Abstract
The ATM/ATR kinases and the Mre11 (Mre11-Rad50-Nbs1) protein complex are central players in the cellular DNA damage response. Here we characterize possible interactions between Aspergillus nidulans uvsBATR and the Mre11 complex (scaANBS1). We demonstrate that there is an epistatic relationship between uvsBATR, the homolog of the ATR/MEC1 gene, and scaANBS1, the homolog of the NBS1/XRS2 gene, for both repair and checkpoint functions and that correct ScaANBS1 expression during recovery from replication stress depends on uvsBATR. In addition, we also show that the formation of UvsC foci during recovery from replication stress is dependent on both uvsBATR and scaANBS1 function. Furthermore, ScaANBS1 is also dependent on uvsBATR for nuclear focus formation upon the induction of DNA double-strand breaks by phleomycin. Our results highlight the extensive genetic interactions between UvsB and the Mre11 complex that are required for S-phase progression and recovery from DNA damage.
The DNA damage response is a protective mechanism that ensures the maintenance of genome integrity during cellular reproduction. DNA damage takes several general forms, including single- and double-strand breaks (DSBs), base damage, and DNA-protein cross-links, which cause replication fork progression blockage and can generate secondary lesions such as DSBs. In complex genomes of higher vertebrates, DNA secondary structures such as hairpins and G4 tetraplexes that spontaneously form at palindromic or repeated sequences can give rise to DSBs as a consequence of the processing of the stalled replication forks (12, 56). DNA synthesis must be restored at sites where replication forks have been damaged or blocked to allow the establishment of a bona fide replication fork to conclude S phase and avoid DSB formation (53). If left unrepaired, DNA damage can result in cell cycle arrest, cell death, and if repaired incorrectly, the loss of genetic information or the accumulation of mutations that lead to cancer in multicellular organisms. DNA replication, gene transcription, DNA repair, and cell cycle checkpoints must all interlink to promote cell survival following DNA damage (38).
The two main signal transduction pathways that respond to DNA damage, namely, the ATM (mutated in ataxia telangiectasia [AT]) and ATR (ATM-Rad3-related; this pathway was recently linked to Seckel syndrome) pathways, are conserved across evolution (1, 46, 55, 74, 76). The ATM pathway responds to the presence of double-strand breaks (DSBs). The ATR pathway also responds to DSBs, but more slowly than ATM. In addition, the ATR pathway can respond to agents that interfere with the function of replication forks, such as hydroxyurea (HU), UV light, and DNA-alkylating agents such as methyl methanesulfonate (MMS) (45, 47). The ATM/ATR kinases phosphorylate and activate proteins in the signal transduction pathways that ultimately interface with the Cdk/cyclin machinery (1). ATR and ATM may possess both overlapping and nonredundant roles in the regulation of the DNA damage response. For instance, the overexpression of ATR complements the radioresistant DNA synthesis phenotype of an AT cell line (9). In the yeast Saccharomyces cerevisiae, the homologues of ATR/ATM, MEC1/TEL1, have functionally redundant roles in both DNA damage repair and telomere length regulation (11). These kinases regulate the activation of two downstream protein kinases, Chk1 and Chk2 (57).
The Mre11 (Mre11-Rad50-Nbs1) protein complex has emerged as a central player in the human cellular DNA damage response, and recent observations suggest that these proteins are at least partially responsible for the linking of DNA damage detection to DNA repair and cell cycle checkpoint functions (51). In humans, the loss of the NBS1 gene is associated with the Nijmegen breakage syndrome, a rare autosomal recessive disorder that belongs to the group of inherited human chromosomal instability syndromes that includes Bloom's syndrome, Fanconi's anemia, and AT. All of these disorders are characterized by spontaneous chromosomal instability, immunodeficiency, and a predisposition to cancer, but they have distinct cytogenetic features and sensitivities to specific DNA-damaging agents (for a review, see references 16, 61, and 62). The NBS1 gene product, nibrin/p95, is part of the Mre11/Rad50 nuclear foci that form at sites of DSBs (6). The Mre11 complex possesses manganese-dependent single-stranded DNA endonuclease and 3′-to-5′ exonuclease activities (65). In humans and yeast, Mre11, Rad50, and Nbs1/Xrs2 assemble into large complexes. These proteins also appear to play a role in telomere maintenance. In addition, they are implicated in the cell's checkpoint response to the presence of DSBs (14). During meiosis, these three proteins are required not only for the resection of DSBs but also to create meiotic DSBs (51). Studies of mammalian and yeast cells have established that the Mre11 complex controls the ATM/Tel1 signaling pathway (4, 13, 32, 52, 66). ATM activation correlates with autophosphorylation on Ser1981 (2), and this phosphorylation requires functions of the Mre11 complex (7, 67). It is possible that the Mre11 complex modulates the substrate recognition of ATM by a direct interaction (37). In addition, the Mre11 complex controls the accumulation of ATM at DSB lesions (36). Recently, Stiff et al. (63) showed that Nbs1 is required for ATR-dependent phosphorylation events. Nakada et al. (43) have demonstrated that the Mre11 complex functions together with exonuclease 1 (Exo1) in the activation of the Mec1ATR signaling pathway after DNA damage and replication blockage. During DSB- and UV-induced checkpoints, the Mre11 complex and Exo1 collaborate to produce long single-stranded DNA tails at DSB ends and to promote Mec1 association with the DSBs.
We have used Aspergillus nidulans as a model system to genetically characterize the cellular response to DNA damage (5, 17, 18, 59; for a review, see references 23, 24, and 34). For this study, we examined possible interactions between A. nidulans uvsBATR, the homolog of the ATR/MEC1 gene, and the Mre11 complex (using scaANBS1, the homolog of the NBS1/XRS2 gene). Our observations suggest the existence of a complex epistatic relationship whereby both UvsBATR and the Mre11 complex contribute to the repair of DSBs and the DNA replication stress response.
MATERIALS AND METHODS
Strains and media.
The A. nidulans strains used for this study are described in Table 1. The media used were of two basic types, i.e., complete and minimal. The complete media comprised the following three variants: YAG (2% glucose, 0.5% yeast extract, 2% agar, trace elements), YUU (YAG supplemented with 1.2 g/liter [each] of uracil and uridine), and liquid YG or YG+UU medium with the same composition (but without agar). The minimal media were a modified minimal medium (MM; 1% glucose, original high-nitrate salts, trace elements, 2% agar, pH 6.5) and minimal medium without glucose (MC). Trace elements, vitamins, and nitrate salts were included as described by Kafer (32) (details are available upon request). Standard genetic techniques for A. nidulans were used for all strain constructions (32).
TABLE 1.
A. nidulans strains used for this study
| Strain | Genotype | Reference or source |
|---|---|---|
| GR5 | pyrG89 wA3 pyroA4 | FGSC A773 |
| AML8 | pyrG89 pabaA1 argB2 yA2 | 30 |
| AAH14 | pyrG89 pabaA1 yA2 ΔuvsB | 30 |
| T20 | pyrG89 wA3 pyroA4 scaA::pyr4 | 5 |
| GG11 | pyrG89 wA3 pyroA4 scaA::pyr4 ΔuvsB::argB | 59 |
| ASH270 | pyrG89 pabaA1 yA2 uvsB110 | 31 |
| sca299-16 | pyrG89 pabaA1 yA2 scaA1 | 5 |
| ASG17 | wA1 uvsC114 uvsC::FLAG | 23 |
| GHG4 | scaA1 uvsC114 uvsC::FLAG | This work |
| ACS2 | pabaA1 yA2 uvsB110 uvsC114 uvsC::FLAG | This work |
| BAGS6 | pabaA1 yA2 ΔuvsB alcA::gfp::scaA | This work |
| UAGS21 | alcA::gfp::scaA | This work |
The viability of growing hyphae was measured as described by Bruschi et al. (5). A suspension of conidiospores (approximately 106 conidia) was spread onto a YUU plate by use of a sterile glass spreader and incubated for 24 h at 37°C to produce a nonsporulating mycelial “mat.” Mycelial “plugs” were cut from the mats with the wide end of a sterile Pasteur pipette. The plugs (four samples of each mutant) were transferred onto YUU plates containing different concentrations of camptothecin (CPT), 4-nitroquinoline oxide (4-NQO), MMS, and HU and then incubated for 72 h at 37°C. The diameters of the resulting colonies were measured, and radial growth was expressed as a percentage of the average diameter of colonies grown on YUU control plates without any drug.
For UV light viability assays, conidiospores (dormant in a quiescent G1 state [3]) were suspended in 0.2% Tween 20 and plated on YUU plates (approximately 100 conidia/plate). The plates were irradiated immediately with UV by use of a UV Stratalinker 1800 (Stratagene) and then incubated at 30°C for 48 h to determine the UV sensitivity of nondividing cells. For determinations of the survival of dividing cells after UV exposure, conidiospores on YUU plates were first allowed to germinate for 4.5 h at 30°C. By this time, the germinated spores had entered the cell cycle and were about to undergo the first mitosis. These germlings were UV irradiated on the plates and then similarly incubated at 30°C for 48 h. Viability was determined as the percentage of colonies on treated plates relative to those on untreated control plates.
