Abstract
Hepatitis B surface antigen (HBsAg) is the major component of the envelope of hepatitis B virus (HBV). As a resident membrane protein in the endoplasmic reticulum, it plays a key role in the viral morphogenesis. Little is known about cellular proteins that interact with HBsAg and thereby contributing to HBV morphogenesis. Using the yeast split-ubiquitin system, a number of cellular membrane proteins have been isolated in this study. These include a resident protein of endoplasmic reticulum (thioredoxin-related transmembrane protein 2), an adaptor protein involved in clathrin-mediated endocytosis and HIV-mediated downregulation of CD4, and a co-receptor of coxsakie B virus. The significance of our findings is suggested by the identification of cellular membrane proteins interacting with other virus proteins. Further functional analysis of these HBsAg- interacting cellular membrane proteins should shed new insights on their role in HBV morphogenesis.
Keywords: HBsAg, Morphogenesis, Cellular Membrane Proteins, Split-ubiquitin Screening System
1. Introduction
Hepatitis B virus (HBV) is a small DNA virus consisting of a nucleocapsid which protects the 3.2 kb viral genome 1. The nucleocapsid is surrounded by an envelope in which the major protein component is the 226 amino acid hepatitis B surface antigen (HBsAg). In addition to its involvement as an envelope protein of the infectious HBV particles, HBsAg is capable of forming into lipoprotein subviral particles (without the nucleocapsid and HBV genome) that are secreted from infected cells. Inappropriate secretion of HBsAg subviral particles with notably its intracellular storage has been clinically implicated in the pathogenesis of chronic hepatitis B 2.
One important step in HBV replication cycle is the viral morphogenesis, particularly the interaction between the nucleocapsid and HBsAg anchored at the endoplasmic reticulum, leading to envelopment and secretion of mature HBV particles 3. Despite the recently reported investigation on the role of chaperones 4, little is known about general network of host cellular proteins that is needed for HBV morphogenesis. While conventional yeast two hybrid screening has been successful in isolating soluble cellular proteins interacting with a bait of interest 5, it can not be applied in the isolation of those interacting with transmembrane bait proteins such as HBsAg.
A yeast split-ubiquitin screening system for the characterization of membrane protein-protein interactions has been described 6-8. We report in this study the application of this system to identify cellular membrane proteins interacting with HBsAg. The significance of our finding in the perspective of better understanding HBV morphogenesis is discussed.
2. Materials and methods
Plasmids and reagents were contained in DUALmembrane Kit (DUALSystems Biotech, Switzerland).
2.1 Amplification and Cloning of HBsAg
The 678bp HBsAg gene was amplified by PCR using full length HBV DNA (adw2 serotype) as the template. The oligonucleotide primers were designed as follows:
5'-TTAGGCCTAAAAATGGAGAACATCACATCAGGA-3'
5'-AAAACAGAGACCCATATGTAAATTGGTACCAATT-3'
The amplified PCR product was cloned into the binding domain vector pTMBV4 in-frame with Cub (the yeast ubiquitin). The construct was then used to transform yeast S. cerevisiae DSY-1 strain by LiOAc method.
2.2 Construction of cDNA Library from HBV-transfected HepG2 Cells
2.2.1 Cell Culture and Transfection
Human hepatomacellular carcinoma cell line, HepG2, was grown in 12ml Dulbecco's Modified Eagle's Medium (DMEM) (Gibco) supplemented with 20% fetal calf serum and maintained in a saturated, humidified environment of 5% CO2 and 95% air at 37oC. The cell density when confluent was approximately 2 x 106 HepG2 cells per 100mm dish. The replicative HBV genome was constructed by cloning a linear form of viral genome into mammalian expression vector pcDNA3.1 9. The linear genome, containing the viral promoter at its 5' end and the region for termination of transcription at its 3' end, was then transfected into these cells as described 9. The ability of this genome in behaving as a replicative virus was assessed by the amount of HBsAg particles produced in the cell culture medium two days after its transfection into HepG2 cells, using ImX semi-quantitative measurement (Abbott Laboratories, USA).
Total RNA was extracted from these HBV-transfected HepG2 cells, and used as template for the synthesis of cDNA. The first strand cDNA was generated using random hexamer primer from BD Matchmaker™ Library Construction & Screening Kits User Manual (BD Biosciences, Clontech, USA). The second strand cDNA was synthesized using Long-Distance PCR (LD-PCR). The PCR mixture containing double stranded cDNA was purified using BD CHROMA SPINTM TE-400 Column.
