Abstract
Nuclear receptors (NRs) are ligand-regulated transcription factors important in human physiology and disease. In certain NRs, including the androgen receptor (AR), ligand binding to the carboxy-terminal domain (LBD) regulates transcriptional activation functions in the LBD and amino-terminal domain (NTD). The basis for NTD–LBD communication is unknown but may involve NTD–LBD interactions either within a single receptor or between different members of an AR dimer. Here, measurement of FRET between fluorophores attached to the NTD and LBD of the AR established that agonist binding initiated an intramolecular NTD–LBD interaction in the nucleus and cytoplasm. This intramolecular folding was followed by AR self-association, which occurred preferentially in the nucleus. Rapid, ligand-induced intramolecular folding and delayed association also were observed for estrogen receptor-α but not for peroxisome proliferator activated receptor-γ2. An antagonist ligand, hydroxyflutamide, blocked the NTD–LBD association within AR. NTD–LBD association also closely correlated with the transcriptional activation by heterologous ligands of AR mutants isolated from hormone-refractory prostate tumors. Intramolecular folding, but not AR–AR affinity, was disrupted by mutation of an α-helical (23FQNLF27) motif in the AR NTD previously described to interact with the AR LBD in vitro. This work establishes an intramolecular NTD–LBD conformational change as an initial component of ligand-regulated NR function.
Keywords: conformation change, FQNLF, FRET, nuclear receptor, estrogen receptor
The nuclear receptor (NR) superfamily consists of a large group of ligand-regulated transcription factors. Several NRs are implicated in human physiology and disease (1, 2) and activation of the estrogen receptors (ER) and androgen receptors (AR) are predisposing factors for breast (3) and prostate cancer (4). Indeed, pharmacologic antagonists of AR and ER are used as antineoplastic agents in these diseases (4–7). It is commonly believed that understanding NR structure and function will facilitate development of specific drugs that can replace or supplement current therapies (2). Ligand binding alters NR structure, cofactor interactions, and transcriptional activity (8). Transcriptional activation functions are present in the amino-terminal domain (NTD; AF-1) and the ligand binding domain (LBD; AF-2) of many NRs, including AR (9) and ER (10). AF-1 is not conserved at the primary sequence level and is poorly characterized functionally (11). In contrast, AF-2 is highly conserved (12) and consists of amino acids that form a coactivator binding pocket on the surface of most NR LBDs (13–16).
In many NRs, both AF-1 and AF-2 activities are suppressed in the absence of ligand and enabled after ligand binding (9, 10), which implies that ligand binding to the LBD somehow unmasks AF-1 activities in the NTD. The molecular/structural basis for LBD communication with AF-1 in full-length molecules remains uncertain. However, an intermolecular interaction between NTD peptides and the agonist-bound LBD has been extensively characterized in vitro and with intracellular two-hybrid assays for the AR (14, 17–21) and ER (22). In the AR NTD, deletion or mutation of a sequence (23FQNLF27) that can bind the AF-2 coactivator pocket of the LBD (14, 19) diminishes activity of the AR at certain promoter elements (21). This finding suggests that an NTD–LBD interaction is functionally important, but it remains unknown whether the NTD interacts with the LBD within one molecule or whether it participates in an intermolecular interaction with the LBD of a second AR molecule.
Of the currently available experimental approaches, FRET (23) uniquely can resolve conformation changes and protein interactions of the intact NR molecule in living cells. FRET allows real-time detection of protein conformation changes based on energy transfer between fluorophores attached to domains of interest. Here, we used FRET to determine the time and subcellular location of ligand-induced conformational changes in AR that underlie its activity as a transcription factor. We contrasted these studies with other members of the NR family, ERα and peroxisome proliferator-activated receptor-γ2 (PPARγ2), and have determined a role for the AR-specific 23FQNLF27 motif in coordinating intramolecular AR conformational changes that precede AR self-association, most likely as a dimer.
Materials and Methods
Plasmid Construction. Plasmids that express AR, ERα, or PPARγ2 as enhanced cyan fluorescent protein (ECFP)–NR, NR–enhanced yellow fluorescent protein (EYFP) or ECFP–NR–EYFP fusions were constructed by inserting PCR-amplified NR cDNAs into ECFP and EYFP-containing expression vectors (Clontech). The AR-AQNAA and ARΔF mutants were constructed by site-directed mutagenesis. AR LBD mutants were subcloned from full-length AR into CFP–AR–YFP. All constructs were sequenced after construction. The mouse mammary tumor virus (MMTV)-luciferase reporter plasmid was kindly provided by K. Yamamoto (University of California, San Francisco).
Cell Culture and Transfection. HeLa cells (n = 200,000) were split into each well of a six-well dish containing a borosilicate glass coverslip and grown in media containing newborn calf serum stripped of androgens. DNA (100 ng per well) was transfected by using Effectene (Qiagen, Valencia, CA). Cells were imaged live at the indicated time points (see Figs. 2, 3, 4, 5 and Figs. 6–9, which are published as supporting information on the PNAS web site). HEK293 cells (n = 1.5 × 106) grown in DMEM-H21 supplemented with 10% FCS were transfected in 3.5-cm dishes by using 1.2 μg of DNA with Lipofectamine Plus (Invitrogen). The day after transfection, 100,000 cells were replated to a 96-well dish in the presence or absence of hormone. Cells were fixed in 4% paraformaldehyde in PBS before reading on the fluorescence plate reader (FPR).