Protein expression and purification and antibody purification.
For ScaANBS1 protein expression, we used the pET-28a (+) (Novagen) vector, which contains a six-His tag. The scaANBS1 open reading frame was amplified from wild-type genomic DNA (the scaANBS1 gene has no introns) with VENT DNA polymerase (BioLabs) and the following primers: SCABHISTART2 (5′-GCGGGATCCATGCTTAGCATAAAATGGG-3′) and SCAHIIISTOP (5′-GGGAAGCTTTCATCGCCTGCGACGA-3′). The target protein was expressed in the engineered Escherichia coli strain BL21(DE3). An overnight culture of BL21(DE3) was diluted 1:100 with Luria-Bertani medium (10 g Bacto tryptone, 5 g Bacto yeast extract, and 10 g sodium chloride per liter, with 1% glucose) supplemented with kanamycin (30 μg/ml) and chloramphenicol (34 μg/ml). The cells were then incubated at 37°C with continuous shaking until reaching an optical density at 600 nm of 0.6, the temperature was changed to 18°C, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 100 μM, and the cells were grown for 24 h. After this period of growth, the cells were harvested and the pellet was resuspended in lysis buffer 1 (50 mM NaH2PO4, 300 mM NaCl, 5 mM imidazole, pH 8.0) plus protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 2.2 μg/ml aprotinin). The cells were sonicated on ice six times for 30 s each time and then centrifuged at 20,000 × g for 1 h at 4°C. The supernatant was collected and the protein concentration was determined by a modified Bradford assay (Bio-Rad). The supernatant containing the target protein was further purified by Ni-nitrilotriacetic acid-agarose chromatography (QIAGEN), and the samples were then analyzed by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Samples were prepared for SDS-PAGE by the addition of 1× sample buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, and 5% bromophenol blue) and heating at 100°C for 3 min. Five micrograms of total protein from each sample was loaded into each lane of a 10% SDS-PAGE gel.
For NpkA protein expression, we used the pGEX2T (QIAGEN) vector, which encodes glutathione S-transferase. The npkA open reading frame was amplified from wild-type genomic DNA (the npkA gene has no introns) with VENT DNA polymerase (BioLabs) and the following primers: NPKBHISTART2 (5′-CGGGATCCCGATGTCGACCTCTAAATCCA-3′) and NPKAECO3′ (5′-CGGAATTCCTAAATTTTGAAGGAGAAAAA-3′). The target protein was expressed in the engineered E. coli strain XL1 Blue. An overnight culture of XL1 Blue was diluted 1:100 with Luria-Bertani medium supplemented with ampicillin (100 μg/ml) and tetracycline (12.5 μg/ml). The cells were then incubated at 37°C with continuous shaking until reaching an optical density at 600 nm of 0.6, the temperature was changed to 30°C, IPTG was added to a final concentration of 100 μM, and the cells were grown for 2 h. After this period of growth, the cells were harvested and the pellet was resuspended in lysis buffer 2 (50 mM HEPES, pH 7.5, 150 mM NaCl) plus the protease inhibitors described above. The cells were sonicated on ice six times for 30 s each time and then centrifuged at 20,000 × g for 1 h at 4°C. The supernatant was collected and the protein concentration was determined by a modified Bradford assay (Bio-Rad). The supernatant containing the target protein was further purified by using glutathione immobilized on cross-linked 4% beaded agarose (Sigma), and the samples were then analyzed by 10% SDS-PAGE.
For anti-ScaA and anti-NpkA production, 2-kg female rabbits were inoculated with 200 μg of either purified ScaA or NpkA. Anti-ScaA and anti-NpkA purification was performed by a modified protocol described by Harlow and Lane (26).
Protein assays and Western blot analysis.
Protein assays were performed by initially growing conidia from the wild-type (GR5), AAH14 (ΔuvsB), and UvsC::FLAG-expressing (ASG19 and ACS2) strains in a reciprocal shaker at 37°C for 16 h in liquid YG medium (plus uridine and uracil if necessary). Mycelia were aseptically transferred to fresh YG medium plus 20 mM HU for 6 h at 37°C. Controls without HU were made for each strain. The mycelia were extensively washed with sterile YG medium and transferred to fresh YG medium at 37°C in a reciprocal shaker. Samples were collected every 10 min for 120 min. Each sample was harvested by filtration through a Whatman no. 1 filter, washed thoroughly with sterile water, quickly frozen in liquid nitrogen, and disrupted by grinding. The total protein was extracted at 4°C with extraction buffer (15 mM p-nitrophenylphosphate, 25 mM Tris, pH 7.5, 15 mM EGTA, pH 7.5, 15 mM MgCl2) plus protease inhibitors (1 μg/ml leupeptin, 10 μg/ml aprotinin, 5 mM benzamidine, 15 mM phenylmethylsulfonyl fluoride). The protein concentration was determined by use of a modified Bradford assay (Bio-Rad), and samples were prepared for 10% SDS-PAGE. After separation of the proteins, the gel was blotted onto a pure nitrocellulose membrane (0.2 μm; Bio-Rad), and after being blocked in 5% dried milk in TBS/T buffer (10 mM Tris-HCl, 150 mM NaCl, pH 8.0, and 0.05% Tween 20), the membrane was probed with anti-ScaA or anti-NpkA polyclonal immunoglobulin G (IgG) antibodies at a 1:100 dilution in TBS/T buffer for 1 h 30 min at room temperature. For UvsC::FLAG experiments, a 1:1,000 dilution of either the monoclonal anti-FLAG M2 monoclonal antibody F-3165 (Sigma, St. Louis, Mo.), the anti-alpha-tubulin clone DM 1A (Sigma, St. Louis, Mo.), or the IgG fraction of goat anti-rabbit muscle aldolase (Rockland, Gilbertsville, Pa.) was used. The membrane was washed four times for 5 min each with TBS/T buffer and then incubated with a 1:7,000 dilution of anti-rabbit IgG-peroxidase for 1 h. After being washed, the blot was developed by use of the SuperSignal ULTRA chemiluminescence detection system (Pierce) and recorded by the use of Hyperfilm ECL (Amersham Biosciences).
Replication checkpoint response.
For mitosis assays, conidiospores were inoculated onto coverslips in YG+UU medium with 0, 6, or 100 mM HU. After 5 to 7 h of incubation at 30°C, coverslips with adherent germlings were transferred to a fixative solution (3.7% formaldehyde, 50 mM sodium phosphate buffer, pH 7.0, 0.2% Triton X-100) for 30 min at room temperature. They were then briefly rinsed with phosphate-buffered saline (140 mM NaCl, 2 mM KCl, 10 mM NaHPO4, 1.8 mM KH2PO4, pH 7.4) and incubated for 5 min in a solution with 100 ng/ml of DAPI (4′,6′-diamidino-2-phenylindole; Sigma Chemical Co.) (27) and 100 ng/ml of calcofluor (fluorescence brightener; Sigma Chemical Co.). After incubation with the dyes, they were washed with phosphate-buffered saline for 5 min at room temperature and then rinsed in distilled water and mounted on the slides. The material was photographed by use of a Zeiss epifluorescence microscope. The number of nuclei was assessed by DAPI staining. Germlings that had two or more nuclei after the HU incubation were scored as having a nonfunctional checkpoint response.
For viability assays, 1.0 × 108 conidia were inoculated into 1.0 ml of YG+UU medium with 0, 6, or 100 mM HU and then incubated in a reciprocal shaker (250 rpm) at 30°C for 6 h. The conidiospores were washed with distilled water, conveniently diluted, plated on YUU, and incubated at 30°C for 48 h. Viability was determined as the percentage of colonies on plates with drug-treated conidiospores relative to those on untreated control plates.
Real-time PCRs.
All PCRs and reverse transcription-PCRs (RT-PCRs) were performed by using an ABI Prism 7700 sequence detection system (Perkin-Elmer Applied Biosystems). Taq-Man EZ RT-PCR kits (Applied Biosystems) were used for RT-PCRs. The thermal cycling conditions comprised an initial step at 50°C for 2 min, followed by 30 min at 60°C for reverse transcription, 5 min at 95°C, and 40 cycles of 94°C for 20 s and 60°C for 1 min. A TaqMan Universal PCR master mix kit was used for PCRs. The thermal cycling conditions comprised an initial step at 50°C for 2 min, followed by 10 min at 95°C and 40 cycles of 95°C for 15 s and 60°C for 1 min. The reactions and calculations were performed according to the method of Semighini et al. (59). The following primers and Lux fluorescent probes (Invitrogen) were used for this work: for β-tubulin (tubC), AntubCprobe (5′-CACTTTATGCCGTCGCCGAAAG[FAM]G-3′) and AntubCprimer (5′-GCAGAATGTCTCGTCGAATG-3′); and for nimA, AnnimAprobe (5′-GACCGGGAGAAGCAGCAAATTCGG[FAM]C-3′) and AnnimAprimer (5′-TGCGGAGCTTGTCTAATTCGTT-3′) (FAM is 6-carboxyfluorescein).
