Abstract
Mutations in mammalian and Drosophila Hel308 and PolQ paralogues cause genome instability but their helicase functions are mysterious. By in vivo and in vitro analysis, we show that Hel308 from archaea (Hel308a) may act at stalled replication forks. Introducing hel308a into Escherichia coli dnaE strains that conditionally accumulate stalled forks caused synthetic lethality, an effect indistinguishable from E.coli RecQ. Further analysis in vivo indicated that the effect of hel308a is exerted independently of homologous recombination. The minimal biochemical properties of Hel308a protein were the same as human Hel308. We describe how helicase actions of Hel308a at fork structures lead specifically to displacement of lagging strands. The invading strand of D-loops is also targeted. Using archaeal Hel308, we propose models of action for the helicase domain of PolQ, promoting loading of the translesion polymerase domain. We speculate that removal of lagging strands at stalled forks by Hel308 promotes the formation of initiation zones, priming restart of lagging strand synthesis.
INTRODUCTION
DNA helicases have crucial roles in maintaining genome stability and stable DNA replication in all organisms. In bacteria and yeasts, the helicases PriA, RecG, RuvAB, RecBCD, UvrD, Srs2, Rep and RecQ are well known for roles in promoting DNA repair and recombination by several possible mechanisms (1–7). However, sequence homologues of nearly all of these proteins seem to be missing from metazoans. The identities of proteins with analogous functions in metazoans are also largely unknown although some helicase activities, such as branch migration during repair by homologous recombination, seem to be conserved with those in bacteria (8,9). RecQ helicases are present in bacteria (RecQ), yeast (Rqh1, Sgs1) and metazoans (BLM, WRN and RecQ4) and their importance, at least in humans, is emphasized by the onset of genome instability syndromes associated with defects in BLM, WRN or RecQ4. Exact mechanisms of how RecQ helicases participate in DNA repair are not fully clear. They vary between members of the family and depend on the choreography of interactions between RecQ helicases and other proteins that function in replication and repair (10). RecQ proteins are proposed to promote replication restart at stalled forks (11,12), suppress homologous recombination (13) and also drive branch migration of Holliday junctions towards resolution reactions that avoid sister chromatid exchange (14). A common theme in all of these processes is the ability of RecQ helicases to unwind DNA strands in branched intermediates both in vivo and in vitro.
Similar to RecQ, the Mus308 family of helicases and putative helicases support genome stability in metazoans (15–17). However, Mus308 proteins are absent from bacteria and yeast, and our knowledge of them trails well behind RecQ. The mus308 locus was identified in Drosophila melanogaster, being required for resistance to DNA crosslinking agents (18). Cloning of the mus308 gene revealed predicted helicase and polymerase domains (19). Subsequently, a human orthologue of Drosophila Mus308 was identified and called PolQ (20). PolQ also comprises predicted helicase and polymerase domains and, like Mus308, nothing is known of its DNA helicase activity. The polymerase function of purified PolQ has been characterized and shown to be highly efficient at bypassing abasic sites in DNA templates, preferentially inserting adenine opposite an AP-site (21). Mouse cells mutated in PolQ and ATM kinase show a synergistic phenotype resulting in high levels of embryonic lethality (16). A subset of the Mus308 family, called Hel308, was identified by homology to the putative helicase domain of Mus308 (22). Hel308 proteins lack a polymerase domain and, like Mus308/PolQ, were identified in multicellular eukaryotes only. Mutations in Drosophila hel308 cause sensitivity to nitrogen mustards and methyl methanesulfonate (MMS) (17). Three pieces of information are available about DNA helicase activity of human Hel308; in vitro it translocates 3′–5′ along single-stranded DNA (ssDNA) on a gapped duplex substrate, its ATPase activity is stimulated by ssDNA and its helicase activity is stimulated by RPA (22). The mechanism of helicase action by Hel308, and how this could fit genetic data from Drosophila and mouse indicating a role in promoting genome maintenance, is not known.
Archaea are attractive for unravelling interplay between proteins during DNA replication in eukaryotes, including metazoans, because they share key protein participants within systems of greatly reduced complexity (23–25). There is a large body of information about replisome proteins in archaea and eukaryotes, but little is known of helicases that may underpin replication through promoting repair or restart at damaged forks. Since sequence homologues of RecQ are absent from the vast majority of archaeal species, we sought to investigate potentially analogous activities to RecQ from putative archaeal helicases of unknown function. Here, we used in vivo screens for RecQ-like phenotypes and unearthed an archaeal orthologue of human Hel308. The biochemical properties of purified archaeal Hel308 are described in detail and tally well with our observations in vivo. They highlight that when acting at fork substrates in vitro, the helicase action of Hel308 specifically dissociates lagging strands.
MATERIALS AND METHODS
General microbiology and molecular cloning
Unless stated, DNA manipulation enzymes were from New England Biolabs. Methanothermobacter thermautotrophicus (Mth) open reading frame (ORF) mth810 was amplified from Mth genomic DNA template prepared according to the methods described previously (26) using Pfx polymerase (Stratagene) and primers containing NdeI (primer 810-1) and HinDIII (primer 810-2) sites for cloning into pT7-7 and pET22b. Restriction sites are underlined: primer 810-1, 5′-GTGTTCATATGAAGTCCCTCCCACC-3′; and primer 810-2, 5′-AGAGTGGCTGCAAGCTTCCTCAGAG. ORF mth810 was cloned into pT7-7 (pEB310) and into pET22b (pEB313). The mth810 sequence of in each case was verified by sequencing. E.coli recQ+ was cloned from genomic DNA using the following primers: 5′-GGAATTCCATATGAATGTGGCGCAGGCG to generate an NdeI site (underlined), and 5′-CGGGATCCCTACTCTTCGTCATCGCC, for a HinDIII site (underlined). Products were cloned into pT7-7 (pEB417) and pET22b (pEB321). Site-directed mutations in mth810 were generated from pEB310 by the Quick-Change procedure (Stratagene) and sequenced to verify desired mutations.
E.coli strains dnaE486 ΔrecQ and ΔrecQ were a generous gift from Dr Takashi Hishida (Osaka University, Japan), and their genotypes are given in ref. (27). Other strains were from the collection of Prof. Bob Lloyd FRS (University of Nottingham, UK), the significant genotypes of which are MG1655 = wild-type; N4583 = (MG1655) Cmr ΔruvABC; N4256 = (MG1655) Kmr ΔrecG263 :: Km; N4971 = (MG1655) (ΔrecG263 :: Km) Cmr ΔruvABC. Spot, streak and UV-killing tests on mth810 used pEB310. For spot tests, cultures were grown to OD600 of 0.8 (Figure 1A) or as overnights (Figure 1B and C) and serially diluted into minimal medium (56/2). Ten microliter of each dilution was spotted onto Luria–Bertani medium (LB) agar plates containing ampicillin (50 μg/ml). Streak tests used 10 μl of overnight culture. Tests using dnaE486 ΔrecQ were incubated at 30, 37 or 42°C as indicated. For UV-killing curves, cultures were grown to OD600 of 0.45 followed by serial dilution and spotting onto LB-ampicillin plates as above.
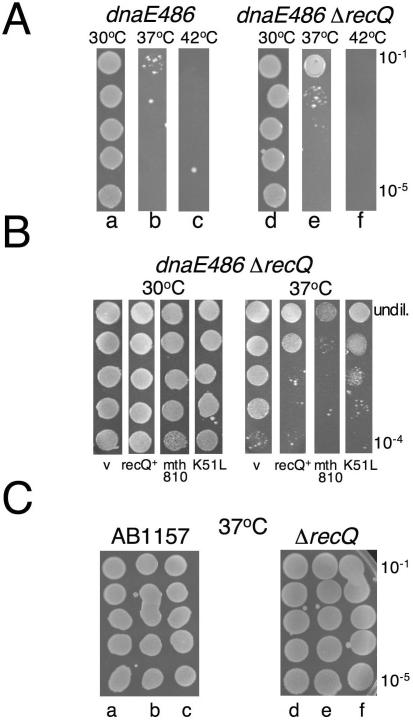
Figure 1.
