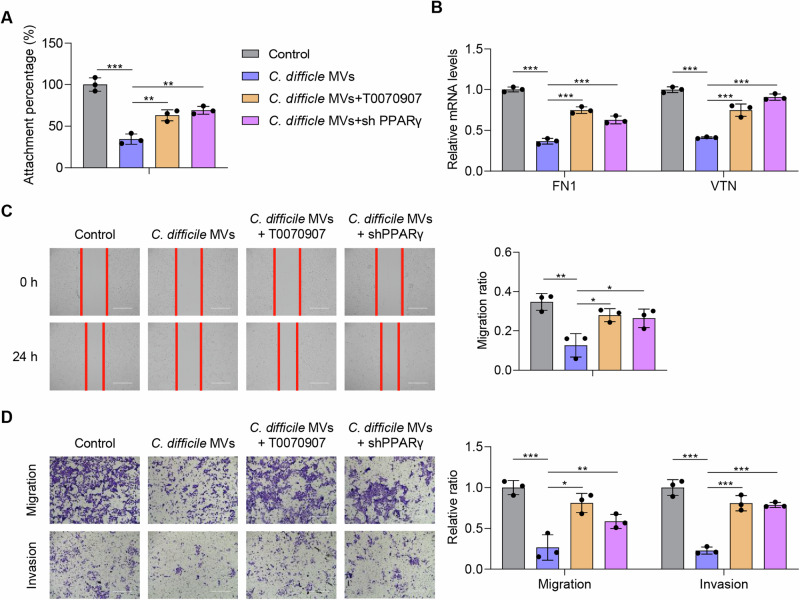
PPARγ is crucial in inhibiting motility of HTR-8/SVneo cells by C. difficile MVs
To clarify the role of PPARγ in C. difficile MVs-mediated inhibition of cell motility, we used T0070907 and PPARγ shRNA for rescue experiments. T0070907 is a potent and selective PPARγ inhibitor. The results of the adhesion assay revealed that the use of T0070907 or PPARγ shRNA effectively enhanced the adhesion ability of HTR-8/SVneo cells, compared with C. difficile MVs group (P < 0.01, Fig. 5A). Both T0070907 or silencing of PPARγ promoted the mRNA expression of FN1 and VTN inhibited by C. difficile MVs in HTR-8/SVneo cells (P < 0.001, Fig. 5B). In the wound healing assay, T0070907 or PPARγ shRNA treatment resulted in an increase in the migration ratio of HTR-8/SVneo cells compared with the C. difficile MVs group (P < 0.05, Fig. 5C). The results of the Transwell assay also showed that T0070907 or PPARγ shRNA treatment increased the migration and invasion rates of HTR-8/SVneo cells (P < 0.05, Fig. 5D). These results strongly indicated that C. difficile MVs inhibited trophoblast adhesion, migration and invasion through the PPAR pathway, and that T0070907 or PPARγ shRNA effectively mitigated this effect in HTR-8/SVneo cells.
Fig. 5. PPARγ is essential for C. difficile MVs to inhibit the motility of HTR-8/SVneo cells.
A The inhibitory effect of C. difficile MVs on HTR-8/SVneo cell adhesion was mitigated by T0070907 and PPARγ shRNA. B Down-regulation of FN and VTN by C. difficile MVs was eliminated by T0070907 and PPARγ shRNA in HTR-8/SVneo cells. C, D The inhibition of C. difficile MVs on the migration and invasion of HTR-8/SVneo cells was alleviated by T0070907 and PPARγ shRNA. Data are represented as mean ± SD including at least three replicates. Data was analyzed using ANOVA with Tukey’s post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001.