Construction of ΔuvsBATR strain.
To replace the uvsBATR gene with the argB marker, we constructed the plasmid pSA1 (30). First, the plasmid pASUB10, which contains the entire uvsB open reading frame, was digested to completion with BamHI. The resulting 6.5-kb BamHI fragment, which contained the vector backbone plus flanking uvsB sequences, was then ligated to a 1.8-kb BamHI fragment containing A. nidulans argB (from pSAL-ArgB). pAS1 was transformed into strain AML8. Arg+ transformants were recovered on MM supplemented with uridine and uracil at 30°C. Transformants were tested for sensitivity to various concentrations of HU and MMS. Transformants exhibiting sensitivity were chosen for further analysis. Southern analysis revealed that the full-length endogenous wild-type copy of uvsB was lacking from the genome of one transformant (30). However, an additional copy of pAS1 was present in the genome of this transformant. Further Southern analysis demonstrated that the additional copy of pAS1 had integrated by a single crossover event at the uvsB locus. The resulting uvsB locus was thus missing 75% of the uvsB coding sequence, which was replaced with argB and an additional copy of the pAS1 plasmid (30).
UvsC subnuclear focus formation and alcA::Gfp::ScaA.
The ASG17 strain containing the UvsC::FLAG fusion protein constructed by Gygax et al. (25) was used in crosses with a sca299-16 mutant and ASH270 to generate GHG4 and ACS2, which introduced the scaA1 and uvsB110 mutations, respectively, into the uvsC::FLAG background (Table 1).
For immunolocalization experiments, a standard protocol described by Harris et al. (28) was used. Mycelia of the ASG17, GHG4, and ACS2 strains were grown on coverslips in YG medium for 12 h. The coverslips were treated with 20 mM HU for 6 h, washed three times with prewarmed YG medium, and grown for 0, 30, 60, and 120 min after drug release. Immunofluorescence was induced by using 10 μg of the mouse anti-FLAG M2 monoclonal antibody F-3165 (Sigma) as the primary antibody and a 1:200 dilution of Cy3-conjugated anti-mouse IgG (Sigma) as the secondary antibody. Nuclei were detected with Hoechst 33258 (Molecular Probes, Eugene, Oreg.). Slides were viewed under an Olympus BX51 fluorescence microscope, and distinct UvsC-FLAG subnuclear foci were counted (see Fig. 4D) (25). Confocal images were obtained with a laser scanning confocal microscope (UNL Microscopy Core Facility) using the following laser lines: 405 nm for Hoechst 33258 and 563 nm for Cy3. Images were subsequently processed with Adobe Photoshop 6.0.
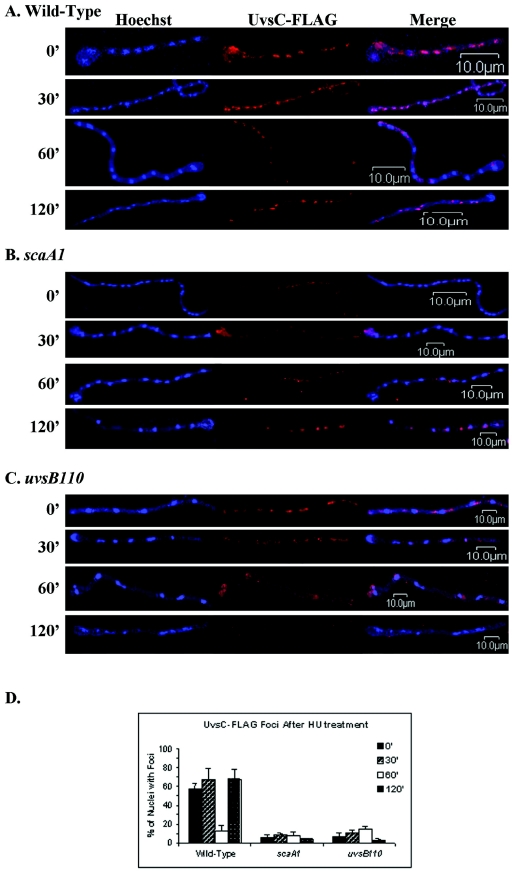
FIG. 4.
Immunolocalization of UvsC::FLAG in germlings during S-phase recovery of the wild-type (ASG17) (A), scaA1 (GHG4) (B), and uvsB110 (ACS2) (C) strains. Mycelia of the ASG17, GHG4, and ACS2 strains were grown on coverslips in YG medium for 12 h. The coverslips were treated with 20 mM HU for 6 h, washed three times with prewarmed YG medium, and grown for 0, 30, 60, or 120 min after HU release. The adherent germlings were fixed, treated with an anti-FLAG antibody, and stained with Hoechst. The samples were analyzed by immunofluorescence microscopy, and distinct UvsC-FLAG subnuclear foci were counted (D). Images were obtained by laser scanning confocal microscopy. Left panels, nuclei stained by Hoechst; middle panels, UvsC::FLAG subnuclear foci; right panels, merged images.
For green fluorescent protein (GFP) experiments, the scaA gene was PCR amplified by the use of Vent DNA polymerase (BioLabs) and the primers SCABHISTART3 (5′-CGGGATCCCGATGCTTAGCATAAATTG-3′) and SCABHISTOP1 (5′-GCGGATCCTCATCGCCTGCGACGA-3′). The PCR product was digested with BamHI and ligated to the previously BamHI-digested and calf intestinal alkaline phosphatase-dephosphorylated vector pMCB17apx (Ampr pyr4 alcA::gfp). This vector construction, pMAGS75, was introduced into the wild-type and AAH14 strains. Germlings of these strains were grown in MC plus 50 mM ethanol in the presence or absence of 10 μg/ml of the radiomimetic drug phleomycin (PHLEO) (Invivogen) for 1 h at 30°C. After the treatment, the germlings were transferred to a fixative GFP solution [50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 6.7, 25 mM EGTA, 5% dimethyl sulfoxide, and 10% formaldehyde] for 15 min at room temperature. The germlings were washed three times with PEM (50 mM PIPES, pH 6.7, 25 mM EGTA, 5 mM MgSO4) and then stained with Hoechst. Slides were analyzed by laser scanning confocal microscopy (UNL Microscopy Core Facility) using the following laser lines: 405 nm for Hoechst 33258 and 488 nm for GFP. Images were subsequently processed with Adobe Photoshop 6.0.
RESULTS
ScaANBS1 and UvsBATR display an epistatic interaction for the response to CPT-induced DNA damage.
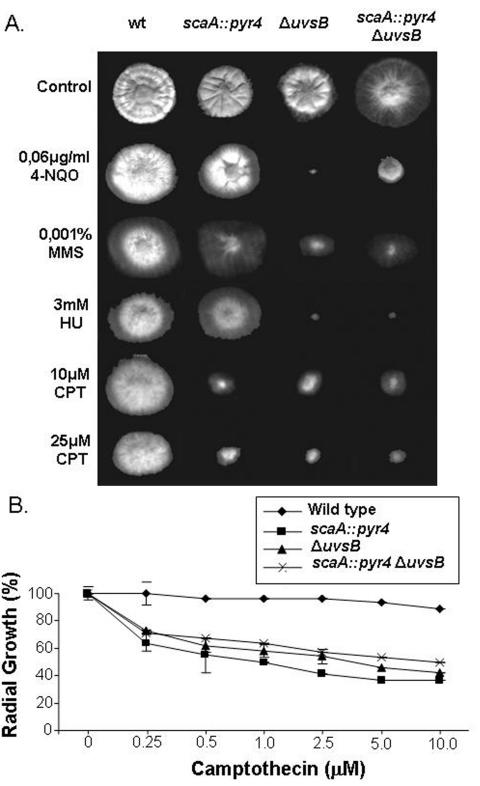
As a first step to look for possible genetic interactions between uvsBATR and scaANBS1, the double mutants and the corresponding parental strains were grown in the presence of different DNA-damaging agents. As shown in Fig. 1, the growth of the double mutant was no slower than that of either of the parental strains in the presence of CPT, suggesting that these two genes function in the same pathway to repair and/or process DNA damage caused by this anti-topoisomerase I drug. Interestingly, the scaA::pyr4 mutation appeared to partially suppress the sensitivity of the ΔuvsBATR mutant to 4-NQO. We previously reported that another mutation that causes defects in recovery from DNA damage, musN227, which affects the A. nidulans MusNRecQ homologue, also partially suppresses the DNA damage sensitivity of uvsBATR mutants (30, 31).
