Abstract
Recently, two types of fatty acid synthases (FASs) have been discovered from apicomplexan parasites. Although significant progress has been made in characterizing these apicomplexan FASs, virtually nothing was previously known about the activation and regulation of these enzymes. In this study, we report the discovery and characterization of two distinct types of phosphopantetheinyl transferase (PPTase) that are responsible for synthesizing holo-acyl carrier protein (ACP) from three apicomplexan parasites: surfactin production element (SFP) type in Cryptosporidium parvum (CpSFP-PPT), holo-ACP synthase (ACPS)-type in Plasmodium falciparum (PfACPS-PPT), and both SFP and ACPS types in Toxoplasma gondii (TgSFP-PPT and TgACPS-PPT). CpSFP-PPT and TgSFP-PPT are monofunctional, cytosolic, and phylogenetically related to animal PPTases. However, PfACPS-PPT and TgACPS-PPT are bifunctional (fused with a metal-dependent hydrolase), likely targeted to the apicoplast, and more closely related to proteobacterial PPTases. The function of apicomplexan PPTases has been confirmed by detailed functional analysis using recombinant CpSFP-PPT expressed from an artificially synthesized gene with codon usage optimized for Escherichia coli. The recombinant CpSFP-PPT was able to activate the ACP domains from the C. parvum type I FAS in vitro using either CoA or acetyl-CoA as a substrate, or in vivo when coexpressed in bacteria, with kinetic characteristics typical of PPTases. These observations suggest that the two types of fatty acid synthases in the Apicomplexa are activated and regulated by two evolutionarily distinct PPTases.
Fatty acids are essential to all organisms, since they are one of the major components in all biomembranes and are a common biological energy resource. The de novo biosynthesis of fatty acids typically starts with the use of acetyl-coenzyme A (CoA) as a precursor and malonyl-CoA as an extender with repetitive addition of two-carbon units for chain elongation (31, 36, 38, 39). Each fatty acyl two-carbon-unit elongation consists of a series of biochemical reactions that are catalyzed by different enzymes (e.g., malonyl/acyl transferase, acyl carrier protein [ACP], ketoacyl synthase [KS], ketoacyl reductase, dehydrase, and enoyl reductase [ER]). In many organisms (e.g., humans, animals, and fungi), these enzymes are fused into one or two multifunctional polypeptides, referred to as type I fatty acid synthases (FASs) (36, 39). In other organisms (e.g., plants and bacteria), these enzymes are discrete, monofunctional proteins, referred to as type II FAS (23, 38). The synthesis of polyketides resembles FAS, except that one or more reduction and/or dehydration steps are ignored during chain elongation, and different starters and extenders may be used, resulting in the retaining of keto groups, hydroxyl moieties, double bonds, and/or branched chains in final products (31, 37).
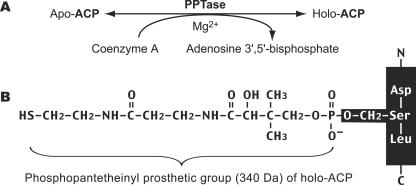
During fatty acid synthesis, the fatty acyl chain needs to be attached to the holo-ACP at the prosthetic 4′-phosphopantetheinyl moiety. However, newly synthesized ACP enzymes or domains are apoproteins that lack the phosphopantetheinyl moiety. The phosphopantetheinyl moiety is posttranslationally transferred from CoA to a Ser residue in the ACP by a phosphopantetheinyl transferase (PPTase) (EC 2.7.8.7) (Fig. 1). There are two major types of PPTases. The holo-ACP synthase (ACPS)-type PPTases are typically specific to the type II FAS or polyketide synthase (PKS) in bacteria, e.g., Escherichia coli AcpS (21, 22). The surfactin production element (SFP)-type enzymes are mostly specific to type I FAS, PKS, or nonribosomal peptide synthase, such as the Bacillus subtilis SFP, yeast Lys5, and human PPTase (19, 26, 28, 30). The FAS α-subunit (FAS2) in fungi also contains an ACPS-type PPTase domain and is sometimes referred to as a third type of PPTase (10, 27).
FIG. 1.
PPTases synthesize holo-ACP by transferring the phosphopantetheinyl moiety from CoA to apo-ACP (A). The phosphopantetheinyl moiety is attached to the Ser residue of ACP (B).
Fatty acid synthesis has been recently discovered for the phylum Apicomplexa, which contains many important protozoon parasites (e.g., Plasmodium, Babesia, Toxoplasma, Sarcosystis, Cyclospora, Eimeria, and Cryptosporidium) (32, 40, 42, 44, 45). As a reflection of their divergent parasitic life styles, the fatty acid metabolism is also diverse among apicomplexans. For example, Plasmodium falciparum possesses only type II enzymes that are targeted to the apicoplast for de novo fatty acid synthesis (32, 41). However, Cryptosporidium parvum employs only a giant, 900-kDa type I FAS (CpFAS1) that is capable of elongating medium- or long-chain fatty acids (42). On the other hand, Toxoplasma gondii and Eimeria tenella contain both type I and II enzymes and are able to synthesize fatty acids de novo and elongate long chain fatty acids (44). In addition, C. parvum also possesses a multifunctional polyketide synthase (CpPKS1) that is not present in any other apicomplexans examined so far (43). Currently, significant progress has been made toward the molecular and functional characterizations of these two types of apicomplexan FASs. However, nothing was known about the PPTases in this group of globally important parasites. It was unclear whether the two distinct apicomplexan ACP proteins were activated by different types of PPTases. Because the synthesis of holo-ACP is essential for fatty acid biosynthesis, PPTases may be considered as potential drug targets in apicomplexans and other pathogens. In addition, we have previously expressed the C. parvum type I CpFAS1 in bacteria. However, the ACP domains within recombinant CpFAS1 proteins were present in a nonfunctional apo form. This prompted us to search for the C. parvum PPTase that could specifically activate the CpFAS1 ACP domains for functional and pharmaceutical exploration.
In this study, we have identified both SFP and ACPS types of PPTases that may be responsible for activating type I and II apicomplexan ACPs, respectively, from three major groups of apicomplexans (i.e., P. falciparum, T. gondii, and C. parvum). Bioinformatics analyses suggest that these two types of apicomplexan PPTases may have different evolutionary origins. The SFP-type PPTases are more closely related to animal and fungal PPTases, while the ACPS-type enzymes were probably acquired from a proteobacterium. To further confirm the function of apicomplexan PPTases, we have also cloned and expressed the PPTase from C. parvum and detailed its enzymatic features in activating recombinant CpFAS1 ACP domains. The expression and protein localization of CpSFP-PPT have been investigated using real-time PCR and immunofluorescence microscopy.