Hel308 from archaea (mth810) interacts genetically with stalled replication forks. (A) Serial dilution of overnight cultures grown at 30°C showing growth of E.coli dnaE486 (lane a) that is severely restricted at 37°C (lane b) is improved by ΔrecQ (lane e). (B) Spot tests from overnight cultures grown at 30°C, showing that plasmid encoded archaeal gene mth810 stunts growth of dnaE486 ΔrecQ at 37°C, the same phenotype as plasmid encoded E.coli recQ. V is empty vector control and K51L denotes the mutation introduced into helicase motif I of mth810 (C) Mth810 has no dominant effect when transformed into wild-type or ΔrecQ strains; lanes a and d are empty vector; lanes b and e are recQ+; and lanes c and f are mth810.
Mth Hel308a protein
MthHel308a was over-produced in E.coli BL21 AI™ (Stratagene) from pEB313, in baffled flasks containing 400 ml of LB-ampicillin (50 μg/ml) that were seeded (1:50 inoculum) with overnight culture and grown at 37°C to OD of 0.5. Cultures were induced with isopropyl-β-d-thiogalactopyranoside (0.8 mM) and allowed to grow for a further 10 min before inducing RNA polymerase by adding 0.2% (w/v) arabinose solution. This sequence of induction seemed to be critical for efficient expression of MthHel308a. Cultures were grown for a further 3–4 h at 37°C and the cells were harvested, resuspended in buffer C (20 mM Tris–HCl, pH 8.0, 1 mM EDTA, 2 mM DTT, 10% glycerol and 100 mM potassium acetate) and flash-frozen in liquid nitrogen.
To purify recombinant Hel308a, thawed cells were lysed by sonication and clarified to obtain soluble cell proteins (S1) that contained MthHel308a. S1 was subjected to salting-out by ammonium sulfate (0–30, 30–50 and 50–80%). MthHel308a precipitated at 50–80% ammonium sulfate and this was resuspended in buffer C containing 1.5 M ammonium sulfate (S2). S2 was loaded onto a HiPrep butyl-sepharose column (Amersham) and MthHel308a eluted at 400–500 mM ammonium sulfate in a gradient of decreasing ionic strength. Pooled fractions containing MthHel308a were dialysed into buffer C and then loaded onto a HiPrep Q-sepharose column (Amersham), eluting at ∼500 mM potassium acetate in a gradient of increasing ionic strength. Fractions containing MthHel308a were pooled and mixed with ammonium sulfate to 0–80% saturation, and the recovered pellet resuspended in 2–3 ml buffer C + 1 M potassium acetate for loading onto S300 gel filtration column (Amersham). Hel308a fractions were dialysed into buffer C and loaded onto a HiPrep heparin column, eluting at 1 M potassium acetate. Pooled fractions were dialysed into buffer C containing 30% glycerol, dispensed into aliquots, flash-frozen in liquid nitrogen and stored at −80°C. The gel filtration step in high salt was necessary to overcome persistent contamination of Hel308a preparations with E.coli RpoB subunit (MW 150 kDa), identified by mass spectrometry. Comparison with known molecular weight standards showed that MthHel308a elutes as a monomer in gel filtration. The identity of pure Hel308a was confirmed by matrix-assisted laser desorption ionization time-of-flight. K51L was purified in exactly the same way as wild-type protein.
ATPase assays
Hydrolysis of ATP by Hel308a was measured spectroscopically using malachite green assays (28). φX174 circular ssDNA and RFII circular double-stranded DNA (dsDNA) substrates were from New England Biolabs. Linear duplex DNA and ssDNA oligonucleotide substrates were the same as in Table 1. RNA was purchased from Sigma–Aldrich. Reactions contained 100 nM Hel308a or its variant K51L, 100 ng DNA or RNA and were at 45°C for the time specified in the figures. The amount of phosphate liberated from ATP in reactions was adjusted against a blank reaction containing buffer, ATP and DNA but no protein. Error bars were generated from standard errors of mean values.
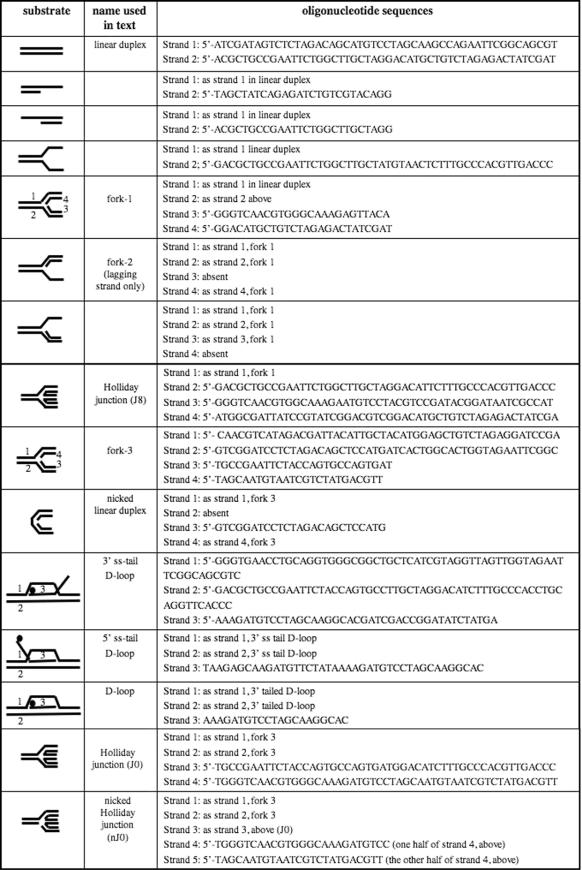
Table 1.
Substrates used in vitro with Hel308a protein
DNA substrates
Table 1 lists DNA substrates and their constituent oligonucleotides described in this study. DNA strands were end-labelled using T4 polynucleotide kinase (PNK) (New England Biolabs) and [γ-32P]ATP (Amersham) (30 min, 37°C). PNK was inactivated (65°C, 30 min) and DNA was separated from unincorporated [γ-32P]ATP using batch gel-filtration columns (BioSpin6, Bio-Rad). 32P-Labelled strand was mixed with appropriate unlabelled DNA strands for annealing in SSC buffer for 6–18 h by reducing the reaction temperature from 95 to 20°C in a heating block. Annealed substrates were purified by gel electrophoresis (10% acrylamide in TBE buffer), visualized on autorad film, excised and eluted from the gel by diffusion at room temperature for 18–36 h into 20 mM Tris, pH 7.5 containing 50 mM NaCl.
DNA binding and unwinding assays
Hel308a-DNA binding assays and unwinding assays were at 45°C in buffer HB (20 mM Tris–HCl, pH 7.5, 8% glycerol, 1 mM DTT and 100 μg/ml BSA) containing 5 mM MgCl2 in 20 μl reaction volumes. Unwinding reactions were supplemented with ATP to 5 mM in standard assays. Hel308a-DNA binding products were loaded in reaction mixture onto 7% acrylamide gels containing 2 mM MgCl2. Gels were run for 2 h at 160 V in 7 mM Tris–HCl, pH 8.0, 6 mM sodium acetate buffer containing 2 mM MgCl2. Unwinding reactions were stopped by adding stop solution (4 mg/ml proteinase K and 1% SDS). Products were analysed on 10% acrylamide TBE gels by electrophoresis for 2 h at 160 V. Time-course reactions used the same conditions but were in 200 μl volumes, removing 20 μl aliquots into stop solution at given time points. For analysis of data, gels were dried using a heated vacuum water pump and developed using phosphorimaging screens (Fuji) on a Storm phosphorimaging device (Molecular Dynamics). DNA moieties were quantified using ImageQuant. Error bars were generated from standard error of means.