FIG. 1.
Growth phenotypes of the wild-type, ΔuvsBATR, scaA::pyr4, and scaA::pyr4 ΔuvsBATR strains. (A) Strains GR5 (wild type), T20 (scaA::pyr4), AAH14 (ΔuvsBATR), and GG11 (scaA::pyr4 ΔuvsBATR) were grown for 72 h at 37°C in YUU medium in the presence or absence of 4-NQO, MMS, HU, and CPT. (B) Viability curves for GR5 (wild type), T20 (scaA::pyr4), AAH14 (ΔuvsBATR), and GG11 (scaA::pyr4 ΔuvsBATR) hyphae grown on YUU containing different concentrations of CPT at 37°C. Growth was measured as the percent change in the radial growth rate compared to that on an untreated YUU control plate.
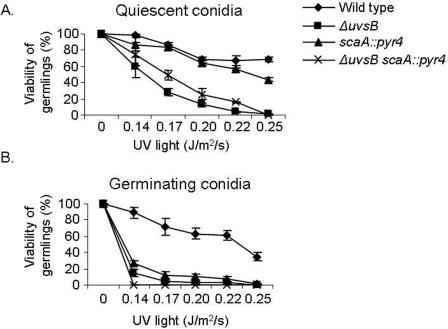
Since the ATR pathway can also respond to UV light, which interferes with the function of replication forks (45, 47), we checked the UV sensitivities of nondividing (quiescent) and dividing (germinating) scaANBS1::pyr4 ΔuvsBATR cells to UV irradiation. When UV irradiation was applied to quiescent conidia, the scaANBS1::pyr4 ΔuvsBATR double mutant strain was as sensitive to UV irradiation as the single mutant ΔuvsBATR strain (Fig. 2A). In contrast, when germinating conidia were exposed to UV light, the scaANBS1::pyr4 ΔuvsBATR double mutant strain was as sensitive to UV irradiation as either the scaANBS1::pyr4 or ΔuvsBATR single mutant strain (P < 0.001) (Fig. 2B). For the latter experiment, conidiospores were first allowed to germinate for 4.5 h before UV irradiation was applied, by which time they had entered the first cell cycle. These results show that scaANBS1 and uvsBATR display epistatic interactions for the detection and/or repair of UV-induced DNA damage in germinating conidiospores.
FIG. 2.
scaANBS1 and uvsBATR interact to monitor and/or repair UV light-induced damage when conidiospores are germinating. Quiescent (A) and germinating (B) conidiospores from different strains were exposed to UV light, and their viability was scored after exposure to this DNA-damaging agent. Viability was determined as the percentage of colonies on treated plates compared to those on untreated control plates. The results are expressed as means ± standard deviations of four independent experiments. Statistical differences were determined by one-way ANOVA followed, when significant, by the Newman-Keuls multiple comparison test, using Sigma Stat statistical software (Jandel Scientific). When germinating conidiospores were exposed to UV light, the ΔuvsB and ΔuvsB scaA::pyr4 strains were not significantly different from the scaA::pyr4 strain.
UvsBATR regulates ScaANBS1 expression during recovery from replication stress.
HU is an inhibitor of ribonucleoside diphosphate reductase, the rate-limiting enzyme in deoxynucleoside triphosphate (dNTP) biosynthesis. The depletion of dNTPs activates the DNA replication checkpoint, which slows the progression through S phase (15). Furthermore, the initiation of DNA replication in the presence of high levels of HU causes DSBs (41). HU is an effective inhibitor of DNA synthesis in A. nidulans (3). Since uvsBATR is involved in the DNA replication checkpoint response (18), we examined the ability of the ΔuvsBATR scaA::pyr4 double mutant to survive a transient period of growth in the presence of HU. Two different assays were used to verify if the DNA replication checkpoint response was impaired in the mutant strains (18). The first assay (i.e., the mitosis assay) monitored mitosis in mutant and wild-type strains incubated in 6 or 100 mM HU for 5 to 7 h. The number of nuclei was assessed by DAPI staining, and if germlings had two or more nuclei after the HU incubation, they were scored as defective in mitosis arrest (Table 2). The second assay (i.e., the viability assay) assessed the germling viability after incubation for 6 h in the presence or absence of 6 or 100 mM HU (Table 3). Both assays measure the state of the replication checkpoint response.
TABLE 2.
Mitosis assay results for A. nidulans wild-type and mutant strains
| Strain | No. of germlings with no mitosis arrest in presence of HUa
|
||
|---|---|---|---|
| 0 mM | 6 mM | 100 mM | |
| GR5 (wild type) | 63.0 ± 12.7 | 8.5 ± 0.7b | 1.5 ± 0.7b |
| AAH14 (ΔuvsB) | 61.5 ± 7.8 | 16.5 ± 2.1 | 15.5 ± 0.7 |
| T20 (scaA::pyr-4) | 69.3 ± 2.1 | 39.0 ± 5.9 | 8.5 ± 2.7 |
| GG11 (ΔuvsB scaA::pyr-4) | 71.3 ± 8.1 | 44.0 ± 7.3 | 11.3 ± 1.5 |
Germlings that had two or more nuclei after the HU incubation were scored as germlings that did not have mitosis arrest. All results are the averages of determinations from three independent experiments, with 100 germlings being evaluated in each. The results are expressed as means ± standard deviations. Statistical differences were determined by one-way ANOVA followed, when significant by the Newman-Keuls multiple comparison test, using Sigma Stat statistical software (Jandel Scientifics). P < 0.05 was considered statistically significant.
Significantly different from all other treatments (P < 0.05).
TABLE 3.
Viability assay results for A. nidulans wild-type and mutant strains
| Strain | % of colonies in presence of HU relative to untreated controlsa
|
|
|---|---|---|
| 6 mM | 100 mM | |
| GR5 (wild type) | 100.0 ± 0b | 100.0 ± 0b |
| AAH14 (ΔuvsB) | 74.6 ± 12.8 | 74.6 ± 10.3 |
| T20 (scaA::pyr-4) | 78.2 ± 8.1 | 70.0 ± 6.2 |
| GG11 (ΔuvsB scaA::pyr-4) | 84.8 ± 4.6 | 85.6 ± 11.2 |
Viability was determined as the percentage of colonies on HU-treated plates compared to those on untreated control plates. All results are averages of determinations from three independent experiments. The data are means ± standard deviations. Statistical differences were determined by one-way ANOVA followed, when significant, by the Newman-Keuls multiple comparison test, using Sigma Stat statistical software (Jandel Scientific). P < 0.05 was considered statistically significant.
Significantly different from all other treatments (P < 0.05).
We previously reported the presence of an intact DNA replication checkpoint in the scaANBS1 inactivation strain (59). However, these results were based on a comparison to the wild-type strain UI224, which has an argB2 mutation. Recently, we observed several interactions between the argB2 mutant and different DNA-damaging agents (data not shown). Kafer (33) also reported that amino acid-requiring A. nidulans mutants were hypersensitive to MMS when assayed by survival and colony formation. Thus, to clarify these results, we repeated the experiments using the original parental wild-type GR5 strain as a control. Interestingly, the scaANBS1 inactivation strain did not have mitosis arrest at 6 mM and 100 mM HU (Table 2), implying that the argB2 mutation suppresses the scaA::pyr4 checkpoint defect. Moreover, as previously reported (18), the ΔuvsB mutant also displayed defects at both concentrations. Although the scaA::pyr4 ΔuvsB double mutant also did not show mitosis arrest at both concentrations, no obvious genetic interaction was noted (Table 2). As previously showed by Fagundes et al. (18), viability was impaired in the ΔuvsBATR strain at both 6 and 100 mM HU (Table 3). The ΔuvsB scaA::pyr4 (GG11) double mutant showed a decrease in viability at 6 mM and 100 mM HU that was comparable to that of the ΔuvsBATR (AAH14) and scaA::pyr4 (T20) strains at 100 mM HU. These results suggest that there is epistasis between uvsBATR and scaANBS1 for germling viability during the replication checkpoint response.