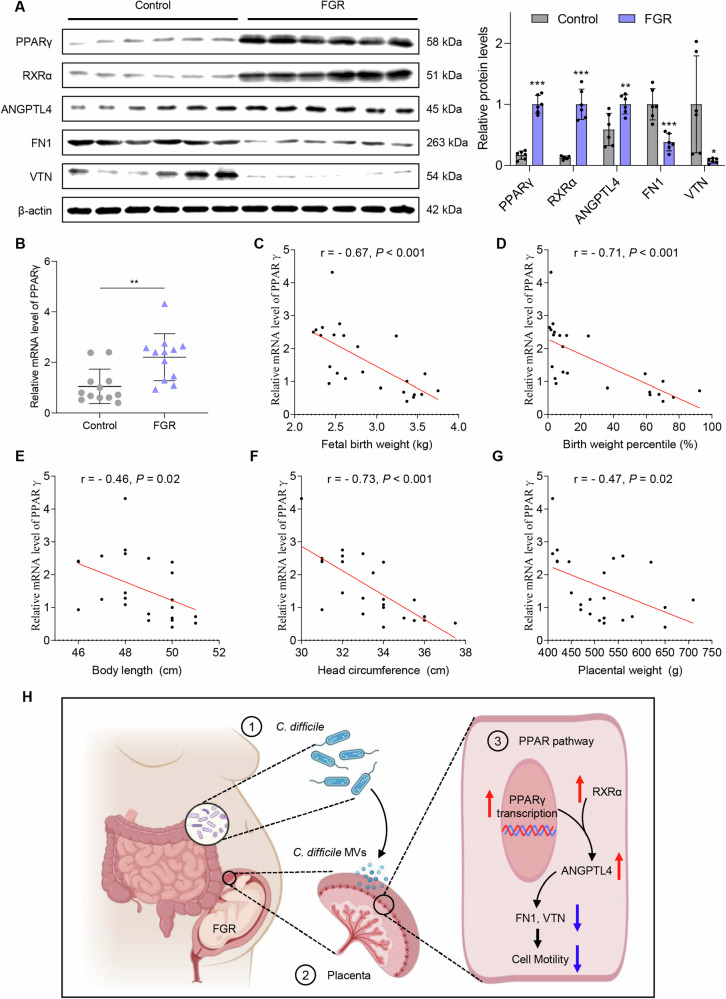
PPARγ is upregulated in the placenta of FGR patients
Compared with healthy pregnant women, the protein expression levels of PPARγ, RXRα and ANGPTL4 were considerably increased in the placenta of FGR pregnant women, while the expression levels of VTN and FN1 was substantially decreased (P < 0.05, Fig. 6A). The relative mRNA expression level of PPARγ was also significantly increased in the placenta of pregnant women with FGR, and was negatively correlated with fetal birth weight, weight percentile, body length, head circumference and placental weight (P < 0.05, Fig. 6B–G). These data demonstrated that PPARγ may exert significant contribution in the development of FGR.
Fig. 6. Expression levels of PPARγ, ANGPTL4, RXRα, VTN and FN1 in the patient’s placenta.
A Protein expression levels of PPARγ, ANGPTL4 and RXRα were elevated in the FGR placenta, and the levels of VTN and FN1 were decreased. B The mRNA expression level of PPARγ was significantly increased in the placenta of FGR. Data are represented as mean ± SD including at least three replicates. Data was analyzed using t-test. *P < 0.05; **P < 0.01; ***P < 0.001. C–G Correlations between the relative mRNA expression level of PPARγ and patient clinical parameters. Spearman’s correlation test was used for statistical analysis. H The schematic diagram of C. difficile and its MVs involvement in FGR.
Discussion
FGR is a common complication of pregnancy, that seriously affects fetal health during the perinatal period and adulthood28. Nevertheless, because its etiology is not fully understood, there is a lack of effective prediction, prevention, and treatment strategies3. Increasing data have suggested that gut microbes are involved in the regulation of maternal and infant health10. In this study, we aimed to explore the mechanism of FGR from the perspective of the gut microbiota. The results showed that C. difficile altered the structure of gut microbiota and was involved in the development of FGR. Moreover, C. difficile MVs inhibited trophoblast motility by activating PPAR pathway, consequently inducing FGR. Finally, elevated levels of PPARγ were also found in the placenta of pregnant women with FGR and were associated with decreased fetal birth weight.
Currently, more and more studies have shown that the imbalance of gut microbiota is closely related to FGR29,30. A study reported that the gut microbiota is disturbed in pregnant women with FGR, where the abundance of clostridium is elevated13. C. difficile is an important part of the Clostridium and is associated with changes in fetal weight17,31. However, whether C. difficile is involved in the development of FGR is unclear. Our results showed for the first time that C. difficile altered the structure of the gut microbiota and participated in the development of FGR.
Bacterial membrane vesicles are an important medium for their interaction with the host32,33. Recent studies have shown the presence of bacterial membrane vesicles in the human placenta, which may result in a low-grade localized inflammation to prime an antigen response in the placenta34. Meanwhile, studies have shown that bacterial membrane vesicles are involved in preeclampsia and FGR by affecting placental function, and are markers of gestational diseases35–38. C. difficile also produces membrane vesicles39. A study has shown that C. difficile MVs induce an inflammatory response in intestinal epithelial cells21. However, the role of C. difficile MVs in FGR remains unclear. Our study showed for the first time that C. difficile MVs induced FGR by inhibiting the motility of trophoblast and damaging placental structure and function.
In order to further explore the mechanism of action of C. difficile MVs in trophoblast cells, we analyzed the target genes regulated by C. difficile MVs via transcriptome. We found for the first time that C. difficile MV enhanced the transcriptional activity of PPARγ and activated the PPAR signaling pathway, thereby inhibiting the trophoblast motility. At present, the effect of PPARγ on trophoblast migration and invasion is still controversial21. A study has shown that IL-17 activates the PPARγ/RXRα signaling pathway, which promotes proliferation, migration and invasion of trophoblast cells40. However, it has also been found that overexpression of PPARγ not only inhibits migration, but also restrains invasion in trophoblast cells41. In addition, PPARγ also inhibits the migration and invasion of other cells, such as hepatocellular carcinoma and pulmonary artery smooth muscle cells42,43.