2.2.2 Construction of cDNA Library in S. cerevisiae DSY-2 Strain
The cDNA library was prepared by transforming the yeast S. cerevisiae DSY-2 strain, in which the cDNA mixture was combined with pDL2-XN vector through homologous recombination. Yeast colony PCR was done for 22 randomly picked colonies to assess the size of cDNA insert. For each colony picked, half was streaked on SD/-Trp plate and the other half inoculated into a 20μl PCR mix.
Three ml of the freezing medium (SD/-Trp, 15% glycerol) was added to each plate and the yeast colonies were dislodged off the plates using a sterile loop. The suspension from each plate was then pooled together into a 1-litre sterile conical flask. The pooled suspension was given a mix by swirling before aliquoting into Falcon tubes proportionally for centrifugation at 700 x g for 5 minutes. The supernatant was discarded and the pellet was resuspended in half the original volume using Freezing medium. 1ml of cells were then aliquoted into ten 1.5ml microcentrifuge tubes, 10ml of cells were aliquoted into five 15ml tubes and the remaining cells aliquoted into 50ml Falcon tubes. The samples were then stored at -80oC.
2.3 Split-Ubiquitin Yeast Two Hybrid Screening
2.3.1 Mating of S. cerevisiae DSY-1 (Bait) and S. cerevisiae DSY-2 (Prey)
The 1ml aliquot (≥2x107 cells) frozen cDNA library was added to 5ml bait cells in a sterile 3-litre conical flask. The mixture was incubated at 30oC for 24 hours with gentle swirling at 30rpm. After 20 hours of mating, a drop of the mating culture was checked under a phase-contrast microscope at 400X magnification to check for the presence of 2 haploids. The culture was split into two 50ml Falcon tubes and centrifuged at 1000 x g for 10 minutes. The 3-litre flask used for mating was rinsed twice with 50ml of 0.5X YPDA/Kanamycin (50μg/ml) per rinsed. Each rinse (50ml) was used to resuspend one pellet. After resuspension, the tubes were centrifuged at 1000 x g for 10 minutes and the supernatant discarded. The pellet was resuspended in 5ml 0.5X YPDA/Kanamycin (50μg/ml) and combined into a total volume of 10ml.
To calculate the mating efficiency, 100μl of a 1: 10 000, 1: 1 000, 1: 100 and 1:10 dilutions of the mating mixture was spread onto SD/-Leu, SD/-Trp and SD/-Leu/-Trp agar plates. The remaining mating mixture was distributed evenly onto 5mM TDO-3AT plates with plating volume of 200μl. The plates were sealed and placed in a 30oC incubator for approximately 7 days.
2.3.2 Selection for S. cerevisiae Diploids Expressing Interacting Proteins
A total of fifteen colonies were selected for the first round of screening. Half of the colony was streaked on SD/-Trp agar plate and the other half inoculated into 20μl of PCR mix. PCR products were sequenced to determine the identity of the interacting proteins.
3. Results and Discussion
3.1 Principle of Split-Ubiquitin Screening System
Studies have shown that the use of yeast two hybrid system, which is a genetic assay for in vivo detection of interactions of proteins, enables identification of protein-protein interactions between a soluble bait and its interacting prey 5. However, such a screening system is not suitable for the analysis of interaction between membrane bound proteins.
The split-ubiquitin yeast two hybrid system has been recently developed for the characterization of membrane protein interactions to overcome this limitation 6-8. In this system, the ubiquitin is split into N-terminal ubiquitin (Nub) and C-terminal ubiquitin (Cub) but the Nub and Cub have high affinity towards each other and is able to reassemble back together spontaneously into ubiquitin protein. When Nub and Cub moieties bind together within a cell, this would activate the ubiquitin-specific protease (UBP) and hence, the transcriptional factor (TF) will be cleaved off. The cleaved TFs such as protein A-LexA-VP16 (PLV) will activate the expression of reporter genes (LacZ and HIS3) in the nucleus. The interaction between various membrane proteins will then be reflected by the activity of these reporter gene products, which can be measured similarly to the conventional yeast two hybrid system.
The bait protein, which is the HBsAg, is fused to a vector (pTMBV4) containing the Cub-reporter protein complex. The prey protein, which is the human malignant liver HepG2 cell transfected with HBV adw2 serotype, is fused to vector (pDL2-XN) which contains a mutated NubG domain. The fusion plasmids are then cloned into the yeast cell and expressed on the cell membrane. If the prey protein interacts with the bait protein, this would bind the NubG and Cub domains together to form the ubiquitin complex.
This complex will then be recognized by UBPs and the TF that is located at the C terminus of Cub will be cleaved off and result in downstream reactions, which is the activation of LacZ and HIS3 reporter genes 7.