Fig. 2.
AR dimerization and NTD-to-LBD folding measured by FRET microscopy. (A) HeLa cells expressing CFP–AR and AR–YFP were imaged at the indicated time points after addition of 100 nM DHT. (Upper) Acceptor signal. (Lower) FRET/donor images calculated from the corrected acceptor, donor, and FRET images (data not shown; see Materials and Methods). (B) Energy transfer of CFP–AR to AR–YFP (black boxes) increased with the relative amount of AR–YFP present to interact with CFP–AR (increasing acceptor/donor measured in the nuclei of 282 DHT-treated HeLa cells). There was no FRET in cells not incubated with DHT (open squares; 358 cells), nor was there FRET between CFP–PPARγ2 and PPARγ2–YFP in cells treated with the agonist ligand GW1929 (gray triangles; 215 cells). (C) Maximal, YFP-saturated FRET/donor values in the nucleus (Nuc) and cytoplasm (Cyto) of HeLa cells coexpressing CFP–AR and AR–YFP after treatment with 10 nM DHT or vehicle for 20 min. (D) FRET/donor values from cells expressing CFP–AR–YFP after treatment with vehicle (n = 173 cells) or 10 nM DHT for 20 min (n = 80 cells). Diagrams in C and D show the inferred changes in dimerization and conformation induced by DHT. Although the diagrams depict the amino and carboxy termini of the monomers in close proximity, our data allow no conclusion about the orientation of ARs within the dimer. Error bars represent the SEM.
Fig. 3.
Intramolecular folding precedes association of AR and ERα monomers but not PPARγ2. FRET was determined in HeLa cells expressing AR (A), ERα (B), or PPARγ2(C) as fusions with CFP–NR and NR–YFP (association) or CFP–NR–YFP (folding). Images were collected every minute at 37°C. DHT (10 nM), estradiol (10 nM), or GW1929 (100 nM) was added 30 sec after the fourth image; an additional 26 images were captured starting 30 sec after the addition of ligand. The mean CFP–NR/NR–YFP or CFP–NR–YFP FRET in the nucleus (nuc; blue diamonds or red squares) and in the cytoplasm (cyto; pink diamonds or green squares) are shown from 12 or 47 cells (AR), 6 or 14 cells (ERα), and 8 or 11 cells (PPARγ2). Error bars represent the SEM.
Fig. 4.
AR NTD–LBD folding correlates with transcriptional activity. (A and B) OH-F antagonizes CFP–AR–YFP FRET in HEK293 cells treated with 10 nM DHT (detected by FPR) (A) and HeLa cells treated with 1 nM DHT with or without 1 μM OH-F (detected by microscopy) (B). (C) 847Y, 877A, and 877S AR mutants found in hormone-refractory prostate cancer allow progesterone (Prog), estradiol (Est), or OH-F to induce CFP–AR–YFP [and AR (32)] transcriptional activation of an MMTV-luciferase reporter in HEK293 cells. All mutants responded normally to DHT. Cells were cultured in quadruplicate in the presence or absence of the indicated ligands for 24 h. (D) FRET assays of the same mutants were performed after 24 h of culture in the presence of the indicated ligands. Error bars represent the SEM.
Fig. 5.
The 23FQNLF27 motif promotes NTD–LBD interactions within a single molecule. (A) Deletion of the 23FQNLF27 motif had no effect on the DHT dose–response (EC50 ≈ 1–3 nM) of CFP–AR–YFP FRET in HEK293 cells (detected by FPR). The overall level of CFP–ARΔ–YFP was lower than CFP–AR–YFP FRET. (B) Comparison of CFP–AR/AR–YFP and CFP–ARΔF/ARΔF–YFP association in HeLa cells. AR and ARΔF achieved maximum nuclear FRET at similar acceptor/donor ratios, indicating an equivalent dimerization affinity. Maximal FRET levels were reduced for the ΔF mutants, indicating an altered dimer structure. (C and D) HeLa cells transfected with CFP–AR–YFP (n = 579 total cells at all time points) or CFP–ARΔF–YFP (n = 347 total cells) were imaged by fluorescence microscopy within the nucleus (C) or cytoplasm (D) at various time points after addition of 100 nM DHT. Error bars represent the SEM.