In summary, this study explored the mechanism of C. difficile and its MVs involved in FGR, providing new insights into the etiology of FGR. However, this study has some limitations. First, although it has been confirmed that C. difficile MVs enhance the transcriptional activity of PPARγ, the molecular mechanism of C. difficile MVs and its components involved in transcriptional regulation of HTR-8/SVneo cells remains to be further explored. Second, the study did not explore the effects of C. difficile MVs on placental transcriptome of mice and the role of silencing PPARγ in vivo. In addition, sample collections are challenging in clinical setting, resulting in a small sample size of human. Finally, the role of C. difficile in FGR by altering the gut microbiota remains to be further explored.
Methods
Patients and specimens
A total of 12 healthy pregnant women and 12 pregnant women with FGR were included in the study (Table 1). All patients delivered at full term. Placenta samples and fetal clinical indicators were collected after delivery. All placental tissues were obtained with informed consent, and this study was approved by the Research Ethics Committee of Nanfang Hospital, Southern Medical University.
Table 1.
Clinical parameters of patients
| Indicators | Control group (N = 12) | FGR group (N = 12) | P | |
|---|---|---|---|---|
| Pregnant woman | Age (year) | 30.7 ± 2.9 | 30.5 ± 2.9 | 0.907 |
| BMI (kg/m²) | 26.5 ± 3.3 | 25.7 ± 3.5 | 0.540 | |
| Gestation (week) | 38.8 ± 1 | 38.9 ± 1.1 | 0.869 | |
| Fetus | Birth weight (Kg) | 3.3 ± 0.4 | 2.5 ± 0.2 | 0.000*** |
| Weight percentile (%) | 53.8 ± 26.3 | 4.4 ± 2.8 | 0.000*** | |
| Body length (cm) | 49.4 ± 1.5 | 47.7 ± 1.2 | 0.005** | |
| Head circumference (cm) | 34.6 ± 1.7 | 32 ± 1.2 | 0.000*** | |
| Placenta | Largest diameter (cm) | 21.6 ± 2.1 | 19.5 ± 1.7 | 0.013* |
| Smallest diameter (cm) | 20.8 ± 2.7 | 18.6 ± 1.4 | 0.018* | |
| Thickness (cm) | 2.9 ± 0.3 | 2.4 ± 0.3 | 0.002** | |
| Weight (g) | 560 ± 84.1 | 469.6 ± 53.3 | 0.005** |
Data are represented as mean ± SD and analyzed using t-test.
*P < 0.05; **P < 0.01; ***P < 0.001.
Bacterial culture
C. difficile strain BNCC 340237 from the BeNa Culture Collection (Guangzhou, CHN) was used in this study. C. difficile BNCC 340237 strain did not produce tcdA or tcdB. C. difficile was cultured in thiolate liquid medium at 37 °C in an anaerobic environment with 80% N2, 10% H2, and 10% CO2 for 24 h. Thiolate liquid medium: casitone 15.0 g, yeast extract 5.0 g, glucose 5.0 g, sodium thiolate 0.5 g, L-cystine 0.5 g, sodium chloride 2.5 g, resazerin 0.001 g, AGAR 0.75 g, distilled water 1.0 L, pH 7.1 ± 0.2.
C. difficile MVs isolation
C. difficile MVs were isolated from the bacterial culture medium supernatant. Briefly, bacterial cells were removed by centrifugation at 6000 × g for 20 min at 4 °C. The bacterial supernatants were filtered through 0.22 μm sterile filters (Millipore, MA, USA). The C. difficile MVs were subsequently collected by twice ultracentrifugation in an SW 32 Ti rotor (Optima XPN-100; Beckman Coulter, USA) at 150,000 × g and 4 °C for 6 h. The final precipitated C. difficile MVs were resuspended in a small amount of PBS, and total protein was quantified using the Pierce BCA Protein Assay Kit (ThermoFisher Scientific, Waltham, MA, USA), after which C. difficile MVs were stored at −80 °C20.
Transmission electron microscope (TEM)
The morphology of the C. difficile MVs was observed using a transmission electron microscope (Hitachi H-7650, Japan). Briefly, the collected C. difficile MVs were mixed with 4% paraformaldehyde and then adsorbed onto carbon-coated copper Electron Microscopy (EM) grids (Agar Scientific) for 20 min. Subsequently, the samples were sequentially washed with PBS, fixed with 1% glutaraldehyde for 2 min, washed with deionized water, and contrasted with 1.5% uranyl acetate for 4 min. Finally, the grids were air-dried and placed in a dark, dust-free environment until observation. Images were acquired at 20,000× magnification with a 100 nm scale bar.
Nanoparticle tracking analysis (NTA)
The size distribution of the C. difficile MVs was analyzed using a NanoSight NS300 instrument (Malvern, UK). The collected C. difficile MVs were diluted to a concentration between 108 and 109/mL with PBS and loaded into a 1 mL syringe. After machine calibration with polystyrene nanospheres, a syringe was used to inject the sample into the sample cell at a rate of 40 μL/s. Finally, the diameters of the observed C. difficile MVs were analyzed using NanoSight Software NTA3.2 Dev Build 3.2.1644.