RESULTS
We sought to develop an in vivo screen for candidate helicases from archaea that may interact with DNA structures formed at sites of stalled DNA replication. The current lack of tools for investigating stalled DNA replication in vivo in homologous archaeal systems led us to heterologous genetics. This approach has been useful for understanding DNA replication in eukaryotes (29), for probing eukaryotic DNA repair pathways with bacterial resolvases (30–32) and for unravelling bacterial repair pathways with an archaeal resolvase (33). We analysed phenotypes of putative helicase genes from the moderately thermophilic archaeon Mth in an E.coli strain carrying a point mutation (dnaE486) in DNA polymerase III. This is a conditional mutation that leads to replication fork stalling at semi-permissive or restrictive temperatures. One gene (mth810) encoding a putative helicase gave a strong synthetically lethal phenotype when introduced into this strain. The purified protein was then subjected to detailed biochemical analysis.
Identification that mth810 interacts genetically with compromised replication forks
E.coli strain dnaE486 carries a point mutation in the α-subunit of DNA polymerase III that makes progression of DNA replication forks sensitive to temperature (34,35). At permissive temperatures (e.g. 30°C) dnaE486 cells replicate normally but at 37°C replication and cell division is impaired, and at 42°C cells do not grow at all. Deletion of recQ in dnaE486 strains partially suppresses this poor growth phenotype, increasing colony-forming units by 10- to 100-fold at 37°C (27) (Figure 1A, compare lanes b and e). We introduced plasmid-encoded E.coli recQ+ (pEB321) into dnaE486 ΔrecQ, resulting in a strongly negative effect on growth at 37°C, measured in spot tests from liquid cultures (Figure 1B). The same system of spot tests was exploited to screen plasmid-encoded, putative helicases from Mth for a negative effect on growth: a phenotype analogous to recQ+. Transformation of E.coli dnaE486 ΔrecQ with gene mth810 had no effect on growth at 30°C, but had a strongly negative effect on growth at 37°C (Figure 1B). Mutation of the mth810 helicase motif I lysine to leucine (K51L) greatly reduced this effect, though did not abolish it (Figures 1B and 2A). This suggested that there is a requirement for ATPase activity by Mth810 for a phenotype in dnaE486 ΔrecQ. However, the fact that growth of K51L was not fully improved, compared with empty vector, suggests that perhaps DNA binding alone by K51L may interfere with processing sites of stalled replication in dnaE486 ΔrecQ. We return to K51L protein later (Figure 4B). Mth810 had no effect on growth of the corresponding wild-type strain (Figure 1C, lane c) or ΔrecQ strain (Figure 1C, lane f), indicting that the phenotype observed in dnaE486 ΔrecQ cannot be attributed to a general dominant negative effect of mth810 on normal growth of E.coli.
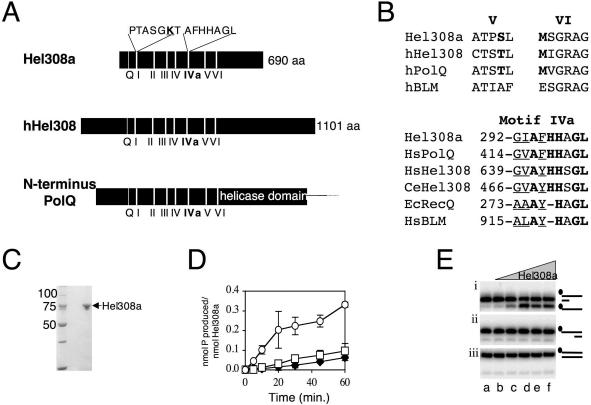
Figure 2.
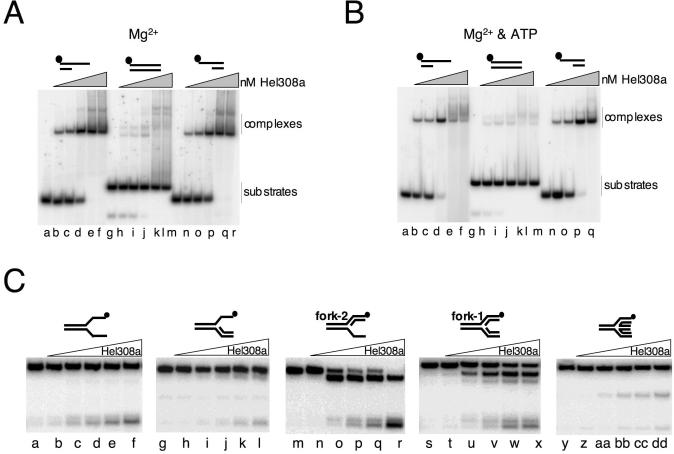
Mth810 is the archaeal orthologue of metazoan Hel308 in sequence and minimal helicase function. (A) Cartoon showing common features of Hel308 from archaea (Hel308a), human (hHel308) and the N-terminal domain of human PolQ. Helicase motifs, including the Q-motif (53), are labelled and the Hel308a sequences are given for motif I and IVa with mutagenized residues in bold and underlined. (B) Sequence details in helicase motifs V and VI that confirm Hel308a as a Hel308/Mus308 family rather than a RecQ helicase. The corresponding motif of human BLM helicase is shown for comparison (hBLM). In each motif peculiar residues conserved in Hel308/Mus308 helicases are in bold. Motif IVa is highly conserved in RecQ and Hel308 proteins. Invariant residues are in bold and highly conserved residues are underlined. Aligned with Hel308a, human Hel308 and human PolQ are Hel308 from Caenorhabditis elegans (CeHel308), E.coli RecQ (EcRecQ) and a human RecQ, BLM (HsBLM). (C) SDS–PAGE gel (10% acrylamide) showing purified recombinant Hel308a (arrowed) from Methanothermobacter. Marker sizes are given on the left of the panel. (D) ATPase activity of Hel308a measured as a function of time in reactions containing no DNA (filled diamond), dsDNA (open square) or ssDNA (open circles). Error bars are derived from the means of three independent experiments. (E) Unwinding reactions of Hel308a on 3′-ssDNA-tailed duplex (i), 5′-ssDNA-tailed duplex (ii) and untailed duplex (iii). Reactions were for 20 min at 45°C containing 2 nM DNA, with 32P-labelled strand indicated by filled circle, 5 mM MgCl2, 5 mM ATP and zero (lane a); 1, 5, 10, 25 and 50 nM Hel308a (lanes b–f).
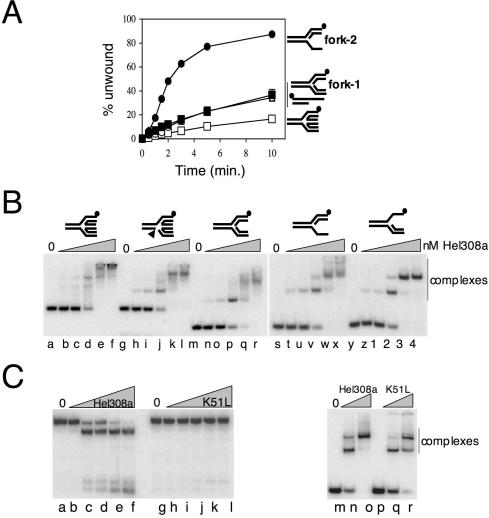
Figure 4.