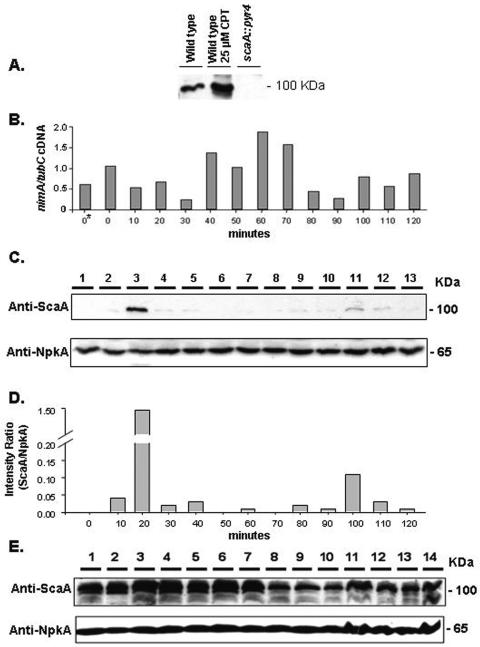
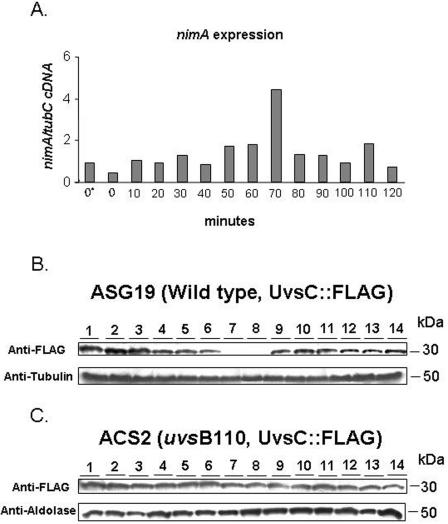
As a preliminary step to characterize the epistasis observed for the DNA replication checkpoint between scaANBS1 and uvsBATR, we attempted to determine if ScaANBS1 expression was dependent on UvsBATR following release from HU-induced replication stress (48). To this end, we raised polyclonal antibodies against ScaA. As shown in Fig. 3A, a single band of approximately 100 kDa was recognized by the antibodies; this band was increased at least 10 times after growth in the presence of CPT (Fig. 3A). As expected, the scaANBS1 deletion mutant had no signal (Fig. 3A, lane 3). To examine ScaA expression upon HU stress and recovery, we incubated wild-type germlings in the presence of 20 mM HU for 6 h, aseptically washed them with sterile water, and allowed them to grow for 120 min. Every 10 min after HU release, samples were taken for the extraction of RNAs and proteins. After HU release, all germlings are still blocked in S phase, and thus, cell division synchrony is maintained (48). We performed real-time RT-PCRs specific for the nimA gene. This gene is preferentially expressed in the G2 phase and at the G2-M transition (48). The expression of the nimA gene was induced 50 to 100% 40 to 70 min after HU release (the difference between these two points and the other time points was statistically significant by one-way analysis of variance [ANOVA]; P < 0.001), which suggests that germlings at these time points are mainly in the G2 and G2-M phases. This implies that the first 30 min after HU release are probably divided between S-phase recovery and progression. For the next step, we examined the expression of scaA; it was about 15 to 20 times more abundant 20 min after HU release than the average expression at the other time points, as quantified by densitometry (Fig. 3D). The same HU block and release experiment was performed again, but with ΔuvsB germlings (Fig. 3E). Interestingly, in the ΔuvsB background, ScaA was expressed at higher levels throughout the time course. In addition, there were several bands of different molecular weights recognized by the anti-ScaA antibody, suggesting that ScaA could be partially degraded and/or subject to posttranslational modification. The incubation of these protein extracts with λ protein phosphatase (dual-specificity serine/threonine/tyrosine phosphatase) did not eliminate the additional bands (data not shown), indicating that phosphorylation is not involved in this phenomenon. Taken together, these results suggest that ScaA expression is increased during recovery from replication stress and that the absence of UvsB leads to an increased expression of ScaA throughout the different phases of the cell cycle though either direct or indirect means.
FIG. 3.
ScaANBS1 temporal expression during S-phase recovery is dependent on UvsBATR. (A) Western blot of total protein extracts from A. nidulans wild-type and scaA::pyr4 strains. The wild-type strain was grown for 30 min in the presence or absence of 25 μM CPT. (B) Real-time RT-PCR specific for A. nidulans nimA gene. Conidia from the wild-type strain (GR5) were grown in a reciprocal shaker at 37°C for 16 h in YG+UU medium. Mycelia were aseptically transferred to fresh YG+UU medium plus 20 mM hydroxyurea (HU) for 6 h (at 37°C). A control without HU was also made (0*). Mycelia were extensively washed with sterile YG medium and then transferred to fresh YG+UU medium at 37°C in a reciprocal shaker. Samples were collected every 10 min for 120 min and then the RNAs were extracted. (C) Western blot of total protein extracts from the A. nidulans wild-type strain (GR5). The protein samples were obtained from mycelia grown as described for panel B (lane 1, time zero; lanes 2 to 13, 10 to 120 min). The membrane was separately labeled with the anti-ScaA and anti-NpkA antibodies. (D) Densitometric analyses of the Western blots are shown in panel C. The results are ratios of the intensities of ScaA signals to the intensities of NpkA signals. (E) The ΔuvsB strain (AAH14) was grown as described for panel B, and a Western blot of the total protein samples was separately labeled with the anti-ScaA and anti-NpkA antibodies (lane 1, time zero; lanes 2 to 13, 10 to 120 min; lane 14, 18 h).
Formation of UvsCRAD51 nuclear foci during recovery from replication stress requires scaANBS1 and uvsBATR.
In A. nidulans, the expression and localization of UvsCRAD51 is regulated by DNA damage (25, 69). Rad51 homologues promote the initial steps of homologous recombination by binding the free ends of the DNA double strands (for a review, see reference 45). In yeast cells, Rad51 forms irradiation-induced subnuclear foci (8, 22). UvsCRAD51 also localizes to similar structures, as determined by using a functional UvsC::FLAG fusion protein expressed under the control of uvsC promoter sequences (25). We determined whether the nuclear localization of UvsCRAD51 during recovery from HU replication stress could be affected by scaANBS1 and uvsBATR inactivation. Thus, we constructed UvsC::FLAG strains in the wild-type, uvsB110, and scaA1 backgrounds, grew them in the presence of HU, washed the germlings from the drug, and immunolocalized UvsCRAD51. As show in Fig. 4A and D, UvsCRAD51 nuclear foci were observed following treatment with HU and during the recovery period. The decrease in foci seen at 60 min correlated with a dramatic decrease in UvsCRAD51 expression (Fig. 5B). Because nimA gene expression peaked at the same time (Fig. 5A), these cells were likely in G2 phase. Thus, the second wave of UvsCRAD51 focus formation observed at 90 min presumably reflected a population of cells entering the next S phase. When the same experiment was carried out with uvsB110 and scaA1 mutants, UvsCRAD51 foci were dramatically reduced to about 80 to 90% of the average levels observed in the wild type. This occurred even through UvsCRAD51 was abundantly expressed during the recovery period in both mutants (Fig. 5C and data not shown). These results demonstrate that UvsCRAD51 focus formation during recovery from replication stress is dependent on scaANBS1 and uvsBATR.
FIG. 5.
UvsCRAD51 expression is absent from G2 phase. (A) Real-time RT-PCR specific for the A. nidulans nimA gene. Conidia from the wild-type (ASG19) or uvsB110 (ACS2) strain were grown in a reciprocal shaker at 37°C for 16 h in YG+UU medium. Mycelia were aseptically transferred to fresh YG+UU medium plus 20 mM HU for 6 h (at 37°C). A control without HU was also made (0*). The mycelia were extensively washed with sterile YG medium and then transferred to fresh YG+UU medium at 37°C in a reciprocal shaker. Samples were collected every 10 min for 120 min, and RNAs and proteins were extracted. Western blots were performed with total protein extracts from the A. nidulans wild-type (ASG19) (B) and uvsB110 (ACS2) (C) strains (lanes 1 and 2, time zero before HU addition and after HU removal, respectively; lanes 3 to 14, 10 to 120 min). The protein samples were obtained from mycelia grown as described for panel A. The membranes were separately labeled with anti-FLAG, anti-tubulin, and anti-aldolase antibodies.
ScaANBS1 localizes to nuclear foci in a UvsBATR-dependent manner.