MATERIALS AND METHODS
Identification and phylogenetic analyses of PPT genes from the apicomplexan genomes.
Four apicomplexan PPTases were identified from three genome databases by BLAST searches (P. falciparum [http://PlasmoDB.org], T. gondii [http://ToxoDB.org], and C. parvum [http://CryptoDB.org]) (5, 20, 29). One SFP-type PPTase was found in C. parvum (CpSFP-PPT), whereas one ACPS type was identified from P. falciparum (PfACPS-PPT). However, both SFP and ACPS types coexist in T. gondii (TgSFP-PPT and TgACPS-PPT). (Note: for clarity, we will use SFP-PPT and ACPS-PPT to refer to the SFP-type and ACPS-type PPTases, respectively). For the CpSFP-PPT gene, the transcript was amplified and cloned from C. parvum sporozoite total RNA by reverse-transcription PCR (RT-PCR). A comparison between cDNA and genomic DNA (gDNA) sequences revealed that CpSFP-PPT contained two introns. For the TgSFP-PPT gene, potential introns were identified with the guide of protein multiple alignments and nucleotide patterns at the intron-exon boundaries of previously annotated genes found in GenBank. The other two apicomplexan PPTases (TgACPS-PPT and PfACPS-PPT) appear to be intronless.
Apicomplexan PPTases were used as queries to search protein databases, including all nonredundant GenBank CDS translations plus RefSeq Proteins plus PDB plus SwissProt plus PIR plus PRF at the National Center for Biotechnology Information using the PSI-BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/) (3). Four iterative PSI-BLAST searches were performed for each protein (matrix = BLOSUM62; gap penalties: existence = 11 and extension = 1). Only sequences with E-values lower than 1 × 10−4 were retrieved for further analysis. Protein multiple sequence alignments were performed using a ClustalW algorithm, and apparent mistakes in alignments were corrected based upon visual inspections. Conserved motifs were determined from protein alignments (see protein alignments in the supplemental material), and block logos were derived from position-specific scoring matrices (PSSM) at the BLOCKS server (http://blocks.fhcrc.org/blocks) (15).
Only amino acid positions that could be unambiguously aligned were selected for phylogenetic analyses (see protein alignments in the supplemental material). Maximum likelihood (ML) trees were constructed using the PROML program distributed in the PHYLIP package (http://evolution.gs.washington.edu/phylip.html). The JTT model of amino acid substitution (18) was used in ML analysis with the consideration of among-site heterogeneity using the fraction of invariance plus a four-rate Γ-distribution model (JTT-f + Γ + Inv). Sequence input orders were randomized with 10 jumps, and global rearrangements were enabled during the tree searches. Missing parameters required by the PROML program were estimated by the Tree-Puzzle v5.2 program (34). In addition, ML trees were also reconstructed using a Bayesian inference method using the same model of amino acid substitutions (i.e., JTT-f + Γ + Inv) (17). A total of 500,000 generations of searches were performed with four chains simultaneously running, and the current trees were saved every 100 generations. Posterior probabilities at tree nodes were obtained by calculating consensus trees from 4,000 Bayesian inference trees written after the ML sums converged.
Gene expression pattern and protein localization of CpSFP-PPT in C. parvum.
The CpSFP-PPT gene expression pattern during the parasite complex life cycle was analyzed by real-time quantitative RT-PCR (qRT-PCR). Total RNA was isolated from different C. parvum (IOWA strain) life cycle stages (i.e., oocysts, sporozoites, intracellular stages cultured in HCT-8 cells for 6, 12, 24, 36, 48, 60, and 72 h) and uninfected host cells using an RNeasy isolation kit (QIAGEN) (7, 24). Since host cell RNA was present in intracellular samples, only the relative level of CpSFP-PPT transcripts was determined in comparison to that of C. parvum 18S rRNA for each sample. Real-time qRT-PCR analyses were performed using the SYBR-green-based iScript One-Step RT-PCR kit (Bio-Rad) and primers specific to CpSFP-PPT (301F, 5′-TCG GAA AGT GAT TCA TTA CTG C-3′; and 421R, 5′-CTC TGG AAG GTG ATA ACT CGG-3′) and 18S rRNA (995F, 5′-TAG AGA TTG GAG GTT GTT CCT-3′; and 1206R, 5′-CTC CAC CAA CTA AGA ACG GCC-3′) (1), respectively. Reactions containing 20 ng of total RNA and 0.2 μM specified primers were first incubated at 48°C for 30 min to synthesize cDNA, heated at 95°C for 15 min to inactivate reverse transcriptase, and then subjected to 45 thermal cycles (95°C, 20 s; 50°C, 30 s; and 72°C, 30 s) of PCR amplification using an iCycler iQ real-time PCR detection system (Bio-Rad). Each reaction contained at least three replicates. Under the specified conditions and based on annealing curve analysis, each pair of primers produced only one amplicon from samples containing parasite RNA but no products from total RNA isolated from uninfected host cells. Standard curves for both pairs of primers were determined using serially diluted DNA templates in real-time PCR analysis, followed by linear regression to determine needed parameters (curve slopes and intersections). Based on the standard curves, the amounts of CpSFP-PPT transcripts and 18S rRNA in every sample were calculated. The level of CpSFP-PPT transcripts in each sample was then normalized using that of 18S as a control (i.e., expressed as a ratio between levels of CpSFP-PPT and 18S rRNA). Finally, the relative level of CpSFP-PPT transcripts in each developmental stage was plotted relative to the mean level of all samples (i.e., ratio between individual normalized level and the overall mean).
For producing polyclonal antibodies to CpSFP-PPT, a short peptide corresponding to the CpSFP-PPT amino acid positions at 81 to 100 (CSPKQVKIIREKGMKPYFKY) was synthesized, cross-linked to keyhole limpet hemocyanin, and used to immunize two pathogen-free rabbits three times according to standard protocols at a commercial source (Alpha Diagnostic International). The preimmune sera and antisera were evaluated using enzyme-linked immunosorbent assay, immunofluorescence microscopy, and Western blot analysis to determine their specificities and titers.