Hel308a targets DNA forks for unwinding and binds to duplex substrates with branchpoints. (A) Time-course unwinding of four substrates acted on by Hel308a: fork-2 (filled circles), fork-1 (filled squares), 3′-ssDNA-tailed duplex (open circles) and Holliday junction (open squares). Substrates used are annotated to the right of the graph. Reactions were at 45°C for the times shown and contained 2 nM DNA, 5 mM MgCl2, 5 mM ATP and 20 nM Hel308a. Error bars are mostly hidden by data points and were derived from means of three independent assays. (B) Gel-retardation binding assays of Hel308a on fully base-paired, static Holliday junction (lanes a–f), static Holliday junction containing a backbone nick indicated by an arrowhead (lanes g–l), fully base-paired fork-3 (lanes m–r), fork-3 lacking a leading strand (lanes s–x) and fork-3 lacking a lagging strand (lanes y–4). Reactions were at 45°C for 10 min in 1 mM magnesium and contained 2 nM DNA and Hel308a at 0, 1, 2, 10, 50 and100 nM. (C) Hel308a K51L protein is unable to unwind fork-2 (lanes g–l), compared with wild-type protein (lanes a–f). Reactions were for 20 min at 45°C containing 2 nM DNA, 5 mM MgCl2, 5 mM ATP and 0, 1, 5, 10, 25 or 50 nM Hel308a. Binding of wild-type Hel308a (lanes m–o) and K51L Hel308a (lanes p–r) to fork 2 were in reactions for 10 min at 45°C containing 5 mM MgCl2, 2 nM fork-2 (labelled on strand 1) and 0, 10 or 100 nM Hel308a protein.
Archaeal Mth810 protein (Hel308a) is a member of the Hel308 helicase family
The mth810 amino acid sequence is conserved in both major branches of archaea and gave robust matches to human Hel308 (9 × 10−30: 25% overall identity) and mouse PolQ (7 × 10−41: 27% overall identity) (Figure 2A). These belong to the Mus308/Hel308 family of putative and known helicases/translocases that are absent from yeast and bacteria. No significant overall homology was detected with eukaryotic or bacterial RecQ helicases. Mth810 matched the N-terminal domain of PolQ (Figure 2A), which is proposed to be a DNA helicase or translocase though no such activity has been reported. We noticed that Mth810 shares sequence characteristics within helicase motifs V and VI that are peculiar to Hel308/Mus308 proteins (22) but are missing from RecQ (Figure 2B). Mth810 also contains a highly conserved motif of unknown function (AF/YHHAGL, often called motif IVa) that is proposed to be diagnostic of Mus308 proteins (19), although it is also conserved throughout RecQ helicases (36) (Figure 2B). Several factors prompted us to call the protein encoded by mth810, Hel308a (Hel308archaea); conservation of motif I (data not shown), V and VI sequence characteristics, presence of motif IVa, high overall sequence identity between Mth810 and Hel308, and shared minimal biochemical properties of Mth810 and human Hel308, shown below.
Purified Hel308a helicase targets DNA fork substrates
The in vivo analysis suggested that Hel308a interacts with DNA structures that arise at stalled replication forks or during their re-setting and repair in the E.coli strain. To investigate these possible interactions biochemically, we purified recombinant Hel308a (Figure 2C). First we compared activities of Hel308a with that reported for human Hel308 (22). ATPase activity of Hel308a was stimulated greatly by ssDNA but not by dsDNA (Figure 2D), the same activity as human Hel308. RNA did not stimulate ATPase activity, and the ATPase activity of purified Hel308a Walker A variant K51L was reduced to 6% of wild-type activity under optimal conditions (data not shown). Hel308a displaced a short (25 nt) oligonucleotide from base paired DNA with a 3′ ssDNA tail (Figure 2E, panel i), but no activity was detected on DNA with 5′ ssDNA tails (panel ii) or with fully base-paired duplex (panel iii). These Hel308a unwinding assays were in 5 mM Mg2+, and we determined that the optimal ATP concentration for unwinding was 5–7.5 mM (data not shown). Unwinding assays on these minimal substrates indicated a 3′ to 5′ polarity for Hel308a, the same as human Hel308 on a similar substrate (22). Binding of Hel308a was indistinguishable between DNA substrates with 3′ or 5′ ssDNA tails (Figure 3A, lanes a–f and m–r) (data not shown) but the fully base-paired duplex was bound poorly if at all (Figure 3A, lanes g–l). Binding to duplex DNA, or duplex with 3′ or 5′ ssDNA tails, was not improved by the addition of ATP to reactions and gels (Figure 3B). We can be sure that ATP was utilized by Hel308a in these reactions because of the apparent dissociation of the labelled strand from the 3′-ssDNA-tailed duplex, the product running off the bottom of the panel (Figure 3B, lanes d–f). These binding reactions suggested that though Hel308a binds to ssDNA or ssDNA–dsDNA transitions, it is ineffective at forming stable complexes with linear duplex DNA.
Figure 3.
Hel308 from archaea preferentially targets fork DNA for unwinding. (A) Gel-retardation binding assays of Hel308a on the substrates in Figure 2E in 1 mM magnesium. Reactions were at 45°C for 10 min and contained 2 nM DNA substrate mixed with 0, 1, 2, 10, 50 and 100 nM Hel308a. (B) The same reactions as in (A), but containing an additional 1 mM ATP in the gel, and all buffers. (C). Unwinding reactions of Hel308a on flayed duplex (lanes a–f), fork with leading strand only (lanes g–l), fork with lagging strand only (fork-2, lanes m–r), fork with both leading and lagging strands (fork-1, lanes s–x) and Holliday junction (lanes y–dd). Reactions were for 20 min at 45°C containing 2 nM DNA, 5 mM MgCl2, 5 mM ATP and zero (lanes a, g, m, s and y) or 1, 5, 10, 25 and 50 nM Hel308a.
Given the effect exerted by hel308a on a strain generating stalled forks, we were intrigued by whether Hel308a could unwind model DNA fork substrates (Figure 3C). Hel308a was very active at unwinding two fork structures (fork-1 and fork-2) (Figure 3C, lanes s–x and m–r, respectively). Very recently, it was reported that Hel308a from the hyperthermophilic archaeal species Pyrococcus furiosus could unwind Holliday junctions (37). We also observed unwinding of Holliday junctions but this activity was weak compared with unwinding of the equivalent forks (Figure 3C, compare lanes y–dd with fork-1 and fork-2). This substrate unwinding preference of Hel308a for forks over Holliday junctions was confirmed in assays measuring unwinding as a function of time (Figure 4A). There was no difference in the ability of Hel308a to unwind J8, containing an homologous core of 8 bp (as in Figure 3C) compared with static Holliday junctions or those with 3 or 12 bp homologous cores (data not shown). Comparisons of the ability of Hel308a to bind to Holliday junction, nicked Holliday junction, the equivalent fork (fork-3) and it derived partial fork substrates, all of which are static making them unable to migrate into other conformations (e.g. fork into Holliday junction), showed no appreciable difference (summarized in Figure 4B). Given the inability of Hel308a to bind to linear duplex DNA (Figure 3A and B), our results for binding to partial forks, forks and junctions suggest a crucial role for DNA branchpoints in substrate recognition by Hel308a. It also suggests that the greater efficiency of unwinding lagging strand only forks (e.g. fork-2) by Hel308a compared with a fork or Holliday junction cannot be explained by more efficient binding to the partial fork, at least in these in vitro reactions. K51L Hel308a protein was also purified and was inactive as a helicase (Figure 4B, lanes a–l), but bound DNA normally (lanes m–r). This was in agreement with our results that ATP-dependent activity of Hel308 is required for the enzyme to exert its effect in vivo (Figure 1B).