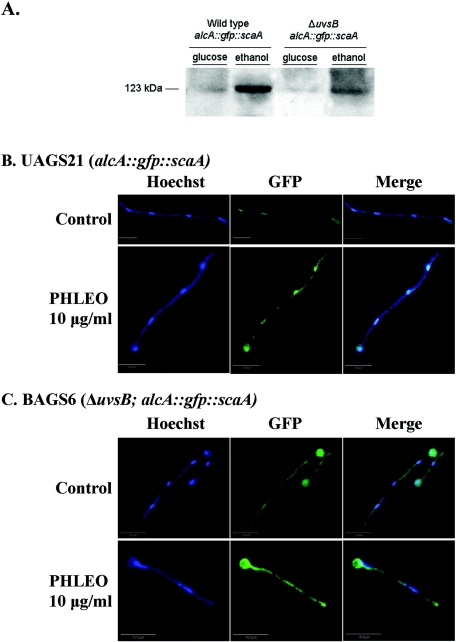
To determine if ScaNBS1 localizes to nuclei in a DNA damage-dependent manner, we constructed ScaA::Gfp and alcAp::Gfp::ScaA strains. In a first set of experiments, we could not observe fluorescence when a wild-type strain expressing ScaA::Gfp driven by its own promoter was challenged with 10 μg/ml of PHLEO (data not shown). As previously observed by Bruschi et al. (5), this could be due to the weak levels of expression of this gene. Thus, we decided to boost the ScaA expression levels by fusing the scaA open reading frame to a GFP construct regulated by the A. nidulans alcA (alcohol dehydrogenase) promoter (19, 72). The alcA promoter is induced to high levels by glycerol, ethanol, and l-threonine and is repressed by glucose (20). This construction (alcAp::Gfp::ScaA) was transformed into the wild-type and AAH14 (ΔuvsB) strains. Several transformants were obtained in which the plasmid had integrated ectopically at different sites and produced large amounts of Gfp::ScaA when induced with ethanol (Fig. 6A). Figure 6B shows wild-type hyphae that were grown in the presence of 50 mM ethanol and exposed for 1 h to 10 μg/ml of PHLEO. Although Gfp::ScaA was barely detectable in untreated wild-type hyphae, it was clearly present within the nuclei of hyphae exposed to PHLEO (Fig. 6B). When the same experiment was performed with the ΔuvsBATR mutant, PHLEO-induced nuclear localization was not observed, despite expression levels that were comparable to those in the wild type (Fig. 6A and B). We observed the same results for two independent transformants obtained for each treatment (data not shown). These results show that ScaANBS1 localizes to nuclei in response to DNA damage (i.e., DSBs) in a manner that requires uvsBATR function. This may account, in part, for the epistatic interaction observed between scaANBS1 and uvsBATR mutations.
FIG. 6.
Gfp::ScaA focus formation is dependent on uvsBATR in response to DSBs caused by PHLEO. (A) Western blot of total protein extracts from A. nidulans wild-type (UAGS21) and ΔuvsB (BAGS6) strains. Conidia from the wild-type (UAGS21) or ΔuvsB (BAGS6) strain were grown in a reciprocal shaker at 37°C for 16 h in MM. Mycelia were aseptically transferred to fresh MM or MM plus 50 mM ethanol for 1 h (at 37°C). Equal amounts of total protein (10 μg) were run in a 10% SDS-PAGE gel and then transferred to a nitrocellulose membrane. The membrane was labeled with the anti-ScaA antibody. Germlings of the UAGS21 (wild-type alcA::gfp::scaA) (B) and BAGS6 (ΔuvsB alcA::gfp::scaA) (C) strains were grown in the presence of MC plus 50 mM ethanol in the presence or absence of 10 μg/ml PHLEO for 1 h at 30°C. After the treatment, the germlings were fixed, washed, and stained with Hoechst. Samples were analyzed by laser scanning confocal microscopy, and Z stacks of 1-μm sections are shown. Bars, 10 μm.
DISCUSSION
We have characterized genetic interactions between A. nidulans uvsBATR and scaANBS1 during the responses to replication stress and DNA damage. The UvsBATR protein is the homolog of ATR/MEC1, whereas we have characterized another phosphoinositide-3-kinase-related protein family (PIKK) that corresponds to the A. nidulans ATM/TEL1 homolog (I. Malavazi and G. H. Goldman, unpublished results). The scaA::pyr4 ΔuvsBATR double mutant was as sensitive to CPT as the corresponding single mutants, suggesting that these two genes are epistatic for the signaling and/or repairing of DNA damage caused by CPT. Cliby et al. (10) have demonstrated that ATR kinase function is necessary for both the G2- and S-phase arrests induced by topoisomerase I poisons and that these cellular responses to topoisomerase poisons appear to be independent of ATM function. Since prolonged exposure to topoisomerase I poisons leads to S-phase slowing (44, 60), we decided to investigate the DNA replication checkpoint response in these mutants. There are at least two S-phase checkpoint mechanisms controlling mitosis in A. nidulans (75). The first S-phase checkpoint is activated when replication is slowed by the addition of HU to a level that does not cause arrest. It responds to the rate of DNA replication and inhibits mitosis via tyrosine phosphorylation of NimXCdc2. If DNA replication is arrested, a second checkpoint involves BimEAPC1 (the homolog of the anaphase-promoting complex subunit APC1). This second S-phase checkpoint occurs when DNA replication is completely inhibited by higher levels of HU than that which stimulates the prolonged DNA replication checkpoint. This information was obtained by characterizing double mutants possessing nimXCdc2AF (a mutated version in which Thr14 is converted to an Ala [A] and Tyr15 is converted to a Phe [F] residue) and the temperature-sensitive mutation bimE7 (49). Either the Cdc2AF or bimE7 mutation alone has a limited capacity to promote mitosis when S phase is arrested, but in combination these two defects allow cells to enter a lethal premature mitosis before the completion of DNA replication. Recently, we demonstrated that the A. nidulans uvsBATR gene is involved in the DNA replication checkpoint responses and that a deletion mutant of npkA, a p34Cdc2-related gene, can suppress its S-phase checkpoint deficiency (18).
scaANBS1 and uvsBATR mutants were epistatic regardless of the HU concentration, suggesting that these genes function in a common pathway that is probably necessary for progression after and/or recovery from replication stress. HU stalls replication forks by depleting the dNTP pool. Another type of replication block might be associated with DNA breaks generated during replication. In theory, DSBs could arise if replication forks pass through nicked DNA or certain repair or recombination intermediates (47). The S-phase checkpoints respond to replication interference by slowing down DNA replication to allow the damage to be repaired before polymerases encounter additional DNA damage (47). Deletion of the ATR orthologs MEC1 and RAD3 in budding (S. cerevisiae) and fission (Schizosaccharomyces pombe) yeasts, respectively, eliminates the DNA replication checkpoint (45). Using a Cre/lox conditional system to study the effect of ATR loss, Brown and Baltimore (4) showed that mammalian ATR is an important regulator of checkpoint signaling pathways that phosphorylates Cdc2 in response to ionizing radiation and stalled replication. D'Amours and Jackson (13) demonstrated that the Mre11 complex is required for Mec1/Rad3-dependent S-phase checkpoint activation. The Mre11 complex could be recruited to DSBs accumulated at stalled replication forks by the ATR-dependent phosphorylation of histone H2AX during S phase (64). In fact, ATR kinase-deficient cells cannot form both γ-H2AX and Mre11 complex foci (21, 71). Similarly, we have found that uvsBATR is required for the formation of ScaA foci upon induction of DSBs by PHLEO. This raises the possibility that uvsBATR plays an important role in the A. nidulans response to DSBs. Curiously, Stiff et al. (63) observed that while replication protein A (RPA) is recruited normally to damage sites in NBS cells, the nuclear retention of ATR is markedly decreased after replication stalling, suggesting that Nbs1 functions to recruit or retain ATR.
The S-phase response of wild-type cells to UV irradiation, i.e., the reduction of the rate of DNA synthesis due to the inhibition of the rate of chain elongation and replicon initiation, can be divided into passive and active inhibition. Passive inhibition of DNA replication is attributed to the physical obstruction of the DNA replication apparatus at sites of DNA damage. Active inhibition is a trans effect mediated through checkpoint signals that emanate from sites of DNA damage and ultimately inhibit the initiation of distant replicons (58). This S-phase checkpoint response imposes transient delays in S-phase progression and provides more time for DNA repair to remove lesions from unreplicated chromatin (29). As previously reported for the uvsB mutant (18, 34), quiescent and germinating conidiospores were more sensitive to UV light than were those of the corresponding wild-type strain. ScaANBS1 and UvsBATR are epistatic for the response to UV irradiation in germinating conidiospores. Although ATR seems to be required for checkpoint responses to replication blocks caused by UV-induced DNA damage (10, 29, 73), the checkpoint pathway that mediates the inhibition of replicon initiation following UV-induced DNA damage is less clear. UV irradiation causes damage to single-stranded DNA, primarily by generating thymine dimers, which are repaired by nucleotide excision mechanisms that do not require a homologous chromatid and therefore should be reparable in G1 (39). S. pombe cells elicit an S-phase checkpoint when they are UV irradiated during G1 phase (54), while in S. cerevisiae, a checkpoint dependent on Mec1p delays progression through S phase in response to UV irradiation (50).