In vivo imaging
First, Cy7 mono NHS ester (Cyanine 7 monosuccinimidyl ester, APExBIO) was used to mark C. difficile MVs. APExBIO was added to the C. difficile MVs-PBS solution at a concentration of 5 μM and incubated at 37 °C for 30 min. Then, PBS was added to wash the mixture. Subsequently, the Cy7-labeled C. difficile MVs were collected, and excess dye was removed by ultracentrifugation, as described above. Cy7-labeled C. difficile MVs (2 × 109) were given to mice by intragastric administration. After 12 h, Cy7 fluorescence in the mice was recorded using a Bruker in vivo fluorescence imaging system (Bruker FX Pro)45.
Animal experiments
All animal experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and the experimental protocols were approved by the local Animal Care and Use Committee of Southern Medical University.
C57BL/6J mice (6 weeks old) were purchased from the Experimental Animal Centre of Southern Medical University. Mice were maintained under pathogen-free conditions and on a 12 h light/12 h dark cycle in a temperature- and humidity-controlled room. Female mice were administered C. difficile 200 μL (1 × 108 CFU/mL) or an equal volume of PBS by intragastric administration every 2 days for 2 weeks. Thereafter, the female mice were housed overnight in a 2:1 ratio with the male mice. The date of gestation was determined by observing the copulation plug and marked as E0.5. The PBS treated mice were divided into DEX group and control group. Mice treated with C. difficile were divided into C. difficile group and C. difficile + DEX group. There were 6 mice in each group. DEX was used to establish the mouse FGR model. Pregnant mice in DEX and C. difficile + DEX groups were intraperitoneally injected with DEX (1 mg/kg/day, Selleck, S1322) for 4 days starting at E14.5. Pregnant mice in the C. difficile and C. difficile + DEX groups continued to be given C. difficile 200 μL (1 × 108 CFU/mL) intragastric administration every 2 days until days E17.5. Mouse feces was collected on days 1 and 14 after intragastric administration of C. difficile for 16S rRNA gene sequencing analysis. Fetal body weight and length were measured on postnatal day 1.
Another batch of mice was administered exogenously with C. difficile MVs (50 μg in 200 μL PBS) or the same volume of PBS by intragastric administration every 2 days until E17.5. At day E17.5, pregnant mice were anesthetized with pentobarbital (1.5% [w/v]) (60 mg/kg). The mouse uterus was then removed. The fetus and placenta were dissected and weighed. Mouse tissues were either snap frozen and stored at −80 °C or immediately fixed with 4% paraformaldehyde and embedded in paraffin according to standard procedures.
16S rRNA gene sequencing
After genomic DNA extraction, the conserved region of rDNA was amplified using specific primers with barcodes. PCR amplification products were recovered and quantified using a QuantiFluorTM fluorometer. The purified amplification products were mixed in equal amounts, connected to a sequencing adapter, and the sequencing library was constructed and sequenced on an Illumina PE250 platform. After raw reads were obtained by sequencing, the low-quality reads were filtered and assembled. The double-end reads were spliced into tags, which were filtered, and the resulting data were called clean tags. Next, clustering was performed based on clean tag to remove the chimera tag detected during the clustering process, and the final data obtained was an effective tag. After OTU were obtained, OTU abundance statistics were performed based on the effective tag. Species annotation, α diversity analysis, β diversity analysis, and community function prediction were performed according to the analysis process. If there was an effective grouping, the differences between the groups were compared and statistical tests were performed.
Cell culture
A human trophoblast cell line (HTR‐8/SVneo cells) was purchased from the American Type Culture Collection (Manassas, VA, USA) and maintained in RPMI 1640 medium (Biological Industries, Israel) supplemented with 10% (v/v) fetal bovine serum (Data Inventory Biotechnology, Hong Kong, China). HTR-8/SVneo cells were derived from chorionic villi explants of human placenta. Cells were identified by the STR (short tandem repeat) authentication and were free of mycoplasma infection. HTR‐8/SVneo cells were grown in a humidified incubator at 37 °C with 5% CO2. HTR‐8/SVneo cells were treated with C. difficile MVs at 5 μg/ml for 24 h, or T0070907 1 μM for 24 h46.
Plasmids and established stable cells
The CDS sequences of the human PPARγ and RXRα genes were subcloned into pSIN-3 × FLAG and pSIN-GFP lentiviral vector, respectively. The PPARγ shRNA sequence was inserted into the PLKO vector. The shRNA targeting sequences were listed in Table 2. All lentiviruses were produced on HEK293T cells. HTR‐8/SVneo cells were infected with lentivirus in the presence of polybrene, and selected with puromycin to establish stably transfected cells47.