Hel308a targets different DNA substrates from RuvABC in vivo
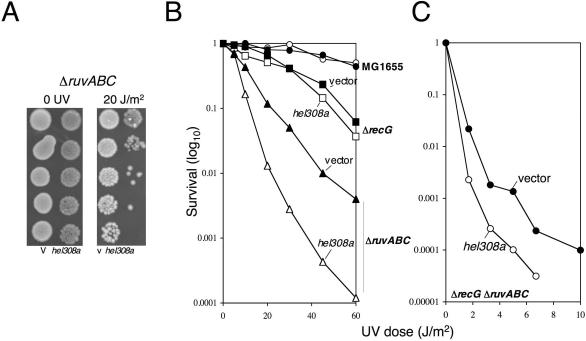
Hel308a unwinding of fork and Holliday junction substrates in vitro led us to investigate whether the helicase was targeting Holliday junctions in vivo, perhaps interfering with homologous recombination. If Hel308a were targeting Holliday junctions in vivo this could explain the phenotype observed in dnaE486 ΔrecQ, since stalled forks are repaired by homologous recombination in bacteria. To detect whether Hel308a can target Holliday junctions in vivo, we tested for phenotypes caused by hel308a in E.coli strains with well-defined DNA repair defects that are unmasked by UV irradiation. The sensitivity to UV irradiation of ΔrecA, Δrep, priA300, ΔuvrD and ΔrecQ strains was unaffected by transformation with hel308a (data not shown). However, a ΔruvABC strain showed 10-fold increased sensitivity to killing by UV light (Figure 5A and B). As expected, the UV phenotype of hel308a in ΔruvABC was the same in ΔruvA and ΔruvC (data not shown). At this point, it would be possible to explain this effect in ΔruvABC if Hel308a was binding to, but not processing, Holliday junctions formed by RecG. However, hel308a also made a ΔruvABC ΔrecG strain more sensitive to UV light (Figure 5C). RuvABC is highly specific for targeting Holliday junction DNA in preference to other DNA substrates (38), so the increased UV sensitivity caused by hel308a in ΔruvABC ΔrecG indicates that Hel308a cannot be targeting Holliday junctions, since their formation and processing is already compromised by the absence of RuvAB. This suggests that in ruv, recG and ruvrecG strains Hel308a interferes with DNA processing by binding to DNA structures other than Holliday junctions or similar repair intermediates that are targeted by RuvABC.
Figure 5.
Hel308 from archaea (mth810) targets different substrates from RuvABC in vivo. (A) Typical spot test plate of the growth phenotype from expression of mth810 in E.coli ΔruvABC exposed to 0 or 20 J/m2 UV light. (B and C) Effect mth810 (pEB310) on UV survival of wild-type E.coli (MG1655, circles), ΔrecG (squares) and ΔruvABC (triangles) strains (B), and a ΔrecG ΔruvABC strain (C). Graphs represent means of three independent experiments.
Hel308a unwinds fork-lagging strands in preference to leading or parental duplex strands
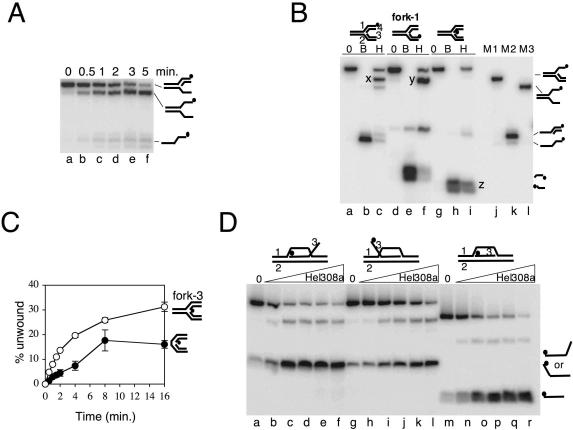
Reactions measuring unwinding of fork-2 by Hel308a suggested that Hel308a displaces the fork-lagging strand in preference to leading strand or strands analogous to the parental duplex (Figure 4C and Figure 6A). We confirmed this by re-visiting fork-1 and 32P-labelling strands in turn to identify directly the strands that are unwound (Figure 6B). In a typical reaction on fork-1 labelled on strand 1 only (Figure 6B, lane c), the vast majority (85%) of total product migrated in the same way as a partial fork marker (compare product x in lane c with marker in lane j). It was significant that unwinding gave negligible product akin to the partial duplex marker in lane k. This indicated removal of either the lagging or the leading strand from the fork and that Hel308a does not unwind well through parental duplex, a reaction that would yield ssDNA-tailed duplex as in marker lane k. In the reaction shown in lane c, it was not possible to ascertain if the product of Hel308a unwinding had a displaced leading or lagging strand since neither is labelled and the partial fork products of either displacement tend to migrate with identical mobilities through these gels. To overcome this, we next labelled fork-1 on strand 3 (Figure 6B, lanes d–f), which gave the same major product as in lane c (product y, lane f). Unwinding of fork-1 labelled on strand 3 by Hel308a gave no short, labelled strand 3 product, illustrated by boiled substrate (lane e). Strand 3 corresponds to a fork-leading strand: we conclude that Hel308a does not unwind this strand. This reaction (lane f) does show a small quantity of partial duplex product (marker lane M2) though this product is much reduced compared with the amount of partial fork (product y), consistent with removal of a fork-lagging strand in preference to unwinding through parental duplex. To confirm this, labelling of fork-1 on strand 4 (lanes g–i) showed displacement of this strand with high efficiency (compare boiled substrate in lane h with product z in lane i). Strand 4 corresponds to a fork-lagging strand. This strand could also be unwound in a duplex substrate that contained a nick in the phosphodiester backbone of one strand only (Figure 6C). This nick is the only difference between this substrate and the duplex that was not unwound (or bound) at all by Hel308a (Figures 2E and 3A). Therefore, a backbone nick is an important structure for substrate recognition by Hel308a. However, unwinding of this nicked duplex was significantly reduced compared with fork-3, which has the same DNA sequence and a nick in the same position, but retains the parental duplex region (Figure 6C). No unwinding activity was detected on the same fork that was ‘sealed’ lacking any gap between lagging and leading strands (data not shown). These results in Figure 6C suggest that Hel308a targets DNA fork substrates most efficiently by engaging both the fork branchpoint, which requires parental duplex, and a strand nick or perhaps ss–dsDNA junctions.
Figure 6.
Hel308a unwinds lagging strands from nicks and the fork branchpoint. (A) Products from unwinding fork-2 by hel308a as a function of time. Reactions contained 2 nM DNA and 20 nM Hel308a in 5 mM MgCl2, 5 mM ATP at 45°C. (B) Reactions showing unwinding of strands in fork-1 by Hel308a. Cartoons of the substrate are shown above the panel in each case with the labelled strand denoted by a filled circle. Strands are numbered on one of these cartoons. Reactions were at 45°C for 20 min containing 5 mM MgCl2, 5 mM ATP, 2 nM DNA and zero (0) or 50 nM Hel308a (H). Lanes marked B contained no Hel308a and the reactions were heated to 95°C for 20 min. Letters X, Y and Z highlight the major product of unwinding in each reaction containing Hel308a. Substrate markers corresponding to fork lacking a lagging strand (M1), flayed duplex (M2) and partial duplex (M3) are annotated beside the panel. (C) Time-course unwinding of fork-3 and its corresponding nicked duplex DNA substrate. Substrates used are annotated to the right of the graph. Reactions were at 45°C for the times shown and contained 2 nM DNA, 5 mM MgCl2, 5 mM ATP and 20 nM Hel308a. Error bars derive from means of two independent assays. (D) Unwinding of the invading strand of D-loop substrates by Hel308a. The D-loop substrates used are annotated above the panel and reactions products are displayed to the right of the panel. Reactions were for 20 min at 45°C containing 2 nM DNA, 5 mM MgCl2, 5 mM ATP and zero or 1, 5, 10, 25 and 50 nM Hel308a.