FRET Collection and Analysis. For FRET detection by microscopy, acceptor, donor, and FRET images were collected as described in refs. 24 and 25. For each cell, three fluorescence channels were collected: the acceptor channel (YFP excited with 496- to 505-nm light; YFP fluorescence collected at 520–550 nm); the donor channel (excited with 431- to 440-nm light; collected at 455–485 nm); and the FRET channel (excited with 431- to 440-nm light; collected at 520–550 nm). Cells separately expressing CFP–AR and AR–YFP established the individual contributions of the donor and acceptor fluorophores to each channel. Following the correction for the amount of background signal and for fluorescence contributed by the acceptor (YFP) to the FRET spectra, the level of FRET was established as the amount of FRET relative to donor fluorescence (FRET/donor). For tracking FRET over time, cells were maintained at 37°C during data collection using a stage warmer (Brooks Industries, Lake Villa, IL). Microscopy data were collected from cells expressing very low amounts of fluorescent protein-tagged AR. High-expressing cells were avoided. Comparing fluorescence values and amounts of a protein estimated by Western blot allowed us to roughly calibrate our equipment. We estimate that <50,000 AR–YFP were present in each of the >5,000 cells imaged in this study. Remarkably similar results were obtained by FPR, which relied on higher expression of AR for sufficient signal.
For FRET detection on the FPR (Safire, Tecan, Durham, NC), cells were cultured in black, clear-bottomed 96-well plates (Costar) as described in ref. 26, fixed, and read on the day of harvest. Measurements were taken from the bottom of the plate with the following settings: YFP, excitation at 485 nm/emission at 527 nm; CFP, excitation at 435 nm/emission at 485 nm; and FRET, excitation at 435 nm/emission at 527 nm. The excitation was performed ±2.5 nm, the emission was recorded ±6 nm. Each plate contained an untransfected cell control (background) and cells transfected with pure CFP and YFP expression plasmids (pECFP-C1 and pEYFP-C1; Clontech). Each data point was collected in quadruplicate. FRET/donor ratios were calculated after background subtraction and correction for acceptor (YFP) contribution into the FRET spectrum.
Luciferase Assays. HEK293 cells (n = 1.5 × 106) were transfected in 3.5-cm dishes with a total 0.5 μg of MMTV-luciferase reporter and 0.05 μg of AR expression plasmid. Cells (n = 100,000) were subsequently replated in quadruplicate into a 96-well plate and cultured overnight with hormone. The following day, cells were washed and lysed. Lysate (5 μl) was read in 50 μlof1× luciferase substrate mix (Pharmingen) on a luminometer (Ultra, Tecan).
Western Blots. HeLa cells transfected with the indicated expression vectors were grown with or without 100 nM dihydrotestorone (DHT). After 24 h, whole-cell lysates or nuclear/cytoplasm extracts were prepared as described in ref. 24. Equivalent amounts of extract (15–20 μg) were resolved with SDS/PAGE and blotted with N-20 antibody (Santa Cruz Biotechnology) at 1:2,000. Bands were quantified with a digital imaging system (Alpha Innotech, San Leandro, CA).
Results
AR-Fluorescent Protein Fusions Preserve Basic Functional Activity. To investigate AR structure and function in the cellular environment, the AR was tagged with fluorescent proteins for expression in cells. The cDNA for CFP or YFP was fused to the amino and/or carboxy termini of AR to create CFP–AR, AR–YFP, and CFP–AR–YFP (Figs. 1A and 6A). The addition of the fluorescent proteins to both ends of the AR did not affect transport induced by the agonist ligand DHT. As expected (27, 28), AR and CFP–AR–YFP transiently expressed in HeLa cells were predominantly but not exclusively cytoplasmic in the absence of ligand and were transported into the nucleus upon DHT addition (Figs. 1B and 6B). Control blots with antibodies against tubulin or histone H1 demonstrated clean separation of the cytoplasmic and nuclear fractions (data not shown). Quantitative fluorescence imaging from multiple cells confirmed that CFP–AR–YFP and the singly fused CFP–AR and AR–YFP migrated at similar rates to the nuclei of HeLa cells after ligand addition (Fig. 6C). Finally, addition of DHT to HEK293 cells expressing the AR–YFP, CFP–AR, and CFP–AR–YFP fusion proteins activated reporter expression from a MMTV promoter similarly to that of unfused AR (Fig. 1C). Thus, basic function was preserved in all three AR fusion proteins, although the effects of CFP or YFP fusion on some other aspects of AR function can never be ruled out.
Fig. 1.
AR-fluorescent protein fusions have normal responses to ligand. (A) Diagram of AR fusions to CFP and YFP. (B) An anti-AR antibody was used to probe nuclear (nuc) and cytoplasmic (cyto) extracts of HeLa cells expressing AR or CFP–AR–YFP (C-AR-Y) after treatment with DHT for different amounts of time. Quantification of band intensities (see Fig. 6B for blot) shows similar rates of ligand-induced nuclear transport for both. (C) MMTV-luciferase reporter activity increased when HEK293 cells expressing AR or AR fluorescent protein fusions were treated for 24 h with 100 nM DHT. The fold activity upon DHT addition is shown for each AR. Error bars represent the SEM.