In Western blot analysis, C. parvum sporozoites were prepared by excystation from fresh oocysts as previously described (24). Sporozoites were lysed in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer containing 1× protease inhibitor cocktail (Sigma), fractionated by 10% SDS-PAGE (∼2 × 107 sporozoites/lane), and transferred to a nitrocellulose membrane. Both rabbit preimmune serum and polyclonal antibody to CpSFP-PPT (1:1,000 dilution) were incubated with membrane blots, which were in turn detected with an Immuno-Star-AP chemiluminescent kit (Bio-Rad).
To detect CpSFP-PPT in parasite cells by immunofluorescence microscopy, C. parvum sporozoites and intracellular stages infecting HCT-8 monolayers (cultured on glass coverslips) were fixed in phosphate-buffered saline (PBS)-formalin (10%), washed with PBS and water, extracted with cold methanol (−20°C for 10 min) and 0.1% Triton X-100 (5 min, all at room temperature unless otherwise specified), respectively, and blocked with 5% bovine serum albumin (BSA)-PBS. Samples were then incubated with rabbit preimmune or antiserum (1:1,000 dilution) in 1% BSA-PBS, washed with PBS, and then incubated with a tetramethyl rhodamine isothiocyanate-labeled monoclonal antibody to rabbit immunoglobulin G (Sigma). Immunolabeled samples were mounted using a SlowFade antifade medium containing 4′,6′-diamidino-2-phenylindole for DNA counterstaining (Molecular Probes).
Expression of recombinant CpSFP-PPT and ACP domains.
The CpSFP-PPT gene was first cloned from parasite cDNA for expression in several commonly used bacterial fusion systems (e.g., pET-based T7-tag, His-tag, and S-tag fusions). However, either the expression was unsuccessful or the expression level was too low to be useful for biochemical assays. The unsuccessful expression of CpSFP-PPT was probably due to the extremely biased codon usages between the AT-rich C. parvum and Escherichia coli genes. To overcome the difficulties in protein expression, we used a PCR-based gene synthesis approach to reconstruct an artificial CpSFP-PPT gene based on E. coli class II codon frequencies. Briefly, a total number of 40 overlapping oligonucleotides (32 to 52 bases) were “reverse translated” from the CpSFP-PPT protein sequence at the “DNAWorks” oligonucleotide design server (http://molbio.info.nih.gov/dnaworks) (16). BamHI restriction sites were avoided in the synthetic gene but added to the 5′ and 3′ ends. A two-step PCR approach was employed. The first PCR contained all 40 oligonucleotides (0.2 μM each), and a 15-cycle amplification by Pfu DNA polymerase produced a smear of products. The second 20-cycle amplification step used the outer two oligonucleotides (0.2 μM) as primers and 1 μl of product from the first PCR as a template and yielded a single product. The synthetic gene was purified from an agarose gel and cloned into the pCR-Blunt II-TOPO vector. After its identity was confirmed by sequencing, the synthetic CpSFP-PPT gene was released by BamHI and inserted into the pET-29a vector with an S tag fused at the N terminus. Expression of S-tag-fused CpSFP-PPT (S-CpSFP-PPT) protein was carried out in a BL21(DE3) E. coli strain as recommended by the manufacturer (Novagen). The S-CpSFP-PPT protein was purified from bacteria using an S-tag thrombin purification kit, in which the fusion protein was isolated by S-protein agarose columns and the “native” CpSFP-PPT portion was released from the columns by digestion with thrombin.
The ACP domains within the loading unit (CpACP1) and elongation module 1 (CpACP2) of CpFAS1 were engineered into the pET24a (Novagen) and pMAL-c2x (New England Biolabs) vectors, respectively, as previously reported (44). The expression of T7-tag-fused CpACP1 (T7-CpACP1) or maltose-binding protein (MBP)-fused CpACP2 (MBP-CpACP2) was carried out in E. coli Rosetta (DE3) or Rosetta strains. Recombinant T7-CpACP1 and MBP-CpACP2 were purified using a T7-tag affinity purification kit (Novagen) or an amylose resin-based affinity purification kit (New England Biolabs), respectively.
Enzyme assays.
The activity of recombinant CpSFP-PPT was assayed using recombinant apo-T7-CpACP1 or apo-MBP-CpACP2 to receive the phosphopantetheinyl moiety from CoA or acetyl-CoA. Typical assay systems were performed in 100 μl Tris-HCl solution (75 mM; pH 7.0) containing MgCl2 (0.0039 to 20 mM), acetyl-CoA (0.39 to 200 μM) (or CoA), apo-ACP (0.85 to 1 μg), and CpSFP-PPT (100 ng) at 37°C for specified time periods (19). After determining that CpSFP-PPT could maintain linear activity for up to 20 min, we assayed the enzyme kinetics with 10-min reactions using [1-14C]acetyl-CoA and MBP-CpACP2 (5.0 μM) as cosubstrates. Reactions were stopped by adding 1 ml trichloroacetic acid (10%) and 25 μl BSA (2%) as a protein carrier (13, 30). Quenched reactions were incubated on ice for 5 min and centrifuged at 10,000 × g for 10 min. Protein pellets were washed three times with 1 ml trichloroacetic acid (10%), dissolved in 150 μl formic acid, and transferred into 10 ml scintillation cocktail (Sigma) for counting of radioactivity. For matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry (MS), reaction mixtures containing 0.1 mM acetyl-CoA (or CoA), 1 μg T7-CpACP1, and 1.0 mM MgCl2 were incubated for 30 min at 37°C, from which 20 μl solution from each reaction was removed for SDS-PAGE (20%) analysis, and the remaining 80 μl solution was desalted with a Bio-Gel P-6DG resin (Bio-Rad) for determining the Mr of T7-CpACP1 by MALDI-TOF MS. Reactions for autoradiography followed similar procedures but used [1-14C]acetyl-CoA (0.1 mM) and T7-CpACP1 or MBP-CpACP2 as substrates. Reactions were fractionated by SDS-PAGE. The radioactivity from the CpSFP-PPT-transferred phosphopantetheinyl moiety on the T7-holo-CpACP1 or MBP-holo-CpACP2 was visualized from dried gels using a FUJI BAS 1800 II PhosphorImager.
Activation of T7-CpACP1 by coexpression with CpSFP-PPT in bacteria.