Hel308a dissociates the ‘invading’ strand from D-loops
We also investigated whether Hel308a can unwind DNA strands in D-loops, DNA structures that resemble replication forks and have been implicated in replication restart in bacteria and viruses (39). D-loop substrates, previously shown to be recognized by the bacterial replication restart helicase PriA in vitro (40), were generated with different polarities of the ‘invading’ strand, in each case called strand 3 (Figure 6D and Table 1). The major product of unwinding by Hel308a in each D-loop substrate was the dissociation of the ‘invading’ strand (Figure 6D, strand 3). This is explained most simply by Hel308a recognizing the branchpoint of strands 1 and 2 that is located proximally to the 5′ (labelled) end of the invading strand 3. Given the 3′–5′ polarity of Hel308a, displacement of the invading strand 3 would result from helicase tracking along strand 2. The possible implications of this reaction on D-loops are discussed below.
DISCUSSION
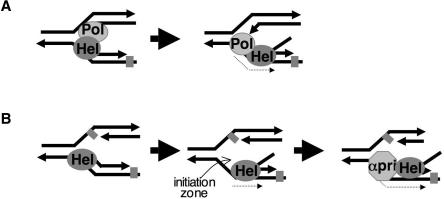
By combining biochemistry with observations made in vivo, we report that archaeal Hel308 helicase targets replication forks for unwinding (Figures 1B, 3C and 4A). At forks, its helicase action is highly preferential in displacing the lagging strand from the branchpoint (Figure 6). It also removes the invading strand from D-loop substrates (Figure 6D). Hel308a may be crucial for maintaining genome stability in archaeal species, especially because most archaeal species lack RecQ homologues. More generally, it was very important to confirm that the archaeal helicase is a bona fide Hel308 family protein. This was realized by the high overall homology between Hel308a and human Hel308, but also through close scrutiny of precise sequence details of Hel308a compared with the metazoan proteins (Figure 2B). During the preparation of this manuscript, it was reported that Hel308a protein from Pyrococcus could unwind Holliday junctions and branch migrate them, leading the authors to propose calling the protein Hjm, for Holliday junction migration (37). As discussed below, our in vivo data and detailed biochemical analysis of the same protein are more in line with biochemical data so far available for human Hel308 and suggests that although Hel308a can unwind Holliday junctions, albeit poorly, it functions most efficiently at forks, especially those possessing a region of ssDNA (e.g. Figure 4A). The activities of archaeal Hel308 shown here, in vivo and in vitro, allow development of models of action for Hel308 helicases in archaea and possibly metazoans (Figure 7). In the case of PolQ, which has a putative Hel308 helicase domain fused to a translesion polymerase, clearance of lagging strands exposing a ssDNA template could promote loading of the polymerase domain, gaining access to AP sites on the lagging strand template (Figure 7A). Lagging strand unwinding by Hel308 could clear the way for loading of proteins required for replication to resume at stalled forks (Figure 7B), discussed more fully below.
Figure 7.
Models proposing how Hel308 helicases may function in archaea and metazoans. (A) Displacement of the lagging strand by the Hel308 helicase (Hel) domain of PolQ may provide access for the translesion polymerase domain to DNA damage (grey square, e.g. AP-sites) located on the lagging strand template. (B) Hel308a/Hel308 (Hel) unwinding of the lagging strand at fork with a compromised leading or lagging strand provides a template for loading of replication restart apparatus, possibly Pol-α-primase (αPri). In each model, translocation of the helicase is indicted by a dotted arrow, away from the fork branchpoint in a 3′–5′ direction with respect to the lagging strand template.
Actions of Hel308a in vivo
The synthetically lethal phenotype from expressing hel308a in E.coli dnaE486 ΔrecQ (Figure 1) was the same as reported for recQ (27) and implied that Hel308a engages with DNA structures that arise at stalled replication forks. This alone cannot discriminate between several possible roles for Hel308a in DNA binding and catalysis on stalling or restart of DNA replication induced by semi-permissive temperatures. However, the growth phenotype of hel308a in replication defective strains fits well with the requirement of hel308 and mus308 in Drosophila for overcoming genome instability caused by cisplatin, MMS and nitrogen mustards, all potent inhibitors of DNA replication (15,17,41). This method of screening from plasmids for growth phenotypes in dnaE486 ΔrecQ may also be useful to rapidly identify functionally important residues in Hel308 and RecQ-family helicases. Hel308a enhanced the UV sensitivity of E.coli ΔruvABC strains, but also a ΔruvABC ΔrecG strain (Figure 5). Bacterial RuvABC underpins repair of replication forks through homologous recombination and fork reversal, each characterized by the generation of Holliday junctions (42,43). RuvABC, a branch migration complex that acts in late stages of homologous recombination, is highly specific for binding, unwinding and resolving Holliday junctions in preference to forks or other branched structures (38). The increased UV sensitivity caused by hel308a in ΔruvABC ΔrecG strongly suggests that Hel308a does not target Holliday junctions but interferes with additional repair processes independently of Ruv. E.coli cells lacking RuvABC and RecG survive through alternative pathways for survival after UV irradiations, which it seems are targeted by Hel308a. These may include replication over damage by translesion polymerases (44) and PriA mediated fork re-setting without recourse to late stages of recombination (45,46). Phenotypes of hel308a expression, though specific to the strains discussed above, in each case negatively affected growth and survival. Fork recovery and DNA repair usually require orchestrated actions of several interacting proteins. This probably explains why Hel308a is unable to complement a defect in repair within E.coli cells, because if it is involved in these processes, it is unlikely to interact with E.coli proteins that direct downstream functions, e.g. bacterial polymerases and replication/primasome assembly proteins.
Actions of Hel308a helicase in vitro
The biochemistry of purified Hel308a is consistent with in vivo observations that hel308a interacts genetically with stalled replication forks. Hel308a translocates 3′–5′ with respect to ssDNA-tracking strand, dissociating annealed strands from the minimal substrate of Hel308a, a 3′-ssDNA-tailed duplex (Figure 2E, panel i). This reaction fits with the only unwinding activity reported for human Hel308, which was on a gapped duplex substrate (22). Although Hel308a is unable to bind to linear duplex DNA (Figure 3A and B), it can bind to and unwind duplex substrates with a nick in one strand (Figure 6C), but Hel308a unwinds the equivalent fork (fork-3) more efficiently (Figure 6C). The protein binds efficiently to fully base-paired static fork and Holliday junction substrates. However, Hel308a also binds comparably well with partial duplex DNA substrates that lack branchpoints available in forks and Holliday junctions. Therefore, the results of band shifts seem to indicate that Hel308a can equally well engage with ss–dsDNA substrates (e.g. partial duplex) and those with branchpoints. Similarly, despite the differences in the ability of Hel308a to unwind partial fork with a lagging strand, compared with fully base paired fork and Holliday junctions, Hel308a seems to bind equally well to all of these substrates, at least in vitro (Figure 4B). However, the helicase activity of Hel308a seems to be more efficient when a branchpoint is present, such as in a partial fork compared with equivalent partial duplex (Figure 4A) or within a fork compared with nicked duplex (Figure 6C). This implies that for helicase activity (but not necessarily DNA binding), two substrate recognition events may be required by Hel308a for most efficient unwinding: recognition of DNA branchpoints as in forks and Holliday junctions, and recognition of ssDNA or ss–dsDNA transitions. This requirement for ssDNA resembles DNA binding characteristics of PcrA, a monomeric DNA helicase in bacteria (47).