Table 2.
shRNAs used in PLKO vector
| Name | shRNA sequence |
|---|---|
| PPARγ shRNA #1 | (Forward) CCGGATGGAGTCCACGAGATCATTTCTCGAGAAATGATCTCGTGGACTCCATTTTTTG |
| (Reverse) AATTCAAAAAATGGAGTCCACGAGATCATTTCTCGAGAAATGATCTCGTGGACTCCAT | |
| PPARγ shRNA #2 | (Forward) CCGGGACAACAGACAAATCACCATTCTCGAGAATGGTGATTTGTCTGTTGTC TTTTTG |
| (Reverse) AATTCAAAAAGACAACAGACAAATCACCATTCTCGAGAATGGTGATTTGTCTGTTGTC | |
| PPARγ shRNA #3 | (Forward) CCGGCTGGCCTCCTTGATGAATAAACTCGAGTTTATTCATCAAGGAGGCCAGTTTTTG |
| (Reverse) AATTCAAAAACTGGCCTCCTTGATGAATAAACTCGAGTTTATTCATCAAGGAGGCCAG |
Hematoxylin and eosin (HE) staining
Placental tissues were fixed in 4% paraformaldehyde, embedded in paraffin and sectioned. Deparaffinization was performed using xylene, ethanol, and water. The sections were successively placed in hematoxylin and eosin staining solutions. Finally, the sections were dehydrated and sealed.
Real-time quantitative PCR
Total RNA was extracted from placental tissues or HTR‐8/SVneo cells, followed by reverse transcription to synthesize cDNA. The resulting cDNA was subjected to qRT-PCR. Quantitative Real-Time PCR was performed using iQ SYBR Green Supermix and an iCycler Real-Time PCR Detection System (Bio-Rad). The sequences of the primers were shown in Table 3, which were synthesized by Sangon Biotech (Shanghai, China). The relative expression of target genes was normalized to the transcription level of 18S rRNA and calculated using the 2 − ΔΔCt method47.
Table 3.
Primers used in qRT-PCR
| Gene name | Primer sequence |
|---|---|
| PPARγ | (Forward) GGGATCAGCTCCGTGGATCT |
| (Reverse) TGCACTTTGGTACTCTTGAAGTT | |
| FN1 | (Forward) CGGTGGCTGTCAGTCAAAG |
| (Reverse) AAACCTCGGCTTCCTCCATAA | |
| VTN | (Forward) GCCTTCACCGACCTCAAGAAC |
| (Reverse) CCCCTGACAGTTGATGCGG | |
| RXRα | (Forward) ATGGACACCAAACATTTCCTGC |
| (Reverse) GGGAGCTGATGACCGAGAAAG | |
| ANGPTL4 | (Forward) GGCTCAGTGGACTTCAACCG |
| (Reverse) CCGTGATGCTATGCACCTTCT | |
| 18s | (Forward) CGGCGACGACCCATTCGAAC |
| (Reverse) GAATCGAACCCTGATTCCCCGTC |
Western blot analysis
Cultured HTR-8/SVneo cells or placental tissue were collected and lysed with RIPA lysis buffer to generate proteins, as described previously. Quantification was performed using a Bradford assay kit. Equal amounts of protein samples were subjected to electrophoresis and transferred to polyvinylidene difluoride membranes. Membranes were incubated with the following antibodies: FN1 (1:2000, 15613-1-AP, Proteintech), VTN (1:10000, 66398-1-Ig, Proteintech), PPARγ (1:1000, 60127-1-Ig, Proteintech), and β-actin (1:10000, 66009-1-Ig, Proteintech). After incubation with HRP-conjugated anti-rabbit or anti-mouse secondary antibodies (1:10,000, Proteintech), the signal was detected using an enhanced chemiluminescence (ECL) kit.
Co-immunoprecipitation (Co-IP)
Cells were lysed using a low-salt IP buffer containing 1 × cocktail (05056489001, Roche) solution. Supernatants were obtained by high-speed centrifugation. The supernatant was then added to 20 µL protein A/G-conjugated beads to wash the sample, quantify the protein, obtain 3 µg protein, and dilute to 1000 µL. The samples from each group were equally divided into IgG and input groups. The corresponding antibody (IgG 1 µg or FLAG 1 µg) was added to the supernatant and incubated overnight. The supernatants were then added to protein A/G-conjugated beads and incubated for 2 h. Finally, the beads were collected, washed, and analyzed by western blotting48.
Attachment assay
The 96-well plate was treated by 200 μl/well of 10 μg/ml fibronectin (Sigma, #F0895) at 4 °C overnight. 200 μl of heat-denatured 1% bovine serum albumin (BSA) was added to each well and incubated at 37 °C for 1 h. Wash the 96-well plate 3 times with PBS. 10,000 cells were plated in each well and incubated for 4 h at 37 °C. The non-adherent cells were removed by washing with PBS. CCK8 (GLPBIO, GK10001) measures the number of cells49.