DHT Induces AR Self-Association Predominantly Within the Nucleus. AR dimerization is required for AR transcription activity (17). Therefore, AR dimers must be present in the cell nucleus, although it is unknown whether they form initially in the cytoplasm. We coexpressed CFP–AR and AR–YFP within HeLa cells and used quantitative FRET microscopy to determine whether the CFP and YFP were brought close enough (<80 Å) by AR interaction or association to allow efficient energy transfer. Transfer of energy from a donor fluorophore (CFP) to an acceptor fluorophore (YFP) results in decreased CFP (donor) fluorescence and increased YFP fluorescence upon CFP excitation (FRET). Thus, if energy is transferred from CFP–AR to AR–YFP, the FRET/donor fluorescence ratio (corrected for the contributions of the acceptor fluorophore to each fluorescence image; see Materials and Methods) increases relative to the FRET/donor fluorescence of CFP alone. Processed FRET/donor images from individual cells are shown in Fig. 2A. Image processing introduces negative number errors, and the FRET data presented in all subsequent figures was more accurately calculated from large nuclear or cytoplasmic regions within the raw images.
Energy transfer was quantified in hundreds of CFP–AR- and AR–YFP-coexpressing cells. In nuclei of cells treated with DHT for 20 min, energy transfer from CFP to YFP increased with YFP amount until sufficient AR–YFP was present to saturate interaction with CFP–AR (Fig. 2B). The relationship of FRET amount to AR–YFP and CFP–AR amount fit well (R2 = 0.8) to an equation (Fig. 2B, black line) that described an interaction between two molecules (24). For cells not treated with ligand (Fig. 2B, dotted line), there was no FRET, and the mathematical relationship suggesting a bimolecular interaction was not observed (R2 = 0.1). Thus, in the absence of ligand, very few AR were detected in which the CFP and YFP were close enough (≈80 Å) to allow efficient energy transfer.
The amounts of FRET at saturating AR–YFP were determined in the nuclei and cytoplasm of cells treated with no ligand or with 10–8 M DHT for 20 min (Fig. 2C). DHT induced a strong increase in FRET that was significantly higher (P < 0.01) in the cell nucleus than in the cytoplasm. Similar analysis of intermolecular interaction was conducted with nuclear PPARγ2, which does not homodimerize (29). CFP–PPARγ2 and PPARγ2–YFP showed no association, even when incubated with ligand (Fig. 2B, gray line). Thus, a highly specific association of AR positions CFP and YFP close enough to permit FRET (<80 Å). Given the small distances involved, this FRET signal represents either direct dimerization between ARs or their simultaneous interaction with another factor that positions AR monomers not much more than a protein domain apart from each other. For simplicity, we shall refer to this self-association as “dimer FRET,” particularly given the demonstrated bimolecular nature of the interaction.
Ligand Repositions the NTD and LBD Within AR. Conformational changes within AR monomers might precede dimerization. To investigate ligand-regulated structural changes within AR, we measured FRET between fluorophores attached to the same AR molecule (CFP–AR–YFP). In the absence of ligand, there was no significant FRET signal in the nucleus or cytoplasm of HeLa cells expressing CFP–AR–YFP (Fig. 2D). This finding indicated that the unliganded AR monomer was in an extended conformation and not self-associated. DHT induced CFP–AR–YFP FRET not only in the nucleus but also in the cytoplasm. Similar DHT-induced FRET in the cytoplasm and nuclei of CFP–AR–YFP-expressing cells (Fig. 2D) contrasted with lower dimer FRET in the cytoplasm (Fig. 2C), suggesting that portions of the DHT-induced cytoplasmic FRET from CFP–AR–YFP arose from an intramolecular event that brought the NTD and LBD into close proximity.
Intramolecular Folding of AR Precedes Association in the Nucleus. To establish whether intramolecular folding induced by DHT precedes or occurs simultaneously with dimerization in the nucleus, we measured the relative rates of ligand-induced folding and dimerization by using time-lapse studies of single cells. Intramolecular CFP–AR–YFP FRET and CFP–AR/AR–YFP dimer FRET were measured within the cytoplasm and nucleus of HeLa cells at 1-min intervals before and after addition of 10 nM DHT (Fig. 3A). To ensure that dimer FRET and intramolecular FRET were detected with equivalent sensitivity, the dimer FRET studies were conducted with cells expressing high amounts of AR–YFP relative to CFP–AR (see Fig. 2B).
CFP–AR–YFP FRET increased rapidly after the addition of DHT, regardless of subcellular localization. The time required to reach half-maximal FRET (t1/2) was ≈3.5 min in the nucleus and in the cytoplasm. The association kinetics of CFP–AR and AR–YFP (Fig. 3A) in the nucleus were significantly slower, with a t1/2 of ≈9.5 min. We observed a similar, slower increase in FRET between CFP–AR-CFP and YFP-AR–YFP relative to CFP–AR–YFP in nucleus (Fig. 7) and cytoplasm. This finding ruled out the possibility that the fast accrual of CFP–AR–YFP FRET arose from a differential dimerization or transport kinetics of the dual-tagged AR in the nuclei of these cells. These results collectively imply that a rapid change in AR monomer structure precedes AR self-association.