For coexpression S-CpSFP-PPT and T7-CpACP1, we first isolated the pRIG-derived pRARE plasmid from Rosetta bacteria (4) and prepared a blunt-ended plasmid by PCR (primers: 5′-ACT AGT AAC GGC CGC CAG TGT GC-3′ and 5′-AGT GGA TCC GAG CTC GGT ACC AAG-3′). Second, the S-CpSFP-PPT construct, including the complete T7 promoter and transcriptional terminator, was amplified from the pET29a-CpSFP-PPT vector (primers: 5′-CTC CTT TCA GCA AAA AAC CCC-3′ and 5′-CCC GCG AAA TTA ATA CGA CTC-3′), phosphorylated, and ligated into the linear pRARE vector. The resulting pRARE-CpSFP-PPT construct and the pET24a-T7-CpACP1 vector were sequentially transformed into BL21(DE3) bacteria for coexpressing S-CpSFP-PPT and T7-CpACP1 proteins. In the control experiments, T7-CpACP1 was coexpressed with plain pRARE vector only. T7-CpACP1 proteins coexpressed with S-CpSFP-PPT or pRARE vector only were purified and desalted for the determination of their molecular masses by MALDI-TOF MS analysis.
Nucleotide sequence accession number.
Nucleotide sequence data for the CpSFP-PPT gene are available in the EMBL, GenBank, and DDJB databases under the accession number AY856092.
RESULTS
Two types of PPTases coexist in the apicomplexan genomes.
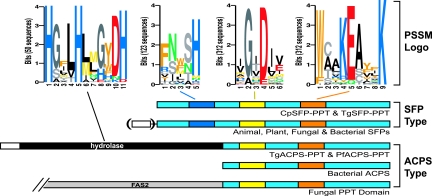
By data mining the apicomplexan genome sequences, we have discovered that apicomplexans possess both SFP-PPT and ACPS-PPT genes in their genomes (Fig. 2; Table 1). However, P. falciparum contains only an ACPS-PPT gene (PfACPS-PPT), while C. parvum has only an SFP-PPT gene (CpSFP-PPT). On the other hand, T. gondii possesses both an ACPS-PPT and an SFP-PPT gene (TgACPS-PPT and TgSFP-PPT). Whether an apicomplexan contains an ACPS-PPT and/or an SFP-PPT gene is fully correlated with the type(s) of FAS/PKS enzymes it possesses (Table 1). Apicomplexan ACPS-PPTs are bifunctional enzymes containing a C-terminal PPTase domain and an N-terminal metal-dependent hydrolase domain of yet-undefined functions (Fig. 2). Such a hydrolase-PPTase fusion is unique, since it has not yet been found in any other taxonomic groups examined. PfACPS-PPT and TgACPS-PPT also contain N-terminal transitional peptide sequences (Fig. 2; Table 1), suggesting that they are likely targeted to the apicoplast for activating type II ACP. On the other hand, SFP-PPTs in C. parvum and T. gondii are monofunctional enzymes that differ structurally from apicomplexan ACPS-PPTs. Both CpSFP-PPT and TgSFP-PPT proteins are predicted to be cytosolic (Table 1). In C. parvum (which lacks a plastid and type II FAS), CpSFP-PPT may function in activating the multiple ACP domains present in both type I CpFAS1 and CpPKS1 that have been shown to be cytosolic (43, 45). However, the type I FAS in T. gondii (TgFAS1) appears to be mainly localized to certain regions of the network-like mitochondrion, particularly in the region surrounding the apicoplast (8), which implies that the activation of its ACP domains by TgSFP-PPT may likely occur in the cytosol before TgFAS1 reaches the mitochondria.
FIG. 2.
Comparison of two types of PPTases among apicomplexans and other groups of organisms. SFP-PPTs are present in bacteria, type I FAS-containing apicomplexans, fungi, animals, and plants. Some plant SFP-PPTs may contain an N-terminal mitochondrium-targeting signal (open box in parenthesis). ACPS-PPTs are present in bacteria, plastid-containing apicomplexans, and fungi. Apicomplexan ACPS-PPTs are bifunctional and contain plastid-targeting signals (open box). Fungal ACPS-PPTs are present as a C-terminal domain in the α-subunit of fatty acid synthase (FAS2). PSSM logos include two motifs for all PPTases, one specific to SFPs and one for hydrolases.
TABLE 1.
Correlation between the types of PPTases and FAS/PKS in apicomplexans
| Organism | Type of FAS/PKS | PPTasea | Signal peptide in PPTases | Prediction method
|
||
|---|---|---|---|---|---|---|
| PlasmoAPb | ChloroPc | PlasMitd | ||||
| P. falciparum | Type II FAS | PfACPS-PPT | Plastid | Yes | No | Nof |
| C. parvum | Type I FAS and PKS | CpSFP-PPT | No | No | No | Nof |
| T. gondii | Type II FAS | TgACPS-PPT | Plastid | Noe | Yes | Nof |
| Type I FAS | TgSFP-PPT | No | No | No | Nof | |
Sources of apicomplexan PPTase protein sequences: CpSFP-PPT (GenBank accession no. AY856092) was annotated from cDNA in this study; PfACPS-PPT (NP_702851) was identified in GenBank by BLAST searches; TgSFP-PPT (contig TGG_995284) and TgACPS-PPT (TgTwinScan_3790) were identified from raw genome sequences at ToxoDB, release 3.0 (http://www.toxodb.org). Also see the supplemental material for conceptually translated TgSFP-PPT and TgACPS-PPT sequences.
PlasmoAP is trained for predicting plastid-targeting sequences in the AT-rich P. falciparum genes and may not be able to accurately predict sequences from other species including T. gondii.
All sequences are predicted to be 99% unlikely to contain mitochondrium-targeting signals.
Both SFP-PPT and ACPS-PPT from apicomplexans and other organisms contain two conserved motifs as determined by PSSM analysis of 312 sequences: [IV]G[ITV]D[ILV][VE] and W[CA][AL]KEAxxK (Fig. 2). The universal conservativeness of these two motifs among all PPTases suggests that some of their residues may be critical to the function of this family of enzymes. In fact, this notion is supported by the recent point-mutational analyses of PPTases in B. subtilis (11, 12). Besides these two motifs, PSSM analyses also identified a motif (FNxSH) present only among SFP-type PPTases (from 123 sequences) (Fig. 2). In addition, the identity of the hydrolase domain in PfACPS-PPT and TgACPS-PPT was also supported by their sequence similarity to other characterized metal-dependent hydrolases and the presence of many conserved residues. The most conserved motif among this group of enzymes is H[GSA]xLH[LM]x[GHE]xDH, in which the three H residues are present in almost all the hydrolase homologues examined here (from 50 sequences) (Fig. 2).