Wound-healing assay
The cells were seeded in a 6-well plate and grown to 90% confluence. Wounds were created using 200 µl pipette tips to scratch a straight line. After that, cells were grown using serum-free medium treated with C. difficile MVs at 5 μg/ml for 24 h, or T0070907 at 1 μM for 24 h. Wound areas were imaged at 0 and 24 h under a microscope. Image J software (NIH, USA) was used to calculate the wound area50.
Transwell assay
The Transwell assay was performed using 12 well plates with 8 µm inserts (Corning, NY, USA). HTR-8/SVneo cells (5 × 105) in 200 µL of serum-free medium were seeded into the inserts and treated with C. difficile MVs at 5 μg/ml for 24 h, or T0070907 at 1 μM for 24 h. Medium (600 µL) with 10% FBS was added to the lower chamber. After being incubated at 37 °C, the migration cells were fixed with 4% formaldehyde, stained with 0.1% crystal violet, and then being counted under a microscope.
Flow cytometry for apoptosis analysis
HTR-8/SVneo cells (1 × 104 /mL) were seeded in 24-well plates. The cells are trypsinized and collected after replacing the fresh medium and treating with C. difficile MVs at 5 μg/ml for 24 h. Cell apoptosis assay was performed by FITC-Annexin V Apoptosis Detection Kit (Biolegend, San Diego, CA, USA) according to the manufacturer’s instructions. All cells were acquired by FACS Calibur fow cytometer (BD Pharmingen, San Diego, CA, USA), and the results were anglicized with Flowjo software and collecting 5000 events for each sample50.
RNA sequencing
Total RNA was isolated from HTR-8/SVneo cells and C. difficile MV-treated HTR-8/SVneo cells using TRIzol reagent and was sent to Novogene for sequencing. mRNA was enriched by removing rRNA using a conventional kit. Eukaryotic mRNA with polyA tails was enriched using magnetic beads with oligo(dT) and disrupted by buffer. The first strand of cDNA was synthesized in the M-MuLV reverse transcriptase system, using fragmented mRNA as a template and random oligonucleotides as primers. Subsequently, the RNA strand was degraded by RNaseH, and the second strand of cDNA was synthesized from the dNTPs using the DNA polymerase I system. The purified double-stranded cDNA was end repaired, A-tailed, and ligated to a sequencing adapter. With ~200 bp AMPure XP beads screening cDNA, PCR amplification and AMPure XP beads were used to purify the PCR products, finally obtaining the library. Based on the HISAT2 alignment results, we reconstructed the transcripts using Stringtie and calculated the expression of all genes in each sample using RSEM. Differential expression analysis was performed using DESeq2 software, with a P ≤ 0.05 and |Log2Ration| ≥ 0. The differentially expressed genes were mapped to each term in the GO database (http://www.geneontology.org/), and the number of differentially expressed genes in each term was calculated to obtain the number of differentially expressed genes with GO function. KEGG annotation was performed using R software’s “phyper” function for enrichment analysis of differentially expressed genes48.
Luciferase reporter gene experiment
HTR-8/SVneo cells were transfected with the negative control plasmid and PPARγ promoter-luciferase recombinant plasmid. HTR-8/SVneo cells were treated with C. difficile MVs at 5 μg/ml for 24 h. After 48 h, luciferase activity was detected using the Dual-Luciferase Reporter Assay System (11402ES60, Yeasen Biotechnology). Promoter activity was normalized to Renilla luciferase activity and expressed as fold-change relative to the control51.
Statistical analysis
Data are presented as the mean ± SD of at least three independent experiments. All statistical analyses were performed using SPSS 24.0 version and GraphPad Prism 8. Student’s t-test (two cohorts), Permanova or ANOVA with Tukey’s post hoc tests (>two cohorts) were used to determine differences among the groups. Correlation analyses were performed based on the Spearman’s rho statistic. In the Figures, p < 0.05 indicates statistical significance (ns, p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001).
Supplementary information
Acknowledgements
We thank Professor Li Liang at Nanfang Hospital, Southern Medical University for technical support. This study was funded by the National Natural Science Foundation of China (2022YFC2704500, 82071669 and 82371690) and Natural Science Foundation of Guangdong Province (2022A1515011730, 2023A1515012094, and 2023A1515011074). The funder played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript.
Author contributions
Z.Q.Z.: conceptualization, data curation, methodology, formal analysis, and writing-original draft. R.S.Z.: data curation, methodology, investigation, and validation. Q.L.Y., Y.H., Z.P.H., L.Y.P., Y.C.Z., X.W.Q., Y.C., Y.W.Z., Y.W., M.L.P., S.J.L.: investigation and methodology. C.H.J., C.S.: supervision, funding acquisition, project administration, review and editing. L.P.H.: conceptualization, review and editing, supervision, funding acquisition, project administration, resources. All authors read and approved the final manuscript.