Several studies have established that the Mre11 complex regulates the activation of the Tel1 signaling pathway (13, 35, 52, 66). When DNA is damaged by UV or chemicals that make bulky base lesions, the main PIKK family damage sensor is the ATR protein (1). Mec1 physically interacts with Ddc2, a protein related to the mammalian ATR-interacting protein ATRIP (45, 57). ATR-ATRIP binds to chromatin and can bind directly to UV-induced lesions or RPA-coated single-stranded DNA generated from the repair or replication of these lesions and then become activated (57). As for the ATM-initiated checkpoint, ATR-initiated signaling also results in the phosphorylation of BRCA1, NBS1, and other targets, promoting the recovery of damaged replication forks and thereby coordinating the inhibition of replication initiation with the recovery of active replication forks by homologous recombination and related processes (57). We have shown that ScaA expression is increased during recovery from replication stress, and its nuclear localization appears to be promoted by UvsBATR. In contrast, Nakada et al. (43) demonstrated that the Mre11 complex controls the Mec1 signaling pathway. They showed that the Mre11 complex collaborates with Exo1 to produce long 3′ single-stranded DNA tails at DSB ends and to promote the Mec1 association with DSBs. The Mre11 complex and Exo1 play overlapping roles in activation in both DSB- and UV-induced checkpoints. The Mre11 complex and Exo1 are also involved in checkpoint responses to stalled DNA replication. However, as emphasized by Uziel et al. (67), the sequence of events at the early stage of the DNA damage response may not necessarily be explained by a simple hierarchy related to the position of each protein in the damage signaling cascade. Although the Mre11 complex regulates the activation of the Tel1 signaling pathway, both the Nbs1 and Mre11 proteins were shown to be phosphorylated in an ATM-dependent manner in response to DSBs (45).
We have shown that ScaANBS1 and UvsBATR promote the formation of UvsCRAD51 nuclear foci during the recovery from replication stress. UvsC expression seems to be cell cycle regulated since UvsC is not present in G2 phase in the wild-type background. Intriguingly, the remarkable loss of UvsC from the G2 phase does not occur in the uvsB110 mutant, raising the attractive possibility that UvsB directly or indirectly regulates the cell cycle-dependent UvsC expression. In S. cerevisiae, Rad51 forms subnuclear foci in mitotic cells that have suffered DNA damage (8, 22). These foci are thought to represent sites of ongoing recombination, and consistent with in vitro observations, their formation requires RPA and Rad52 (42, 68, 70). Lisby et al. (40) have shown that only a subset of the proteins that are recruited to DSBs are also recruited to HU-stalled replication forks. Interestingly, these authors found that when HU induces replication fork collapse, Rad52 can still be recruited to nuclear foci even in the absence of Mec1 function. Our contrasting observation that Rad51 recruitment requires ATR function may reflect differences in the ways that yeast and filamentous fungi cope with DNA damage at HU-stalled replication forks.
Acknowledgments
This research was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil.
We thank Vladimir Efimov for providing the plasmid pMCB17apx, Terri Fangman and Joe Zhao (Microscopy Core, University of Nebraska Center for Biotechnology) for their invaluable assistance with confocal microscopy, Amy Hofmann for providing the uvsB deletion mutant, and Everaldo dos Reis Marques for helping with the real-time RT-PCR experiments.
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer association. Nature 421:499-506. [DOI] [PubMed] [Google Scholar]
- 3.Bergen, L. G., and N. R. Morris. 1983. Kinetics of the nuclear division cycle of Aspergillus nidulans. J. Bacteriol. 156:155-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, E. J., and D. Baltimore. 2003. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 17:615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruschi, G. C. M., C. C. de Souza, M. R. Z. K. Fagundes, M. A. C. Dani, M. H. S. Goldman, N. R. Morris, L. Liu, and G. H. Goldman. 2001. Sensitivity to camptothecin in Aspergillus nidulans identifies a novel gene, scaA, related to the cellular DNA damage response. Mol. Genet. Genomics 265:264-275. [DOI] [PubMed] [Google Scholar]
- 6.Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau, J. R. Yates III, L. Hays, W. F. Morgan, and J. H. Petrini. 1998. The hMRE11/hRAD50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477-486. [DOI] [PubMed] [Google Scholar]
- 7.Carson, C. T., R. A. Schwartz, T. H. Stracker, C. E. Lilley, D. V. Lee, and M. D. Weitzman. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22:6610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caspari, T., J. M. Murray, and A. M. Carr. 2002. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 16:1195-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cliby, W. A., C. J. Roberts, K. A. Cimprich, C. M. Stringer, J. R. Lamb, S. L. Schreiber, and S. H. Friend. 1998. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 17:159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cliby, W. A., K. A. Lewis, K. K. Lilly, and S. H. Kaufmann. 2002. S phase and G2 arrests induced by topoisomerase I poisons are dependent on ATR kinase function. J. Biol. Chem. 277:1599-1606. [DOI] [PubMed] [Google Scholar]
- 11.Craven, R. J., P. W. Greenwell, M. Dominska, and T. D. Petes. 2002. Regulation of genome stability by TEL1 and MEC1, yeast homologs of the mammalian ATM and ATR. Genetics 161:493-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cromie, G. A., C. B. Millar, K. H. Schmidt, and D. R. Leach. 2000. Palindromes as substrates for multiple pathways of recombination in Escherichia coli. Genetics 154:513-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Amours, D., and S. P. Jackson. 2001. The yeast Xrs2 complex functions in S-phase checkpoint regulation. Genes Dev. 15:2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Amours, D., and S. P. Jackson. 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3:317-327. [DOI] [PubMed] [Google Scholar]
- 15.Desany, B. A., A. A. Alcasabas, J. B. Bachant, and S. J. Elledge. 1998. Recovery from DNA replicational stress is the essential function of the S phase checkpoint pathway. Genes Dev. 12:2956-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Digweed, M. 1993. Human genetic instability syndromes: single gene defects with increased risk of cancer. Toxicol. Lett. 67:259-281. [DOI] [PubMed] [Google Scholar]
- 17.Fagundes, M. R. Z. K., L. Fernandes, M. Savoldi, S. D. Harris, M. H. S. Goldman, and G. H. Goldman. 2003. Identification of a topoisomerase I mutant, scsA1, as an extragenic suppressor of a mutation in scaANBS1, the apparent homolog of human nibrin in Aspergillus nidulans. Genetics 164:935-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fagundes, M. R. Z. K., J. F. Lima, M. Savoldi, I. Malavazi, R. E. Larson, M. H. S. Goldman, and G. H. Goldman. 2004. The Aspergillus nidulans npkA gene encodes a Cdc2-related kinase that genetically interacts with the UvsBATR kinase. Genetics 167:1629-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez-Abalos, J. M., H. Fox, C. Pitt, B. Wells, and J. H. Doonan. 1998. Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus, Aspergillus nidulans. Mol. Microbiol. 27:121-130. [DOI] [PubMed] [Google Scholar]
- 20.Flipphi, M., J. Kocialkowska, and B. Felenbok. 2002. Characteristics of physiological inducers of the ethanol utilization (alc) pathway in Aspergillus nidulans. Biochem. J. 15:25-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franchitto, A., and P. Pichierri. 2002. Bloom's syndrome protein is required for correct relocalization of RAD50/MRE11/NBS1 complex after replication fork arrest. J. Cell Biol. 157:19-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasior, S. L., H. Olivares, U. Ear, D. M. Hari, R. Weichselaum, and D. K. Bishop. 2001. Assembly of RecA-like recombinases: distinct roles for mediator proteins in mitosis and meiosis. Proc. Natl. Acad. Sci. USA 98:8411-8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldman, G. H., S. L. McGuire, and S. D. Harris. 2002. The DNA damage response in filamentous fungi. Fungal Genet. Biol. 35:183-195. [DOI] [PubMed] [Google Scholar]
- 24.Goldman, G. H., and E. Kafer. 2004. Aspergillus nidulans as a model system to characterize the DNA damage response in eukaryotes. Fungal Genet. Biol. 41:428-442. [DOI] [PubMed] [Google Scholar]
- 25.Gygax, S. E., C. P. Semighini, G. H. Goldman, and S. D. Harris. 2005. SepCTF4 is required for the formation of DNA damage-induced UvsCRAD51 foci in Aspergillus nidulans. Genetics 169:1391-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harlow, E., and D. Lane. 1999. Using antibodies. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Harris, S. D., and P. R. Kraus. 1998. Regulation of septum formation in Aspergillus nidulans by a DNA damage checkpoint pathway. Genetics 148:1055-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris, S. D., A. F. Hofmann, H. W. Tedford, and M. P. Lee. 1999. Identification and characterization of genes required for hyphal morphogenesis in the filamentous fungus Aspergillus nidulans. Genetics 151:1015-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heffernan, T. P., D. A. Simpson, A. R. Franck, A. N. Heinloth, R. S. Paules, M. Cordeiro-Stone, and W. K. Kaufmann. 2002. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol. Cell. Biol. 22:8552-8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofmann, A. F. 2001. Regulation of the Aspergillus nidulans DNA damage response by uvsB and musN. Ph.D. thesis. University of Connecticut, Farmington.