Hel308a was highly active at unwinding model fork DNA substrates that possess a lagging strand (Figures 3C and 4A). Activity was low on a fork lacking both lagging and leading strands, or on a fork with only a leading strand. Efficient unwinding of fork substrates by Hel308a reconciles well with its helicase activity requiring a DNA branchpoint and a point of ss–dsDNA transition. Recognition of DNA branchpoints allows Hel308a to unwind Holliday junctions as well as forks in vitro, though forks are unwound far more efficiently (Figures 3C and 4A). Low activity of Hel308a on Holliday junctions may highlight the need for interaction with ssDNA via at least a nick for efficient catalysis, as is presented in fork substrates. Recently, it was reported (37) that equivalent of Hel308a from the archaeon P.furiosus can dissociate α-structures, implying Holliday junction branch migration. Alternatively, this reaction can be explained without the need for the helicase recognising Holliday junctions: the α-structure can be dissociated by 3′–5′ translocation of Hel308a from the ssDNA gap or nick remote from the Holliday junction, displacing the annealed strand from linear duplex. This unwinding activity is akin to that on a duplex with exposed 3′ ssDNA (Figure 2E, panel i) or a nicked duplex substrate (Figure 6C). It is also worth noting that Hel308a does not show the high specificity for Holliday junction binding, in vivo or in vitro, exhibited by bacterial RuvABC, the only well-characterized branch migration helicase.
Most significantly, Hel308a targets removal of the lagging strand from forks (Figure 6B). Hel308a was unable to unwind a fork that comprised a leading strand but no lagging strand (Figure 3C). The ability of Hel308a to dissociate the lagging strand in preference to parental duplex strands cannot be ascribed to the unknown processivity of Hel308a because in each case strands are annealed by 25 bp. The most likely explanation for how Hel308a unwinds the lagging strand is summarized in Figure 7. By targeting the branchpoint of a fork and gaining access to ssDNA, either at a nick or on exposed ssDNA, Hel308a can translocate 3′–5′ along the single strand displacing the lagging strand as it does so. This mechanism does not require template switching by the helicase to unwind the lagging strand, and fits with our in vivo and in vitro data, and information known about human Hel308 (22). Hel308a also displaced the invading strand from model D-loop substrates (Figure 6D). This activity can arise from Hel308a tracking along strand 2 in a 3′–5′ direction, and in doing so removing strand 3, the invading strand. In bacteria replication restart is primed from D-loops by the actions of PriA and interacting partners (45,48,49). The invading strand primes leading strand synthesis while DnaB helicase loads on the ssDNA lagging strand template. The actions of Hel308a we describe in vitro would remove the capacity to prime replication on the leading strand by displacing the invading strand, and therefore would act counter to replication restart, at least within models from bacteria. If this activity were to be reproduced in vivo it would potentially involve Hel308a in anti-recombinagenic functions. In a purely mechanistic sense, displacement of the invading strand by Hel308a is analogous to removal of fork lagging strands if Hel308a associates with the substrate D-loop at the branchpoint located at the 5′ end of the invading strand (e.g. model D-loop corresponding to lanes m–r, Figure 6D). However, we conclude that in vivo Hel308a is more likely to function in clearing lagging strands at fork structures, in preference to dissociation of parental duplex DNA or destabilization of D-loops.
Possible roles for Hel308 helicases in genome stability
Inactivating Hel308 helicase was recently linked to a mutant phenotype for the first time, whereby Drosophila homozygous for insertions in hel308 became hypersensitive to cisplatin (17). This implicates Hel308 in some aspect of DNA repair, possibly linked to replication since cisplatin is a potent inhibitor of replication forks. In removing lagging strands at forks, Hel308 proteins may function in clearing lagging strand templates for loading proteins required to restart DNA replication. Removal of the lagging strand could provide a template for loading any of the primase, replicative helicase or polymerases required for replication. In this way, in archaea and metazoans Hel308 could generate an okazaki initiation zone by removal of lagging strands, promoting loading of polymerase-α-primase, also active in archaea (50), to resume lagging strand synthesis (Figure 7B). The absence of this helicase function could explain why hel308 mutant strains of Drosophila are very sensitive to agents that block DNA replication, as they are perhaps unable to resume replication efficiently.
Under other circumstances, the same action by the helicase domain of human PolQ (Mus308) could orientate loading of the translesion polymerase domain of the same polypeptide giving the polymerase access to AP-sites (Figure 7A) (21). Another possible important consequence of Hel308a removing lagging strands at stalled forks could be to expose ssDNA, acting as a signal to DNA damage checkpoints. Mechanisms of ssDNA signalling for repair are not yet known in archaea, but are in eukaryotes (51,52) and in the bacterial SOS response, where RecQ may have a major role (27), in each case relying to some extent on the detection of ssDNA. The similarity between archaea and eukaryotes in DNA replication makes it likely that they use similar mechanisms to repair and restart replication forks in response to damage signalling mechanisms at sites remote from replication origins. Hel308a/Hel308 may participate in this in archaeal cells and in metazoans to provide interactions with other replication and repair proteins after exposing a template DNA strand. Hel308 from archaea may offer guidance in how this family of helicases function in genome stability.
Acknowledgments
The authors are grateful to Bob Lloyd for continuing valuable discussions. The authors thank Takashi Hishida for kindly providing E.coli strains and anonymous referees for helpful observations. The Wellcome Trust funded this work through a RCDF to E.L.B. Funding to pay the Open Access publication charges for this article was provided by JISC funding.
Conflict of interest statement. None declared.
REFERENCES
- 1.Aylon Y., Kupiec M. DSB repair: the yeast paradigm. DNA Repair (Amst.) 2004;3:797–815. doi: 10.1016/j.dnarep.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 2.Cox M.M., Goodman M.F., Kreuzer K.N., Sherratt D.J., Sandler S.J., Marians K.J. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 3.Michel B. Replication fork arrest and DNA recombination. Trends Biochem. Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- 4.McGlynn P., Lloyd R.G. Recombinational repair and restart of damaged replication forks. Nature Rev. Mol. Cell Biol. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 5.Kowalczykowski S.C., Dixon D.A., Eggleston A.K., Lauder S.D., Rehrauer W.M. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singleton M.R., Wigley D.B. Modularity and specialization in superfamily 1 and 2 helicases. J. Bacteriol. 2002;184:1819–1826. doi: 10.1128/JB.184.7.1819-1826.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolodner R.D., Putnam C.D., Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 8.Constantinou A., Chen X.B., McGowan C.H., West S.C. Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO J. 2002;21:5577–5585. doi: 10.1093/emboj/cdf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y., Masson J.Y., Shah R., O'Regan P., West S.C. RAD51C is required for Holliday junction processing in mammalian cells. Science. 2004;303:243–246. doi: 10.1126/science.1093037. [DOI] [PubMed] [Google Scholar]