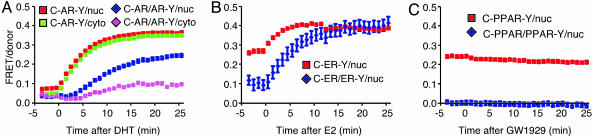
Specificity of NTD–LBD Interactions Among NRs. To investigate whether ligand-induced rapid intramolecular folding was a common feature of NRs, we studied the intramolecular and dimerization kinetics for two other NRs. Unlike AR, ERα and PPARγ2 are predominantly nuclear in the absence of ligand. In the absence of estradiol, almost no ERα dimers were detected (Fig. 3B), just as nuclear-localized AR did not produce significant dimer FRET before ligand binding. However, unliganded CFP–ERα–YFP produced significant intramolecular FRET, indicating that, in contrast to AR, the fluorescent proteins attached to the NTD and LBD of unliganded ERα were closer than 80 Å. Upon estradiol addition, there was a rapid, further increase in intramolecular CFP–ERα–YFP FRET (t1/2 ≈ 1.2 min) that significantly preceded the acquisition of dimer CFP–ERα/ERα–YFP FRET (t1/2 ≈ 4.7 min). Thus, as with AR, ligand binding to ERα induced a rapid intramolecular fold and a slower dimerization. In contrast, PPARγ2, which biochemical evidence indicates does not form homodimers (29), showed no ligand-induced FRET from intramolecular folding or association (Fig. 3C), even though ligand addition caused CFP–PPARγ2- and PPARγ2–YFP-dependent activation of a PPAR-responsive reporter (data not shown). Thus, rapid intramolecular folding of the AR NTD and LBD followed by their close association within a presumed dimer pair is a feature shared with some but not all NRs.
NTD–LBD Folding Is Blocked by an AR Antagonist Ligand. To examine the functional significance of the NTD–LBD interactions, we tested the effects of a well characterized antagonist of AR transcription on intramolecular and dimer FRET. HEK293 cells were transfected with CFP–AR–YFP, plated in quadruplicate in a 96-well dish, and exposed to 10 nM DHT and increasing amounts of hydroxyflutamide (OH-F). After 24 h, cells were fixed and read on a FPR as described in ref. 26. The FPR rapidly measures thousands of cells and complements the more laborious microscopy-based technique. The strong FRET signal induced by DHT was competed effectively by excess OH-F in a dose-dependent manner (Fig. 4A). When measured by FRET microscopy, OH-F also inhibited CFP–AR–YFP FRET in the nuclear and cytoplasmic compartments of HeLa cells (Fig. 4B). Thus, OH-F binding prevents association of the NTD and LBD within a single molecule. OH-F also reduced cytoplasm-tonucleus transport of CFP–AR–YFP (Fig. 8A) and did not promote FRET between AR–YFP and CFP–AR (Fig. 8B). Having observed that a transcriptional antagonist blocked intramolecular folding, we next examined the extent to which the FRET signal would predict AR transcriptional activity.
FRET Measurement of NTD–LBD Interaction Correlates with AR Transcriptional Activity. AR mutations have been isolated from hormone-refractory prostate tumors that permit AR transcriptional regulation in response to heterologous ligands, including OH-F, progesterone, and estrogen (30–33). Three of these AR mutants (H874Y, T877A, and T877S) were subcloned into CFP–AR–YFP and transiently expressed in HEK293 cells together with an MMTV-luciferase reporter. Transfected cells were cultured in the presence of 0, 10, or 100 nM DHT, progesterone, estrogen, or OH-F. As expected, wild-type CFP–AR–YFP responded strongly to DHT not only in the transcription assay (Fig. 4C) but also in the FPR-based FRET assay (Fig. 4D), whereas its responses to heterologous ligands were moderate (estrogen and progesterone) or minimal (OH-F). In contrast, the AR mutants induced parallel estrogen, progesterone, and OH-F activations of reporter gene activity and FRET. FRET microscopy studies in HeLa cells confirmed that these AR mutants partially restored AR NTD–LBD folding in the nucleus and in the cytoplasm (Fig. 9). The close parallel between the FRET-based biophysical readout (a direct measure of ligand-induced conformational change) and the reporter gene activation (an indirect measure of ligand-induced conformational change) suggested that the FRET signal reflected a transcriptionally competent AR conformation.
The 23FQNLF27 Motif Orients the AR LBD and NTD Within Monomers. AR intramolecular folding may involve the previously described in vitro interaction between NTD and LBD fragments. That NTD–LBD interaction is mediated largely by the 23FQNLF27 motif and other motifs in the AR NTD (20). However, it remains unclear whether 23FQNLF27 participates in an intramolecular (folding) interaction or an intermolecular interaction.
We deleted the five amino acids comprising 23FQNLF27 and tested the effect of this mutation (ARΔF) on AR structure. First, we used the FPR to compare the DHT dose–response for cells expressing CFP–AR–YFP or CFP–ARΔF–YFP. The EC50 of DHT for the induction of AR and ARΔF FRET was 1–3 nM (Fig. 5A). Introduction of alanine residues in place of phenylalanine and leucine (23AQNAA27) within the 23FQNLF27 motif also reduced FRET in a manner similar to the ΔF mutation (Fig. 10, which is published as supporting information on the PNAS web site). Thus, disruption of 23FQNLF27did not impact the concentration of DHT required to achieve a steady-state conformational change in cells incubated for prolonged times with ligand.