Different evolutionary origins of apicomplexan ACPS-PPT and SFP-PPT.
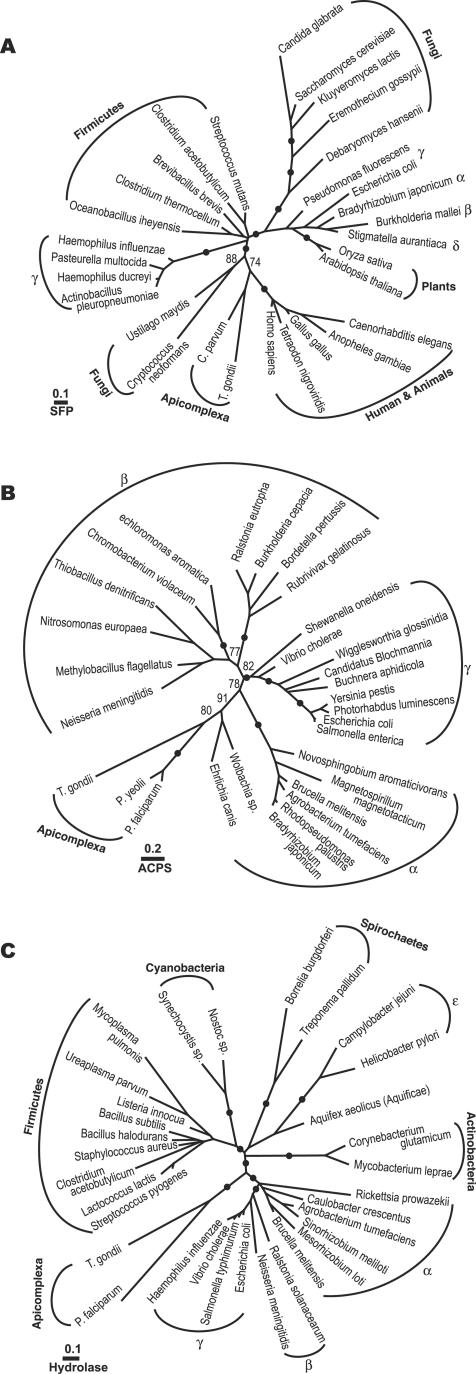
Although both SFP-PPT and ACPS-PPT are present in the Apicomplexa or even within one species (e.g., T. gondii), these enzymes may have different evolutionary origins based on the phylogeny individually performed for SFP-PPT, ACPS-PPT, and hydrolase protein sequences. Apicomplexan SFP-PPTs are phylogenetically related to animal and some fungal PPTases in maximum-likelihood trees inferred from 30 closely related protein sequences (Fig. 3A). The animals plus apicomplexans plus fungi cluster was subsequently joined by a group of γ-proteobacteria. It is noted that plants and some fungi formed two clades separated from the animal clade (Fig. 3A). However, because the resolving power of the SFP-PPT data set was limited by the small number of alignable positions (i.e., 52 amino acids), one could not firmly determine whether the separation among eukaryotic SFP-PPTs (more intriguingly, the separation of different fungal sequences) is caused by the long branch attraction artifact or indeed a result of multiple evolutionary origins of these eukaryotic SFP-PPTs.
FIG. 3.
ML trees inferred from (A) SFP-PPT sequences (30 sequences, 52 positions; −lnL = 2132.5); (B) ACPS-PPT sequences (30 sequences, 83 positions; −lnL = 3327.5); and (C) metal-dependent hydrolase sequences (32 sequences, 88 positions; −lnL = 4264.4). Statistical supporting values are posterior probabilities based on a Bayesian inference method. Only posterior probability values of >70% are shown, and solid dots represent PP values of ≥95%.
On the other hand, apicomplexan ACPS-PPTs are more closely related to bacterial PPTases. ML phylogeny based on 30 protein sequences placed PfACPS-PPT and TgACPS-PPT within the α-proteobacterial cluster (Fig. 3B). The proteobacterial origin of apicomplexan ACPS-PPTs was further supported by the phylogenetic analysis of the hydrolase domains, in which the apicomplexan clade formed a sister group to the α-, β-, and γ-proteobacterial clade (though paraphyletic to the α-proteobacterial hydrolases) (Fig. 3C).
The unusual two-intron-containing CpSFP-PPT gene is expressed in all parasite life cycle stages.
Among apicomplexan PPTase genes, PfACPS-PPT (NP_702851) has recently been annotated by the P. falciparum genome-sequencing consortium. TgACPS-PPT (TgTwinScan_3790) and TgSFP-PPT (contig TGG_995284) were identified from the ToxoDB (Release 3.0; 10× coverage) raw genome sequence data (http://www.toxodb.org) (see TgSFP-PPT and TgACPS-PPT protein sequences in the supplemental material). CpSFP-PPT has been annotated by the C. parvum genome project (EAK89734) as an intronless gene. However, our sequence analysis of CpSFP-PPT cDNA indicated that this gene is actually interrupted by two introns. The first intron contains multiple stop codons in all three open reading frames. However, the second intron is cryptic, since it contains no stop codon in the open reading frame extended from the second exon, thus misleading the computational prediction of C-terminal amino acids. Introns are not common in C. parvum, and only a few of them are reported (6, 7, 9). The genome-wide analysis suggests that only ∼5% of C. parvum genes may contain introns (2). Therefore, the presence of two introns in CpSFP-PPT appears to be unusual for this parasite.