Data availability
The data analyzed in this study are available within the article or from the corresponding author upon request. The raw sequencing datasets have been deposited into China National GeneBank Sequence Archive (CNSA) of China National GeneBank DataBase (CNGBdb) with the accession numbers CNP0005528 and CNP0005529.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhiqiang Zha, Chunhong Jia, Ruisi Zhou.
Contributor Information
Chao Sheng, Email: shengchao3@126.com.
Liping Huang, Email: lphuang2006@126.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-024-00630-5.
References
- 1.Fetal growth restriction: ACOG practice bulletin, number 227. Obstet. Gynaecol. 137, e16–e28 (2021). [DOI] [PubMed]
- 2.Pels, A. et al. Interventions affecting the nitric oxide pathway versus placebo or no therapy for fetal growth restriction in pregnancy. Cochrane Database Syst. Rev.7, CD014498 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gumina, D. L. & Su, E. J. Mechanistic insights into the development of severe fetal growth restriction. Clin. Sci.137, 679–695 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’Agostin, M. et al. Long-term implications of fetal growth restriction. World J. Clin. Cases11, 2855–2863 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giouleka, S. et al. Fetal growth restriction: a comprehensive review of major guidelines. Obstet. Gynecol. Surv.78, 690–708 (2023). [DOI] [PubMed] [Google Scholar]
- 6.Yang, J., Liu, Y. & Dong, M. Integrated bioinformatics analysis to screen hub gene signatures for fetal growth restriction. Genet. Res.2023, 3367406 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melamed, N. et al. FIGO (international Federation of Gynecology and obstetrics) initiative on fetal growth: best practice advice for screening, diagnosis, and management of fetal growth restriction. Int. J. Gynaecol. Obstet.152(Suppl 1), 3–57 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, X. et al. Gut dysbiosis induces the development of pre-eclampsia through bacterial translocation. Gut69, 513–522 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Farinella, R. et al. Maternal anthropometric variables and clinical factors shape neonatal microbiome. Sci. Rep.12, 2875 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian, M. et al. Maternal microbe-specific modulation of the offspring microbiome and development during pregnancy and lactation. Gut Microbes15, 2206505 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferretti, P. et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe24, 133–145.e135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stepan, H. et al. Clinical utility of sFlt-1 and PlGF in screening, prediction, diagnosis and monitoring of pre-eclampsia and fetal growth restriction. Ultrasound Obstet. Gynecol.61, 168–180 (2023). [DOI] [PubMed] [Google Scholar]
- 13.Tu, X. et al. Characteristics of the gut microbiota in pregnant women with fetal growth restriction. BMC Pregnancy Childbirth22, 297 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Schwartzenberg, R. J. et al. Caloric restriction disrupts the microbiota and colonization resistance. Nature595, 272–277 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pichler, K. et al. Impact and time course of clostridium difficile colonization in very low birth weight infants. Neonatology114, 149–154 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Vu, K. et al. From birth to overweight and atopic disease: multiple and common pathways of the infant gut microbiome. Gastroenterology160, 128–144.e110 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Ferraris, L. et al. Carriage and colonization of C. difficile in preterm neonates: a longitudinal prospective study. PloS ONE14, e0212568 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliva Chávez, A. S. et al. Tick extracellular vesicles enable arthropod feeding and promote distinct outcomes of bacterial infection. Nat. Commun.12, 3696 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, S. et al. Microbial collaborations and conflicts: unraveling interactions in the gut ecosystem. Gut Microbes16, 2296603 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, Y. et al. Extracellular vesicles derived from Akkermansia muciniphila promote placentation and mitigate preeclampsia in a mouse model. J. Extracell. Vesicles12, e12328 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholas, A. et al. Clostridium difficile-derived membrane vesicles induce the expression of pro-inflammatory cytokine genes and cytotoxicity in colonic epithelial cells in vitro. Microb. Pathog.107, 6–11 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Chen, W. et al. Fetal growth restriction impairs hippocampal neurogenesis and cognition via Tet1 in offspring. Cell Rep.37, 109912 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Colson, A. et al. Adaptations of the human placenta to hypoxia: opportunities for interventions in fetal growth restriction. Hum. Reprod. Update27, 531–569 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Hiden, U. et al. Membrane-type matrix metalloproteinase 1 regulates trophoblast functions and is reduced in fetal growth restriction. Am. J. Pathol.182, 1563–1571 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Abbas, Y. et al. Investigation of human trophoblast invasion in vitro. Hum. Reprod. Update26, 501–513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan, S. et al. FN1 mRNA 3’-UTR supersedes traditional fibronectin 1 in facilitating the invasion and metastasis of gastric cancer through the FN1 3’-UTR-let-7i-5p-THBS1 axis. Theranostics13, 5130–5150 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruzha, Y. et al. Role of vitronectin and its receptors in neuronal function and neurodegenerative diseases. Int. J. Mol. Sci.23, 12387 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang, Z. et al. Altered distribution of fatty acid exerting lipid metabolism and transport at the maternal-fetal interface in fetal growth restriction. Placenta139, 159–171 (2023). [DOI] [PubMed] [Google Scholar]