- 10.Hickson I.D. RecQ helicases: caretakers of the genome. Nature Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 11.Courcelle J., Hanawalt P.C. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol. Gen. Genet. 1999;262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- 12.Cobb J.A., Bjergbaek L., Shimada K., Frei C., Gasser S.M. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003;22:4325–4336. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harmon F.G., Kowalczykowski S.C. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 1998;12:1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu L., Hickson I.D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 15.Boyd J.B., Sakaguchi K., Harris P.V. mus308 mutants of Drosophila exhibit hypersensitivity to DNA cross-linking agents and are defective in a deoxyribonuclease. Genetics. 1990;125:813–819. doi: 10.1093/genetics/125.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shima N., Munroe R.J., Schimenti J.C. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol. Cell. Biol. 2004;24:10381–10389. doi: 10.1128/MCB.24.23.10381-10389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laurencon A., Orme C.M., Peters H.K., Boulton C.L., Vladar E.K., Langley S.A., Bakis E.P., Harris D.T., Harris N.J., Wayson S.M., et al. A large-scale screen for mutagen-sensitive loci in Drosophila. Genetics. 2004;167:217–231. doi: 10.1534/genetics.167.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyd J.B., Golino M.D., Nguyen T.D., Green M.M. Isolation and characterization of X-linked mutants of Drosophila melanogaster which are sensitive to mutagens. Genetics. 1976;84:485–506. doi: 10.1093/genetics/84.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris P.V., Mazina O.M., Leonhardt E.A., Case R.B., Boyd J.B., Burtis K.C. Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol. Cell. Biol. 1996;16:5764–5771. doi: 10.1128/mcb.16.10.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seki M., Marini F., Wood R.D. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seki M., Masutani C., Yang L.W., Schuffert A., Iwai S., Bahar I., Wood R.D. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marini F., Wood R. A human DNA helicase homologous to the DNA cross-link sensitivity protein mus308. J. Biol. Chem. 2002;277:8716–8723. doi: 10.1074/jbc.M110271200. [DOI] [PubMed] [Google Scholar]
- 23.Robinson N.P., Dionne I., Lundgren M., Marsh V.L., Bernander R., Bell S.D. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell. 2004;116:25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- 24.Kelman L.M., Kelman Z. Archaea: an archetype for replication initiation studies? Mol. Microbiol. 2003;48:605–615. doi: 10.1046/j.1365-2958.2003.03369.x. [DOI] [PubMed] [Google Scholar]
- 25.Bell S.P. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16:659–672. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- 26.Guy C.P., Majernik A.I., Chong J.P., Bolt E.L. A novel nuclease-ATPase (Nar71) from archaea is part of a proposed thermophilic DNA repair system. Nucleic Acids Res. 2004;32:6176–6186. doi: 10.1093/nar/gkh960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hishida T., Han Y.W., Shibata T., Kubota Y., Ishino Y., Iwasaki H., Shinagawa H. Role of the Escherichia coli RecQ DNA helicase in SOS signaling and genome stabilization at stalled replication forks. Genes Dev. 2004;18:1886–1897. doi: 10.1101/gad.1223804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bird L.E., Hakansson K., Pan H., Wigley D.B. Characterization and crystallization of the helicase domain of bacteriophage T7 gene 4 protein. Nucleic Acids Res. 1997;25:2620–2626. doi: 10.1093/nar/25.13.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee M.G., Nurse P. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature. 1987;327:31–35. doi: 10.1038/327031a0. [DOI] [PubMed] [Google Scholar]
- 30.Doe C., Dixon J., Osman F., Whitby M. Partial suppression of the fission yeast rqh1− phenotype by the expression of a bacterial Holliday junction resolvase. EMBO J. 2000;19:2751–2762. doi: 10.1093/emboj/19.11.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doe C.L., Ahn J.S., Dixon J., Whitby M.C. Mus81-eme1 and rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- 32.Odagiri N., Seki M., Onoda F., Yoshimura A., Watanabe S., Enomoto T. Budding yeast mms4 is epistatic with rad52 and the function of Mms4 can be replaced by a bacterial Holliday junction resolvase. DNA Repair (Amst.) 2003;2:347–358. doi: 10.1016/s1568-7864(02)00234-3. [DOI] [PubMed] [Google Scholar]
- 33.Bolt E.L., Lloyd R.G., Sharples G.J. Genetic analysis of an archaeal Holliday junction resolvase in Escherichia coli. J. Mol. Biol. 2001;310:577–589. doi: 10.1006/jmbi.2001.4791. [DOI] [PubMed] [Google Scholar]
- 34.Vandewiele D., Fernandez de Henestrosa A.R., Timms A.R., Bridges B.A., Woodgate R. Sequence analysis and phenotypes of five temperature sensitive mutator alleles of dnaE, encoding modified alpha-catalytic subunits of Escherichia coli DNA polymerase III holoenzyme. Mutat. Res. 2002;499:85–95. doi: 10.1016/s0027-5107(01)00268-8. [DOI] [PubMed] [Google Scholar]
- 35.Wechsler J.A., Gross J.D. Escherichia coli mutants temperature-sensitive for DNA synthesis. Mol. Gen. Genet. 1971;113:273–284. doi: 10.1007/BF00339547. [DOI] [PubMed] [Google Scholar]
- 36.Bernstein D.A., Zittel M.C., Keck J.L. High-resolution structure of the E.coli RecQ helicase catalytic core. EMBO J. 2003;22:4910–4921. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujikane R., Komori K., Shinagawa H., Ishino Y. Identification of a novel helicase activity unwinding branched DNAs from the hyperthermophilic archaeon, Pyrococcus furiosus. J. Biol. Chem. 2005;280:12351–12358. doi: 10.1074/jbc.M413417200. [DOI] [PubMed] [Google Scholar]
- 38.Ingleston S.M., Sharples G.J., Lloyd R.G. The acidic pin of RuvA modulates Holliday junction binding and processing by the RuvABC resolvasome. EMBO J. 2000;19:6266–6274. doi: 10.1093/emboj/19.22.6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones J.M., Nakai H. PriA and phage T4 gp59: factors that promote DNA replication on forked DNA substrates microreview. Mol. Microbiol. 2000;36:519–527. doi: 10.1046/j.1365-2958.2000.01888.x. [DOI] [PubMed] [Google Scholar]
- 40.McGlynn P., Al-Deib A.A., Liu J., Marians K.J., Lloyd R.G. The DNA replication protein PriA and the recombination protein RecG bind D-loops. J. Mol. Biol. 1997;270:212–221. doi: 10.1006/jmbi.1997.1120. [DOI] [PubMed] [Google Scholar]
- 41.Tosal L., Comendador M.A., Sierra L.M. The mus308 locus of Drosophila melanogaster is implicated in the bypass of ENU-induced O-alkylpyrimidine adducts. Mol. Gen. Genet. 2000;263:144–151. doi: 10.1007/s004380050041. [DOI] [PubMed] [Google Scholar]
- 42.Seigneur M., Bidnenko V., Ehrlich S.D., Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 43.McGlynn P., Lloyd R.G. Genome stability and the processing of damaged replication forks by RecG. Trends Genet. 2002;18:413–419. doi: 10.1016/s0168-9525(02)02720-8. [DOI] [PubMed] [Google Scholar]
- 44.Walker G.C. SOS-regulated proteins in translesion DNA synthesis and mutagenesis. Trends Biochem. Sci. 1995;20:416–420. doi: 10.1016/s0968-0004(00)89091-x. [DOI] [PubMed] [Google Scholar]
- 45.Xu L., Marians K.J. PriA mediates DNA replication pathway choice at recombination intermediates. Mol. Cell. 2003;11:817–826. doi: 10.1016/s1097-2765(03)00061-3. [DOI] [PubMed] [Google Scholar]
- 46.Jaktaji R.P., Lloyd R.G. PriA supports two distinct pathways for replication restart in UV-irradiated Escherichia coli cells. Mol. Microbiol. 2003;47:1091–1100. doi: 10.1046/j.1365-2958.2003.03357.x. [DOI] [PubMed] [Google Scholar]
- 47.Velankar S.S., Soultanas P., Dillingham M.S., Subramanya H.S., Wigley D.B. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 48.Jones J.M., Nakai H. Duplex opening by primosome protein PriA for replisome assembly on a recombination intermediate. J. Mol. Biol. 1999;289:503–516. doi: 10.1006/jmbi.1999.2783. [DOI] [PubMed] [Google Scholar]
- 49.Gregg A.V., McGlynn P., Jaktaji R.P., Lloyd R.G. Direct rescue of stalled replication forks via the combined action of PriA and RecG helicase activities. Mol. Cell. 2002;9:241–251. doi: 10.1016/s1097-2765(02)00455-0. [DOI] [PubMed] [Google Scholar]
- 50.Liu L., Komori K., Ishino S., Bocquier A.A., Cann I.K., Kohda D., Ishino Y. The archaeal DNA primase: biochemical characterization of the p41–p46 complex from Pyrococcus furiosus. J. Biol. Chem. 2001;276:45484–45490. doi: 10.1074/jbc.M106391200. [DOI] [PubMed] [Google Scholar]
- 51.Garvik B., Carson M., Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nyberg K.A., Michelson R.J., Putnam C.W., Weinert T.A. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 53.Tanner N.K., Cordin O., Banroques J., Doere M., Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]