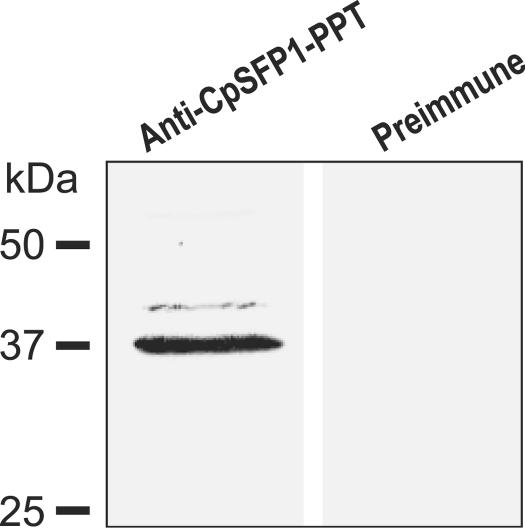
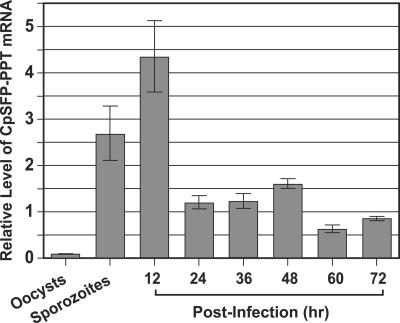
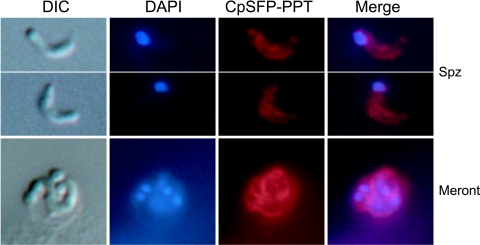
In Western blotting analysis of fractionated proteins from C. parvum sporozoites, the rabbit anti-CpSFP-PPT antibodies recognized a major band at ∼37 kDa (Fig. 4), although a slightly higher minor band (probably caused by nonspecific reactions) was also observed. The detected molecular weight agrees well with the theoretical mass of the CpSFP-PPT protein. Real-time qRT-PCR analysis indicated that the CpSFP-PPT gene is expressed in all C. parvum life cycle stages, suggesting that the activation of ACP domains is a continuous process during parasite development. However, the relative level of CpSFP-PPT transcripts (normalized using the level of 18S rRNA) was slightly higher in sporozoites and intracellular parasites at 12 h postinfection than other stages (Fig. 5). The lowest level of CpSFP-PPT expression was in the oocysts, and correlated with the overall low metabolic activity in this environmental parasite stage (Fig. 5). The low-level expression of CpSFP-PPT in unexcysted oocysts is unlikely to be an experimental artifact caused by poor isolation of oocyst RNA, because a comparable level of 18S rRNA was detected from this sample. The CpSFP-PPT protein was also detected in the cytosol of all parasite life cycle stages by indirect immunofluorescence microscopy (Fig. 6), which agrees with the lack of any signal peptide sequence and is congruent with the cytosolic nature of its substrates (CpFAS1 and CpPKS1) (44).
FIG. 4.
Western blot detection of CpSFP-PPT in total protein extracted from C. parvum sporozoites using a rabbit polyclonal antibody against a synthetic peptide specific to CpSFP-PPT. The negative control used rabbit preimmune serum.
FIG. 5.
Relative transcriptional levels of the CpSFP-PPT gene among various C. parvum developmental stages as determined by real-time quantitative RT-PCR. The amount of CpSFP-PPT transcript in each sample was first normalized with that of 18S rRNA. The relative levels of transcripts in all samples were then expressed by comparing their normalized values with the mean value.
FIG. 6.
Indirect immunofluorescence microscopic detection of CpSFP-PPT in C. parvum free sporozoites (Spz) and an intracellular developmental stage (a merozoite-containing meront) cultured in HCT-8 cells for 24 h. Samples were labeled with rabbit polyclonal antibody to CpSFP-PPT and TRITC-conjugated antirabbit immunoglobulin G monoclonal antibody that gave a diffused pattern in the cytosol of all samples. Preimmune serum did not label the parasite (data not shown). The nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI). Parasite morphology is shown as differential interference constrast (DIC) images taken from the same microscopic fields. A similar pattern of cytosolic distribution of CpSFP-PPT was observed in other intracellular stages (e.g., 12 to 72 h postinfection) but is not shown here.
CpSFP-PPT is capable of activating CpFAS1 ACP domains in vitro or when coexpressed in E. coli.
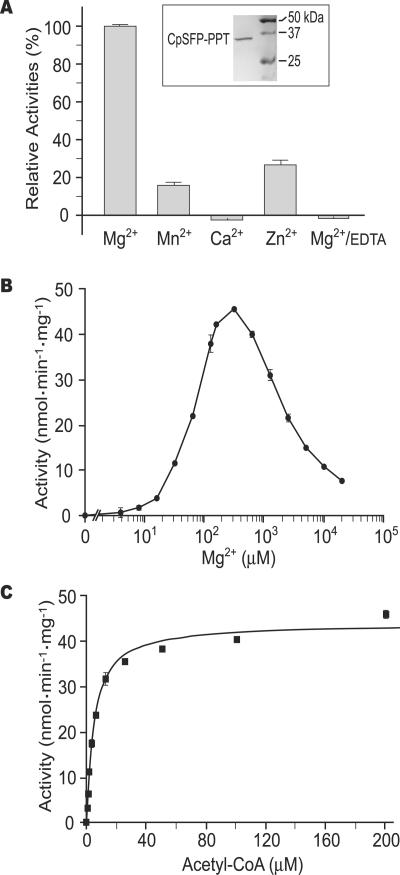
The S-tag-fused CpSFP-PPT protein was successfully expressed from a synthetic gene in bacteria. Using an S-protein-based affinity purification procedure, the “native” CpSFP-PPT portion was cleaved from the S tag and purified to homogeneity (Fig. 7A, inset). Radiochemical assays revealed that like human PPTases (19), CpSFP-PPT is able to utilize both CoA and acetyl-CoA as substrates for transferring the phosphopantetheine moiety to recombinant T7-CpACP1 and MBP-CpACP2 proteins. The activity of CpSFP-PPT is Mg2+ dependent and can be inhibited by EDTA (Fig. 7A). However, the optimal Mg2+ concentration for CpSFP-PPT (0.1 to 1 mM) differs dramatically from that for the human PPTase (5 to 20 mM) (19). Furthermore, although Mg2+ at low concentrations could cooperatively activate CpSFP-PPT, it also inhibited enzyme activity at higher-than-optimal concentrations (Fig. 7B). The detrimental effect of Mg2+ at higher-than-optimal concentrations has not been reported for other PPTases. However, such an “inhibitory effect” of a metal cofactor at high concentrations has been reported for other Mg2+-dependent enzymes, e.g., the smooth muscle myosin phosphatase and some rubber transferases (33, 35). In the latter case, it was speculated that metal, substrate, and metal-substrate complex could independently bind to the enzyme active site with different affinities. At low Mg2+ concentrations, metal predominantly bound to substrate, thus activating metal-dependent enzyme activity. However, when Mg2+ concentrations reached too high, it started to compete with the substrate for the active site, thus inhibiting the enzyme activity (35).
FIG. 7.