The 23FQNLF27 motif might participate in an NTD–LBD interaction between separate ARs to stabilize dimerization. We tested this possibility directly by measuring the intracellular association of AR and ARΔF using CFP–AR/AR–YFP FRET. No FRET was detected in the absence of DHT, but FRET was detected in DHT-treated cell nuclei for AR and ARΔF. Comparison of the donor/acceptor ratios to FRET indicated that both sets of data fit well with a curve that describes an interaction between two molecules (Fig. 5B), and the amounts of YFP-labeled AR required to achieve half-maximal FRET were statistically the same for the wild-type AR (2.14 ± 0.24, 95% CI) and ARΔF (1.72 ± 0.51), indicating similar self-affinities. Thus, deletion of the 23FQNLF27 motif did not affect intracellular AR dimerization, and models of AR dimerization that assume a reliance on the interaction of FQNLF with AF-2 between monomers are not supported by our findings. However, the maximal FRET level was significantly higher for the wild-type AR than for ARΔF, which demonstrates that the FQNLF motif is required to properly orient the NTD and LBD domains between members of a dimer. We also found that deletion of 23FQNLF27 reduced FRET from CFP–AR–YFP in the cytoplasm (Fig. 5C) and in the nucleus (Fig. 5D). Thus, the 23FQNLF27 motif is required for maximal association of the NTD and LBD within AR and between AR molecules within a dimer but does not affect dimerization affinity.
Discussion
Live-cell FRET was used to precisely identify and characterize a ligand-induced NTD–LBD association within full-length AR. The agonist-induced conformation correlated well with transcriptional activity of the AR, in particular with those activities associated with acquisition of response to heterologous ligands in hormone-refractory prostate cancers (32, 34). The results are consistent with a model in which the unliganded AR exists in a relatively unfolded state in the cytoplasm and nucleus. After agonist binding, the AR rapidly converts to an active form in which the NTD and LBD within a single AR come into close association. The significant time periods (≈3.5 min) required for ligand-induced conformation changes may reflect the transit time of the ligand to the receptor or may suggest the involvement of other accessory factors in this process. In addition, the conformational change is followed ≈6 min later by AR association, which occurs more rapidly and efficiently in the nucleus. These findings imply that dimerization requires additional and compartment-specific molecular events.
Distinct NTD-LBD Interactions Among NRs. The NTD–LBD interactions of AR that follow ligand binding constitute one type of conformation change within the NR family. Estradiol also induced an NTD–LBD fold within CFP–ERα–YFP. In contrast, agonist had no effect on the interdomain structure of CFP–PPARγ2–YFP. Both ERα and PPARγ2 exhibited intramolecular FRET in the absence of ligand, whereas AR did not. It is possible that the higher baseline FRET levels of the unliganded ERα and PPARγ2 simply reflect the reduced distance between the NTD and LBD of ERα (595 aa) and PPARγ2 (505 aa) relative to AR (920 aa). Conversely, these different FRET levels might also represent distinct conformations specific to each unliganded NR.
It was recently reported that estradiol did not induce a conformational shift in CFP–ERα–YFP expressed in U2OS cells, whereas tamoxifen did (35). This study contrasts with our findings in HeLa cells and may indicate distinct ER structures or interactions under different cell environments and conditions. For AR, which we studied in detail, FRET microscopy yielded similar results in HeLa and HEK293 cells. However, it remains to be determined whether analyses in other cell types might reveal distinct, cell-specific AR conformers.
23FQNLF27-Dependent and Independent NTD–LBD Associations. Prior work indicated that a NTD–LBD interaction in AR depended on the 23FQNLF27 motif (14, 17–21). The data described here extends those studies by distinguishing between intramolecular and intermolecular events occurring within or between full-length AR molecules and by providing temporal and subcellular resolution of these processes. Prior in vitro studies suggested that the 23FQNLF27 motif may not be the only contributor to NTD–LBD interaction (20). Indeed, we found that deletion of 23FQNLF27 reduced but did not eliminate intramolecular FRET (Fig. 5), and it is likely that additional NTD–LBD interactions may aid or partially compensate the 23FQNLF27-dependent fold. Moreover, we observed a similar NTD–LBD fold for ERα, which does not contain an obvious FQNLF motif, supporting the idea that other NR domains can participate in ligand-activated folding. Because the 23FQNLF27 motif is well described to interact with the coactivator binding pocket of the AR LBD (14), an intriguing possibility is that the rapid and relatively stable intramolecular fold prevents or modulates cofactor binding to the AR LBD, as has been recently suggested (14). This idea may help account for prior studies suggesting that, unique among the NRs, transcriptional activation by AR at certain promoters does not require cofactor binding to the LBD (9, 19). Given the association of this intramolecular fold with promiscuous responses in hormone-refractory AR LBD mutants, this intramolecular event may be a therapeutic target of considerable importance.
Conformation as a Measure of NR Activity. Traditional analysis of NR function has been based on biochemistry (e.g., ligand binding, DNA binding, cofactor binding), cell trafficking studies, and measures of transcriptional activity using reporter genes. Crystallographic studies of isolated NR LBDs have identified ligand-specific conformational changes in the LBD (13), but little is known of the structural basis of domain interactions within a full-length NR. An important new therapeutic strategy could be to target NR conformation through the allosteric modulation of domain interactions, distinct from competitive pharmacologic agents. Yet until now it has not been possible to directly measure NR conformational changes in the relatively physiologic context of an intact cell.