3.2 Identification of Cellular Membrane Proteins Interacting with HBsAg
A number of cellular proteins have been isolated using the conventional yeast two hybrid screening 10, 11. However, these reported HBV interacting proteins have all been isolated from normal liver cells that had not been exposed to HBV, which may reflect physical but physiologically not meaningful interactions. There is therefore a need to comprehensively isolate and characterize, in an environment, cellular proteins interacting not only with soluble HBV proteins, but also with membrane bound HBsAg.
To this end, a linearised form of HBV genome has been constructed in the mammalian expression vector pcDNA3.1 9. The linear HBV genome contained the viral promoter at its 5' end and the region for termination of transcription at its 3' end 9. The ability of this genome in behaving as a replicative virus after its transfection in HepG2 cells was then measured by the amount of HBsAg particles secreted into cell culture medium two days after its transfection. Using the ImX semi-quantitative measurement, results in Table 1 suggested that HBsAg produced in the culture medium (rate of 65.8) was significantly higher than the negative control (normal culture medium with a rate of 4.3), but lower than the positive control (rate of 221.4). Our results suggested that a cellular environment exposed to HBV could be generated by transfecting a replicative HBV genome.
Table 1.
Semi-Quantitative Measurement of HBsAg Following HBV-Transfection
| Reaction Rate / Reactivity | |
|---|---|
| Replicative HBV Genome | 65.8 / Reactive |
| Positive Control | 221.4 / Reactive |
| Negative Control | 4.3 / Non-Reactive |
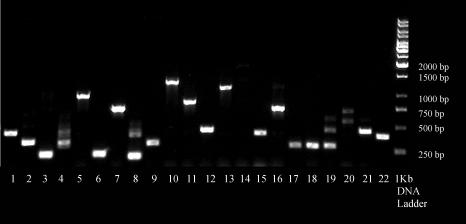
A cDNA library was therefore constructed from HBV-transfected HepG2 cells, using homologous recombination in yeast. PCR analysis on randomly selected colonies revealed a wide range of inserts from 250 bp (lanes, 2, 6 and 8, Fig. 1) to 1,500 bp (lane 10 and 13, Fig. 1). In addition, cDNA insert was detected in all selected colonies except colony 14. The wide range of cDNA inserts and absence of empty vector as shown in Fig. 1 indicated that the cDNA library constructed in this study was suitable for the screening of cellular proteins interacting with HBsAg.
Figure 1.
Determination of Size of Inserts in cDNA Library Constructed from HBV-transfected HepG2 Cells. The construction of this cDNA library from HBV-infected HepG2 cells was described in MATERIALS AND METHODS. Yeast colonies were selected randomly for PCR amplification. Size of individual inserts reflected by the size of the amplified PCR product was analysed on an agarose gel. Lane 1 to 22, PCR product from each of the 22 randomly selected colonies. Lane 1 kb DNA Ladder, DNA molecular weight marker
With HBsAg as bait, fifteen colonies were obtained from screening the above cDNA library using the yeast split-ubiquitin system. Sequence analysis revealed three interacting cellular membrane proteins as summarized in Table 2.
Table 2.
Summary of Cellular Membrane Proteins Interacting with HBsAg
| Clone | Cellular Membrane Protein | Function | Reference |
|---|---|---|---|
| 1 | Thioredoxin-related transmembrane protein 2 (trx2) | Stimulate cell growth and prevent apoptosis | 12, 13 |
| 3 | Adaptor-related protein complex 2 (AP-2) | Clathrin-mediated endocytosis and protein sorting | 14, 15 |
| 4 | Decay accelerating factor for complement (DAF) | Regulation of complement activation and virus co-receptor | 16, 17 |
While the role of the ER resident protein trx2 13 in HBV morphogenesis remains to be elucidated, AP-2 and DAF have been implicated in virus infections 15, 17. A major component of clathrin-associated adaptor protein complex that is involved in the clathrin-mediated endocytosis 14, AP-2 has also been involved in the downregulation of CD4 and MHC class I molecules by nef (a regulatory protein of HIV) thereby resulting in an increased HIV virulence 15. On the other hand, the decay-accelerating factor (DAF) with its well-established role in the inhibition of activation of complement cascade 16 has recently been shown to be a co-receptor for coxackie B virus and involved in the transcytosis of the virus 17. Further functional analysis of these HBsAg-interacting cellular membrane proteins should shed new insights on their role in HBV morphogenesis.