Enzymatic activity of recombinant CpSFP-PPT using [1-14C]acetyl-CoA and MBP-CpACP2 as cosubstrates. (A) Effects of cations (1.0 mM each) on the CpSFP-PPT activities. Inset shows the purified CpSFP-PPT (without S tag) analyzed by SDS-PAGE. (B) Effects of Mg2+ concentrations on the activity of CpSFP-PPT. (C) Enzyme kinetics of CpSFP-PPT determined using various concentrations of [1-14C]acetyl-CoA and 5.0 μM MBP-CpACP2 as cosubstrates. Note: In all samples, bars represent standard-error-of-the-mean values derived from at least three replicated reactions. The data presented here were generated from at least two independent experiments.
In the absence of Mg2+, CpSFP-PPT might utilize Mn2+ or Zn2+ (albeit with much lower enzymatic activities) but not Ca2+ (Fig. 7A). Using [1-14C]acetyl-CoA as a substrate and MBP-CpACP2 as a cosubstrate, the Vmax of CpSFP-PPT was determined as 44.1 (±0.675) nmol · mg−1 · min−1 (Fig. 7C), which is much lower than the reported Vmax values for human PPTase using human FAS ACP (288 nmol · mg−1 · min−1) or mitochondrial ACP (186 nmol · mg−1 · min−1) as cosubstrates (19). The Km was 4.9 (±0.325) μM (Fig. 7C), which is comparable to those of humans (3.1 to 7.5 μM). The Kcat (∼1.6 min−1) and Kcat/Km (0.324 min−1 · μM−1) were at the low end among the PPTases reported so far (19, 25, 26).
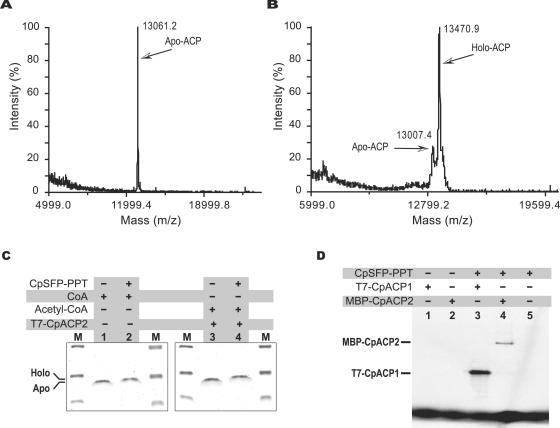
The successful transfer of a phosphopantetheine moiety from both acetyl-CoA and CoA to T7-CpACP1 was ultimately confirmed by three different approaches. The presence of a phosphopantetheinyl moiety in holo-ACP proteins was made evident by the differences in molecular masses between holo- and apo-ACP proteins as determined by MALDI-TOF MS analysis (Table 2; Fig. 8A and B) and SDS-PAGE fractionation (Fig. 8C). Radioactive phosphopantetheinyl moieties transferred from acetyl-CoA were detected in T7-CpACP1 and MBP-ACP2 fusion proteins that were treated with CpSFP-PPT by autoradiography but not in the proteins that received no CpSFP-PPT treatment (Fig. 8D).
TABLE 2.
Determination of molecular masses of recombinant T7-CpACP1 by MALDI-TOF MSa
| Preparation of T7-CpACP1 | Theoretical mass (Da) | Determined mass (Da) |
|---|---|---|
| Treated with CoA and Acetyl-CoA, but without CpSFP-PPT | 13,066 | 13,060 |
| Treated with CoA and CpSFP-PPT | 13,405 | 13,400 |
| Treated with acetyl-CoA and CpSFP-PPT | 13,448 | 13,484b |
| Untreated, isolated from bacteria with plain pRARE vector only | 13,065 | 13,061 |
| Untreated, isolated from bacteria co-expressed with CpSFP-PPT | 13,405 | 13,007 (22%) 13,470 (78%)c |
The 339-Da (or 382-Da) difference between apo- and holo-ACP following modification by CpSFP-PPT is due to the transferred phosphopantetheinyl moiety from CoA (or acetyl-CoA).
The discrepancy between theoretical and determined mass (≈36 Da) is probably caused by the N acetylation or carbamoylation of T7-CpACP1 as described for human ACP (19).
About 78% of the T7-CpACP1 coexpressed with CpSFP-PPT is in holo form (also see Fig. 8A and B for the mass spectrometry profiles). The discrepancy between theoretical and determined masses (65 Da) here indicates that a fatty acyl group is probably added to the holo-CpACP1, a phenomenon that has also been described for the bacterially expressed P. falciparum type II ACP (41).
FIG. 8.
Detection of the phosphopantetheinyl moiety from CpSFP-PPT-activated CpFAS1 ACP domains. (A) MALDI-TOF mass spectrum of T7-CpACP1 expressed in bacteria. A single peak at 12,061 Da indicates that all C. parvum T7-CpACP1 proteins were present in apo form. (B) MALDI-TOF mass spectrum of T7-CpACP1 coexpressed with CpSFP-PPT in bacteria. The major peak at 13,471 Da represents holo-T7-CpACP1 (∼78%), while the minor peak with lower molecular mass represents apo-T7-CpACP1 (∼22%). (C) Differentiation between apo- and holo-CpACP1 by SDS-PAGE (20% gel) stained with Coomassie blue. Apo-T7-CpACP1 (lanes 1, 3) migrated slightly faster than holo-T7-CpACP1 that received a phosphopantetheinyl moiety from CoA (lane 2) or acetyl-CoA (lane 4) after CpSFP-PPT treatment. (D) CpSFP-PPT-catalyzed transfer of a phosphopantetheinyl moiety from [1-14C]acetyl-CoA (included in all reactions) to either T7-CpACP1 (lane 3) or MBP-CpACP2 (lane 4) was detected by autoradiography. No radioactivity was detected from these two proteins receiving no CpSFP-PPT treatment (lanes 1 and 2) or from CpSFP-PPT itself (lane 5).
In addition, we also tested whether S-tag-fused CpSFP-PPT could activate T7-CpACP1 by coexpressing both proteins in bacteria. When the T7-CpACP1 protein coexpressed with S-CpSFP-PPT in bacteria was affinity purified and subjected to MALDI-TOF MS analysis, we observed that approximately 78% of the protein was present in holo form (Table 2; Fig. 8A and B). This observation suggests that S-CpSFP-PPT is not 100% efficient in activating coexpressed T7-CpACP1 in bacterial cells, probably due to the limited supply of CoA and acetyl-CoA in bacterial cells.