- 29.Yang, J. et al. Unfavourable intrauterine environment contributes to abnormal gut microbiome and metabolome in twins. Gut71, 2451–2462 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao, Z. et al. Alterations in the gut microbiome and metabolisms in pregnancies with fetal growth restriction. Microbiol. Spectr.11, e0007623 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheepers, L. E. et al. The intestinal microbiota composition and weight development in children: the KOALA Birth Cohort Study. Int. J. Obes.39, 16–25 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Doré, E. & Boilard, E. Bacterial extracellular vesicles and their interplay with the immune system. Pharmacol. Ther.247, 108443 (2023). [DOI] [PubMed] [Google Scholar]
- 33.Xiao, M., Li, G. & Yang, H. Microbe-host interactions: structure and functions of Gram-negative bacterial membrane vesicles. Front. Microbiol.14, 1225513 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menon, R. et al. Amplification of microbial DNA from bacterial extracellular vesicles from human placenta. Front. Microbiol.14, 1213234 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farrelly, R. et al. Extracellular vesicles as markers and mediators of pregnancy complications: gestational diabetes, pre-eclampsia, preterm birth and fetal growth restriction. J. Physiol.601, 4973–4988 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barnes, M. V. C., Pantazi, P. & Holder, B. Circulating extracellular vesicles in healthy and pathological pregnancies: A scoping review of methodology, rigour and results. J. Extracell. Vesicles12, e12377 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aplin, J. D. et al. Tracking placental development in health and disease. Nat. Rev. Endocrinol.16, 479–494 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Lara, B. et al. Porphyromonas gingivalis outer membrane vesicles shape trophoblast cell metabolism impairing functions associated to adverse pregnancy outcome. J. Cell. Physiol.238, 2679–2691 (2023). [DOI] [PubMed] [Google Scholar]
- 39.Caballano-Infantes, E. et al. Membrane vesicles of toxigenic clostridioides difficile affect the metabolism of liver HepG2 cells. Antioxidants12, 818 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, Z. et al. Interleukin-17 promotes proliferation, migration, and invasion of trophoblasts via regulating PPAR-γ/RXR-α/Wnt signaling. Bioengineered13, 1224–1234 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, J. & Bai, W. P. C1q/tumor necrosis factor related protein 6 (CTRP6) regulates the phenotypes of high glucose-induced gestational trophoblast cells via peroxisome proliferator-activated receptor gamma (PPARγ) signaling. Bioengineered13, 206–216 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuo, Q. et al. PPARγ coactivator-1α suppresses metastasis of hepatocellular carcinoma by inhibiting Warburg effect by PPARγ-dependent WNT/β-catenin/pyruvate dehydrogenase kinase isozyme 1 Axis. Hepatology73, 644–660 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Wang, M. et al. FGF21 attenuates pulmonary arterial hypertension via downregulation of miR-130, which targets PPARγ. J. Cell. Mol. Med.26, 1034–1049 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bachurski, D. et al. Extracellular vesicle measurements with nanoparticle tracking analysis—an accuracy and repeatability comparison between NanoSight NS300 and ZetaView. J. Extracell. Vesicles8, 1596016 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu, L. et al. Extracellular vesicles of Fusobacterium nucleatum compromise intestinal barrier through targeting RIPK1-mediated cell death pathway. Gut Microbes13, 1–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Armistead, B. et al. Induction of the PPARγ (peroxisome proliferator-activated receptor γ)-GCM1 (glial cell missing 1) syncytialization axis reduces sFLT1 (soluble fms-like tyrosine kinase 1) in the preeclamptic placenta. Hypertension78, 230–240 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zha, Z. et al. Neuron specific enolase promotes tumor metastasis by activating the Wnt/β-catenin pathway in small cell lung cancer. Transl. Oncol.14, 101039 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu, L. et al. Neuron-specific enolase promotes stem cell-like characteristics of small-cell lung cancer by downregulating NBL1 and activating the BMP2/Smad/ID1 pathway. Oncogenesis11, 21 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai, Y. C. et al. ANGPTL4 overexpression inhibits tumor cell adhesion and migration and predicts favorable prognosis of triple-negative breast cancer. BMC Cancer20, 878 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen, X. et al. Knockdown of ENTPD5 inhibits tumor metastasis and growth via regulating the GRP78/p-eIF-2α/CHOP pathway in serous ovarian cancer. J. Ovarian Res.15, 69 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang, S. S. et al. Fusobacterium nucleatum-derived succinic acid induces tumor resistance to immunotherapy in colorectal cancer. Cell Host Microbe31, 781–797.e789 (2023). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analyzed in this study are available within the article or from the corresponding author upon request. The raw sequencing datasets have been deposited into China National GeneBank Sequence Archive (CNSA) of China National GeneBank DataBase (CNGBdb) with the accession numbers CNP0005528 and CNP0005529.