The FRET-based method described here offers opportunities to study the regulation of NR conformation in different cell types and possibly in different tissues of live animals. Combined with genetic manipulations in model organisms, FRET-band analyses may prove very useful for identifying cellular events that modify NR structure or protein interactions within the intracellular environment. Our finding that analysis of FRET via FPR faithfully reproduced many of the details uncovered by more laborious microscopic analyses suggests that FRET may provide a complementary, high-throughput method for detecting cellular or pharmacological events that specifically inhibit or enhance NR ligand-induced conformation change. This high-throughput capability may help identify drugs that operate differently in specific cell types or that induce alterations in the kinetics of conformation changes or protein interactions. Any or all of these approaches could have profound impact on understanding previously unrecognized pathways involved in NR action and may speed the discovery of new therapies for human diseases (2).
Supplementary Material
Acknowledgments
We thank Junlian Hu for expert technical assistance and Robin Chedester for assistance in manuscript preparation. This work was supported by U.S. Department of Defense Grants DAMD17-01-1-0190 and PC040777 (to F.S.), National Institutes of Health Grants R21 062782 (to F.S.) and R21 NS45350 (to M.I.D.), the Prostate Cancer Foundation (M.I.D.), Muscular Dystrophy Association Grant MDA3408 (to M.I.D.), and the Sandler Family Supporting Foundation (M.I.D.).
Author contributions: F.S. and M.I.D. designed research; F.S., X.C., M.G., A.A.K.M., and M.I.D. performed research; F.S., X.C., S.B., M.S.C., and M.I.D. contributed new reagents/analytic tools; F.S., X.C., M.S.C., J.N.M., and M.I.D. analyzed data; and F.S., M.S.C., J.N.M., and M.I.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AR, androgen receptor; CFP, cyan fluorescent protein; DHT, dihydrotestorone; ER, estrogen receptor; FPR, fluorescence plate reader; LBD, carboxy-terminal domain; MMTV, mouse mammary tumor virus; NR, nuclear receptor; NTD, amino-terminal domain; OH-F, hydroxyflutamide; PPARγ2, peroxisome proliferator-activated receptor-γ2; YFP, yellow fluorescent protein.
References
- 1.Katzenellenbogen, J. A., O'Malley, B. W. & Katzenellenbogen, B. S. (1996) Mol. Endocrinol. 10, 119–131. [DOI] [PubMed] [Google Scholar]
- 2.Gronemeyer, H., Gustafsson, J.-A. & Laudet, V. (2004) Nat. Rev. Drug. Discovery 3, 950–964. [DOI] [PubMed] [Google Scholar]
- 3.Chlebowski, R. T., Hendrix, S. L., Langer, R. D., Stefanick, M. L., Gass, M., Lane, D., Rodabough, R. J., Gilligan, M. A., Cyr, M. G., Thomson, C. A., et al. (2003) J. Am. Med. Assoc. 289, 3243–3253. [DOI] [PubMed] [Google Scholar]
- 4.Taplin, M. E. & Balk, S. P. (2004) J. Cell Biochem. 15, 483–490. [DOI] [PubMed] [Google Scholar]
- 5.Osborne, C. K. (1998) N. Engl. J. Med. 339, 1609–1618. [DOI] [PubMed] [Google Scholar]
- 6.Geller, J. (1993) Cancer 71, S1039–S1045. [Google Scholar]
- 7.Baum, M., Budzar, A. U., Cuzick, J., Forbes, J., Houghton, J. H., Klijn, J. G., Sahmoud, T. & Group., A. T. (2002) Lancet 359, 2131–2139. [DOI] [PubMed] [Google Scholar]
- 8.McKenna, N. J. & O'Malley, B. W. (2002) Cell 108, 465–474. [DOI] [PubMed] [Google Scholar]
- 9.Simental, J. A., Sar, M., Lane, M. V., French, F. S. & Wilson, E. M. (1991) J. Biol. Chem. 266, 510–518. [PubMed] [Google Scholar]
- 10.Metzger, D., Ali, S., Bornert, J. M. & Chambon, P. (1995) J. Biol. Chem. 270, 9535–9542. [DOI] [PubMed] [Google Scholar]
- 11.Kumar, R. & Thompson, E. B. (2003) Mol. Endocrinol. 17, 1–10. [DOI] [PubMed] [Google Scholar]
- 12.Danielian, P. S., White, R., Lees, J. A. & Parker, M. G. (1992) EMBO J. 11, 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng, W., Ribeiro, R. C., Wagner, R. L., Nguyen, H., Apriletti, J. W., Fletterick, R. J., Baxter, J. D., Kushner, P. J. & West, B. L. (1998) Science 280, 1747–1749. [DOI] [PubMed] [Google Scholar]