- 31.Hofmann, A. F., and S. D. Harris. 2000. The Aspergillus nidulans uvsB gene encodes an ATM-related kinase required for multiple facets of the DNA damage response. Genetics 154:1577-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kafer, E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19:33-131. [DOI] [PubMed] [Google Scholar]
- 33.Kafer, E. 1987. MMS sensitivity of all amino acid-requiring mutants in Aspergillus and its suppression by mutations in a single gene. Genetics 115:671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kafer, E., and G. S. May. 1998. Toward repair pathways in Aspergillus nidulans, p. 477-502. In J. A. Nickoloff and M. F. Hoekstra (ed.), DNA damage and repair, vol. 1. Humana Press, Totowa, N.J.
- 35.Kastan, M. B., and D. S. Lim. 2000. The many substrates and functions of ATM. Nat. Rev. Mol. Cell Biol. 1:179-186. [DOI] [PubMed] [Google Scholar]
- 36.Kitagawa, R., C. J. Bakkenist, P. J. McKinnon, and M. B. Kastan. 2004. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 18:1423-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, J. H., and T. T. Paull. 2004. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 304:93-96. [DOI] [PubMed] [Google Scholar]
- 38.Levitt, N. C., and I. D. Hickson. 2002. Caretaker tumour suppressor genes that defend genome integrity. Trends Mol. Med. 8:179-186. [DOI] [PubMed] [Google Scholar]
- 39.Lindahl, T. 1982. DNA repair enzymes. Annu. Rev. Biochem. 51:61-87. [DOI] [PubMed] [Google Scholar]
- 40.Lisby, M., J. H. Barlow, R. C. Burgess, and R. Rothstein. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118:699-713. [DOI] [PubMed] [Google Scholar]
- 41.Merrill, B. J., and C. Holm. 1999. A requirement for recombinational repair in Saccharomyces cerevisiae is caused by DNA replication defects of mec1 mutants. Genetics 153:595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyazaki, T., D. A. Bressan, M. Shinohara, J. E. Haber, and A. Shinohara. 2004. In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair. EMBO J. 23:939-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakada, D., Y. Hirano, and K. Sugimoto. 2004. Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol. Cell. Biol. 24:10016-10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson, W. G., and M. B. Kastan. 1994. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol. Cell. Biol. 14:1815-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nyberg, K. A., R. J. Michelson, C. W. Putnam, and T. A. Weinert. 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36:617-656. [DOI] [PubMed] [Google Scholar]
- 46.O'Driscoll, M., V. L. Ruiz-Perez, C. G. Woods, P. A. Jeggo, and J. A. Goodship. 2003. A splicing mutation affecting expression of ataxia-telangiectasia and Rad-3 related protein (ATR) results in Seckel syndrome. Nat. Genet. 33:497-501. [DOI] [PubMed] [Google Scholar]
- 47.Osborn, A. J., S. J. Elledge, and L. Zou. 2002. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 12:509-516. [DOI] [PubMed] [Google Scholar]
- 48.Osmani, S. A., G. S. May, and N. R. Morris. 1987. Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans. J. Cell Biol. 104:1495-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osmani, S. A., and X. S. Ye. 1997. Targets of checkpoints controlling mitosis: lessons from lower eukaryotes. Trends Cell Biol. 7:283-288. [DOI] [PubMed] [Google Scholar]
- 50.Paulovich, A. G., and L. H. Hartwell. 1995. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell 82:841-847. [DOI] [PubMed] [Google Scholar]
- 51.Petrini, J. H. 1999. The mammalian Mre11-Rad50-Nbs1 protein complex: integration of functions in the cellular DNA-damage response. Am. J. Hum. Genet. 64:1264-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petrini, J. H. 2000. The Mre11 complex and ATM: collaborating to navigate S phase. Curr. Opin. Cell Biol. 12:293-296. [DOI] [PubMed] [Google Scholar]
- 53.Pichierri, P., and A. Franchitto. 2004. Werner syndrome protein, the MRE11 complex and ATR: ménage-à-trois in guarding genome stability during DNA replication? BioEssays 26:306-313. [DOI] [PubMed] [Google Scholar]
- 54.Rhind, N., and P. Russell. 1998. The Schizosaccharomyces pombe S-phase checkpoint differentiates between different types of DNA damage. Genetics 149:1729-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rotman, G., and Y. Shiloh. 1998. ATM: from gene to function. Hum. Mol. Genet. 7:1555-1563. [DOI] [PubMed] [Google Scholar]
- 56.Samadashwily, G. M., G. Raca, and S. M. Mirkin. 1997. Trinucleotide repeats affect DNA replication in vivo. Nat. Genet. 17:298-304. [DOI] [PubMed] [Google Scholar]
- 57.Sancar, A., L. A. Lindsey-Boltz, K. Unsal-Kaçmaz, and S. Linn. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 7:39-85. [DOI] [PubMed] [Google Scholar]
- 58.Santocanale, C., and J. F. X. Diffley. 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395:615-618. [DOI] [PubMed] [Google Scholar]
- 59.Semighini, C. P., M. R. von Zeska Kress Fagundes, J. C. Ferreira, R. C. Pascon, M. H. S. Goldman, and G. H. Goldman. 2003. Different roles of the Mre11 complex in the DNA damage response in Aspergillus nidulans. Mol. Microbiol. 48:1693-1709. [DOI] [PubMed] [Google Scholar]
- 60.Shao, R. G., T. Shimizu, and Y. Pommier. 1997. Abrogation of an S-phase checkpoint and potentiation of camptothecin cytotoxicity by 7-hydroxystaurosporine (UCN-01) in human cancer cell lines, possibly influenced by p53 function. Cancer Res. 57:4029-4035. [PubMed] [Google Scholar]
- 61.Shiloh, Y. 1997. Ataxia-telangiectasia and the Nijmegen breakage syndrome: related disorders but genes apart. Annu. Rev. Genet. 31:635-662. [DOI] [PubMed] [Google Scholar]
- 62.Stewart, G., and S. J. Elledge. 2002. The two faces of BRCA2, a FANCtastic discovery. Mol. Cell 10:2-4. [DOI] [PubMed] [Google Scholar]
- 63.Stiff, T., C. Reis, G. Alderton, L. Woodbine, M. O'Driscoll, and P. A. Jeggo. 2004. Nbs1 is required for ATR-dependent phosphorylation events. EMBO J. 24:199-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tauchi, H., S. Matsuura, J. Kobayashi, S. Sakamoto, and K. Komatsu. 2002. Nijmegen breakage syndrome gene, NBS1, and molecular links to factors for genome stability. Oncogene 21:8967-8980. [DOI] [PubMed] [Google Scholar]
- 65.Trujillo, K. M., S. S. Yuan, E. Y. Lee, and P. Sung. 1998. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 34:21447-21450. [DOI] [PubMed] [Google Scholar]
- 66.Usui, T., H. Ogawa, and J. H. Petrini. 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7:1255-1266. [DOI] [PubMed] [Google Scholar]
- 67.Uziel, T., Y. Lerenthal, L. Moyal, Y. Andegeko, L. Mittelman, and Y. Shiloh. 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 22:5612-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Dick, E., A. Z. Stasiak, A. Stasiak, and S. C. West. 1999. Binding of double-strand breaks in DNA by human Rad52 protein. Nature 398:728-731. [DOI] [PubMed] [Google Scholar]
- 69.Van Heemst, D., K. Swart, E. F. Holub, R. van Dijk, H. H. Offenberg, T. Goosen, H. W. van den Broek, and C. Heyting. 1997. Cloning, sequencing, disruption, and phenotypic analysis of uvsC, an Aspergillus nidulans homologue of yeast RAD51. Mol. Gen. Genet. 254:654-664. [DOI] [PubMed] [Google Scholar]
- 70.Wang, X., and J. E. Haber. 2004. Role of Saccharomyces cerevisiae single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair. PLoS Biol. 2:E21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ward, I. M., and J. Chen. 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276:47759-47762. [DOI] [PubMed] [Google Scholar]
- 72.Waring, R. B., G. S. May, and N. R. Morris. 1989. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-coding genes. Gene 79:119-130. [DOI] [PubMed] [Google Scholar]
- 73.Wright, J. A., K. S. Keegan, D. R. Herendeen, N. J. Bentley, A. M. Carr, M. F. Hoekstra, and P. Concannon. 1998. Protein kinase mutants of human ATR increase sensitivity to UV and ionizing radiation and abrogate cell cycle checkpoint control. Proc. Natl. Acad. Sci. USA 95:7445-7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang, J., Y. Yu, H. E. Hamrick, and P. J. Duerksen-Hugues. 2003. ATM, ATR and DNA-PK: initiators of the cellular genotoxic stress responses. Carcinogen 24:1571-1580. [DOI] [PubMed] [Google Scholar]
- 75.Ye, X. S., R. R. Fincher, A. Tang, K. O'Donnell, and S. A. Osmani. 1996. Two S-phase checkpoint systems, one involving the function of both BIME and Tyr15 phosphorylation of p34Cdc2, inhibit NIMA and prevent premature mitosis. EMBO J. 15:3599-3610. [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou, B. B., and S. J. Elledge. 2002. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]