Acknowledgments
We thank Dr. Peter Lee Peng Foo for constructing the bait plasmid. This work was supported by grant 03/1/22/18/229 (WN Chen) from the Biomedical Research Council, Agency for Science, Technology and Research, Singapore. TL Tan is a recipient of a graduate research scholarship from Nanyang Technological University, Singapore. QC Toh and WQ Teo are from Ngee Ann Polytechnic, Singapore. CY Ho and S Parida are undergraduate students from School of Biological Sciences, Nanyang Technological University, Singapore.
References
- 1.Chen WN, Oon CJ. Human hepatitis B virus mutants: significance of molecular changes. FEBS Lett. 1999;453:237–242. doi: 10.1016/s0014-5793(99)00704-8. [DOI] [PubMed] [Google Scholar]
- 2.Foo NC, Ahn BY, Ma X, Hyun W, Yen TS. Cellular vacuolization and apoptosis induced by hepatitis B virus large surface protein. Hepatology. 2002;36:1400–1407. doi: 10.1053/jhep.2002.36819. [DOI] [PubMed] [Google Scholar]
- 3.Khan N, Guarnieri M, Ahn SH. et al. Modulation of hepatitis B virus secretion by naturally occurring mutations in the S gene. J Virol. 2004;78:3262–3270. doi: 10.1128/JVI.78.7.3262-3270.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prange R, Werr M, Loffler-Mary H. Chaperones involved in hepatitis B virus morphogenesis. Biol Cem. 1999;380:305–314. doi: 10.1515/BC.1999.042. [DOI] [PubMed] [Google Scholar]
- 5.Fields S, Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 6.Stagljar I, Korostensky C, Johnsson N, te Hessen S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc Natl Acad Sci USA. 1998;95:5187–5192. doi: 10.1073/pnas.95.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stagljar I, Fields S. Analysis of membrane protein interactions using yeast-based technologies. Trends Biochem Sci. 2002;27:559–563. doi: 10.1016/s0968-0004(02)02197-7. [DOI] [PubMed] [Google Scholar]
- 8.Thaminy S, Auerbach D, Arnoldo A, Stagljar I. Identification of novel ErbB3-interacting factors using the split-ubiquitin membrane yeast two-hybrid system. Genome Res. 2003;13:1744–1753. doi: 10.1101/gr.1276503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen WN, Oon CJ, Toh I. Altered antigenicities of hepatitis B virus surface antigen carrying mutations outside the common 'a' determinant. Am J Gastroenterol. 2000;95:1098–1099. doi: 10.1111/j.1572-0241.2000.01958.x. [DOI] [PubMed] [Google Scholar]
- 10.Huang CJ, Chen YJ, Ting LP. Hepatitis B virus core protein interacts with the C-terminal region of actin-binding protein. J Biomed Sci. 2000;7:160–168. doi: 10.1007/BF02256623. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, Torii N, Furusaka A, Malayaman N, Hu Z, Liang TJ. Structural and functional characterization of interaction between hepatitis B virus X protein and the proteasome complex. J Biol Chem. 2000;275:15157–15165. doi: 10.1074/jbc.M910378199. [DOI] [PubMed] [Google Scholar]
- 12.Powis G, Mustacich D, Coon A. The role of the redox protein thioredoxin in cell growth and cancer. Free Rad Biol Med. 2000;29:312–322. doi: 10.1016/s0891-5849(00)00313-0. [DOI] [PubMed] [Google Scholar]
- 13.Meng X, Zhang C, Chen J. et al. Cloning and identification of a novel cDNA coding thioredoxin-related transmembrane protein 2. Biochem Genet. 2003;41:99–106. doi: 10.1023/a:1022073917044. [DOI] [PubMed] [Google Scholar]
- 14.Motley A, Bright NA, Seaman MN, Robinson MS. Clathrin-mediated endocytosis in AP2-depleted cells. J Cell Biol. 2003;162:909–918. doi: 10.1083/jcb.200305145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piguet V, Trono D. The Nef protein of primate lentiviruses. Rev Med Virol. 1999;9:111–120. doi: 10.1002/(sici)1099-1654(199904/06)9:2<111::aid-rmv245>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 16.Harris CL, Rushmere NK, Morgan BP. Molecular and functional analysis of mouse decay accelerating factor (CD55) Biochem J. 1999;341:821–829. [PMC free article] [PubMed] [Google Scholar]
- 17.Selinka HC, Wolde A, Sauter M, Kandolf R, Klingel K. Virus-receptor interactions of coxsackie B viruses and their putative influence on cardiotropism. Med Microbiol Immunol. 2004;193:127–131. doi: 10.1007/s00430-003-0193-y. [DOI] [PubMed] [Google Scholar]