- 14.He, B., Gampe, R. T., Kole, A. J., Hnat, A. T., Stanley, T. B., An, G., Stewart, E. L., Kalman, R. I., Minges, J. T. & Wilson, E. M. (2004) Mol. Cell 16, 425–438. [DOI] [PubMed] [Google Scholar]
- 15.Matias, P. M., Carrondo, M. A., Coelho, R., Thomaz, M., Zhao, X. Y., Wegg, A., Crusius, K., Egner, U. & Donner, P. (2002) J. Med. Chem. 45, 1439–1446. [DOI] [PubMed] [Google Scholar]
- 16.Sack, J. S., Kish, K. F., Wang, C., Attar, R. M., Kiefer, S. E., An, Y., Wu, G. Y., Scheffler, J. E., Salvati, M. E., Krystek, S. R. J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 4904–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong, C. I., Zhou, Z. X., Sar, M. & Wilson, E. M. (1993) J. Biol. Chem. 268, 19004–19012. [PubMed] [Google Scholar]
- 18.Kemppainen, J. A., Langley, E., Wong, C. I., Bobseine, K., Kelce, W. R. & Wilson, E. M. (1999) Mol. Endocrinol. 13, 440–454. [DOI] [PubMed] [Google Scholar]
- 19.He, B., Kemppainen, J. A., Voegel, J. J., Gronemeyer, H. & Wilson, E. M. (1999) J. Biol. Chem. 274, 37219–37225. [DOI] [PubMed] [Google Scholar]
- 20.He, B., Kemppainen, J. A. & Wilson, E. M. (2000) J. Biol. Chem. 275, 22986–22994. [DOI] [PubMed] [Google Scholar]
- 21.He, B., Lee, L. W., Minges, J. T. & Wilson, E. M. (2002) J. Biol. Chem. 277, 25631–25639. [DOI] [PubMed] [Google Scholar]
- 22.Kraus, W. L., McInerney, E. M. & Katzenellenbogen, B. S. (1995) Proc. Natl. Acad. Sci. USA 92, 12314–12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, J., Campbell, R. E., Ting, A. Y. & Tsien, R. Y. (2002) Nat. Rev. Mol. Cell. Biol. 3, 906–918. [DOI] [PubMed] [Google Scholar]
- 24.Schaufele, F., Wang, X., Liu, X. & Day, R. N. (2003) J. Biol. Chem. 278, 10578–10587. [DOI] [PubMed] [Google Scholar]
- 25.Weatherman, R. V., Chang, C.-Y., Clegg, N. J., Carroll, D. C., Day, R. N., Baxter, J. D., McDonnell, D. P., Scanlan, T. S. & Schaufele, F. (2002) Mol. Endocrinol. 16, 487–496. [DOI] [PubMed] [Google Scholar]
- 26.Pollitt, S. K., Pallos, J., Shao, J., Desai, U. A., Ma, A. A., Thompson, L. M., Marsh, J. L. & Diamond, M. I. (2003) Neuron 40, 685–694. [DOI] [PubMed] [Google Scholar]
- 27.Gregory, C. W., Johnson, R. T. J., Mohler, J. L., French, F. S. & Wilson, E. M. (2001) Cancer Res. 61, 2892–2898. [PubMed] [Google Scholar]
- 28.Nightingale, J., Chaudhary, K. S., Abel, P. D., Stubbs, A. P., Romanska, H. M., Mitchell, S. E., Stamp, G. W. & Lalani, e.-N. (2003) Neoplasia 5, 347–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tontonoz, P., Graves, R. A., Budavari, A. I., Erdjument-Bromage, H., Lui, M., Hu, E., Tempst, P. & Spiegelman, B. M. (1994) Nucl. Acids Res. 22, 5628–5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veldscholte, J., Ris-Stalpers, C., Kuiper, G. G., Jenster, G., Berrevoets, C., Claassen, E., van Rooij, H. C., Trapman, J., Brinkmann, A. O. & Mulder, E. (1990) 173, 534–540. [DOI] [PubMed]
- 31.Taplin, M. E., Rajeshkumar, B., Halabi, S., Werner, C. P., Woda, B. A., Picus, J., Stadler, W., Hayes, D. F., Kantoff, P. W., Vogelzang, N. J., et al. (2003) J. Clin. Oncol. 21, 2673–2678. [DOI] [PubMed] [Google Scholar]
- 32.Taplin, M. E., Bubley, G. J., Shuster, T. D., Frantz, M. E., Spooner, A. E., Ogata, G. K., Keer, H. N. & Balk, S. P. (1995) N. Engl. J. Med. 332, 1393–1398. [DOI] [PubMed] [Google Scholar]
- 33.Gottlieb, B., Beitel, L. K., Wu, J. H. & Trifiro, M. (2004) Hum. Mutat. 23, 527–533. [DOI] [PubMed] [Google Scholar]
- 34.Fenton, M. A., Shuster, T. D., Fertig, A. M., Taplin, M. E., Kolvenbag, G., Bubley, G. J. & Balk, S. P. (1997) Clin. Cancer Res. 3, 1383–1388. [PubMed] [Google Scholar]
- 35.Michalides, R., Griekspoor, A., Balkenende, A., Verwoerd, D., Janssen, L., Jalink, K., Floore, A., Velds, A., van`t Veer, L. & Neefjes, J. (2004) Cancer Cell 5, 597–605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.