DISCUSSION
Apicomplexan parasites differ from their human and animal hosts in fatty acid synthesis by possessing either a type II pathway for de novo synthesis of long-chain fatty acids (e.g., P. falciparum) (32, 40, 41) or uniquely organized type I FAS for making very long chain fatty acids from long-chain fatty acids (e.g., C. parvum) (42, 44) or both (e.g., T. gondii and E. tenella) (8). The ACP proteins can be either discrete monofunctional enzymes in a type II pathway or functional domains fused within multifunctional FASs or PKSs. The types of PPTase present in an apicomplexan correlated well with the types of FASs possessed by the organism (Table 1), which supports the hypothesis that two distinct PPTases may be responsible for the activation of two different types of ACP in apicomplexans: the SFP-PPTs for type I ACP domains and the ACPS-PPTs for type II ACP enzymes.
The two types of apicomplexan PPTases appear to have different evolutionary origins: the SFP-PPTs in C. parvum and T. gondii are closely related to animal and fungal PPTases (Fig. 3A), whereas the ACPS-PPTs in P. falciparum and T. gondii are bacterium-like, probably having been acquired from a proteobacterial ancestor (Fig. 3B and C). The presence of two conserved motifs in both ACPS-PPT and SFP-PPT indicates that the two types of PPTases must have evolved from a common ancestor. The separation between ACPS-PPT and SFP-PPT might have occurred before the emergence of eukaryotes, based on the fact that prokaryotic and eukaryotic sequences are intermixed in both SFP-PPT and ACPS-PPT trees. In addition, apicomplexan SFP-PPTs appear to be more ancient than ACPS-PPTs, since the former (clustered with animals and fungi) were likely eukaryotic descendants, while the latter (bacterium-like) were probably acquired from a proteobacterium by lateral gene transfer after the Apicomplexa diverged from other eukaryotes.
The ability to obtain holo-ACP is critical to the biochemical and pharmaceutical studies of FASs. Bacterial expression systems are commonly used in producing recombinant apicomplexan proteins for functional analysis. Previous studies have shown that recombinant P. falciparum ACP enzymes are mostly in holo form when expressed in E. coli, suggesting bacterial ACPS is capable of activating apicomplexan type II ACP (41). In our previous studies, the 900-kDa CpFAS1 protein has been successfully expressed in bacteria as five individual modules (44). However, the ACP domains were found to be apoenzymes, suggesting that the E. coli ACPS was unable to activate the parasite ACP during expression. This technical obstacle apparently has limited our ability in functional dissection of the unique type I CpFAS1 and CpPKS1 using recombinant proteins. Therefore, the discovery and expression of a functional CpSFP-PPT enzyme clearly indicates that recombinant parasite ACP domains could be activated by CpSFP-PPT, thus clearing the path for the future functional analyses, including the determination of the final product(s), of CpFAS1 and CpPKS1 using recombinant proteins.
Fatty acid synthesis has recently been explored as a novel drug target (14, 32, 40, 42). The ultimate dependency of both type I and II FAS (and PKS) on PPTases for synthesizing holo-ACP, together with the high sequence divergence between apicomplexan and animal PPTases, indicates that PPTases may be explored as promising drug targets for this group of globally important parasites.
Supplementary Material
Acknowledgments
We thank Co-May Nguyen for technical assistance.
This research was supported by a grant from National Institutes of Health (R01 AI44594).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Abrahamsen, M. S., and A. A. Schroeder. 1999. Characterization of intracellular Cryptosporidium parvum gene expression. Mol. Biochem. Parasitol. 104:141-146. [DOI] [PubMed] [Google Scholar]
- 2.Abrahamsen, M. S., T. J. Templeton, S. Enomoto, J. E. Abrahante, G. Zhu, C. A. Lancto, M. Deng, C. Liu, G. Widmer, S. Tzipori, G. A. Buck, P. Xu, A. T. Bankier, P. H. Dear, B. A. Konfortov, H. F. Spriggs, L. Iyer, V. Anantharaman, L. Aravind, and V. Kapur. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441-445. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baca, A. M., and W. G. Hol. 2000. Overcoming codon bias: a method for high-level overexpression of Plasmodium and other AT-rich parasite genes in Escherichia coli. Int. J. Parasitol. 30:113-118. [DOI] [PubMed] [Google Scholar]
- 5.Bahl, A., B. Brunk, J. Crabtree, M. J. Fraunholz, B. Gajria, G. R. Grant, H. Ginsburg, D. Gupta, J. C. Kissinger, P. Labo, L. Li, M. D. Mailman, A. J. Milgram, D. S. Pearson, D. S. Roos, J. Schug, C. J. Stoeckert, Jr., and P. Whetzel. 2003. PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res. 31:212-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caccio, S., G. Larosa, and E. Pozio. 1997. The beta-tubulin gene of Cryptosporidium parvum. Mol. Biochem. Parasitol. 89:307-311. [DOI] [PubMed] [Google Scholar]
- 7.Cai, X., C. A. Lancto, M. S. Abrahamsen, and G. Zhu. 2004. Intron-containing beta-tubulin transcripts in Cryptosporidium parvum cultured in vitro. Microbiology 150:1191-1195. [DOI] [PubMed] [Google Scholar]
- 8.Crawford, M. J., G. Zhu, and D. S. Roos. 2003. Both type I and type II fatty acid synthases in Toxoplasma gondii. Molecular Parasitology Meeting XIV, abstr. 14C.
- 9.Deng, M., T. J. Templeton, N. R. London, C. Bauer, A. A. Schroeder, and M. S. Abrahamsen. 2002. Cryptosporidium parvum genes containing thrombospondin type 1 domains. Infect. Immun. 70:6987-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fichtlscherer, F., C. Wellein, M. Mittag, and E. Schweizer. 2000. A novel function of yeast fatty acid synthase. Subunit alpha is capable of self-pantetheinylation. Eur. J. Biochem. 267:2666-2671. [DOI] [PubMed] [Google Scholar]
- 11.Finking, R., M. R. Mofid, and M. A. Marahiel. 2004. Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in protein-protein recognition. Biochemistry 43:8946-8956. [DOI] [PubMed] [Google Scholar]
- 12.Finking, R., J. Solsbacher, D. Konz, M. Schobert, A. Schafer, D. Jahn, and M. A. Marahiel. 2002. Characterization of a new type of phosphopantetheinyl transferase for fatty acid and siderophore synthesis in Pseudomonas aeruginosa. J. Biol. Chem. 277:50293-50302. [DOI] [PubMed] [Google Scholar]
- 13.Gehring, A. M., K. A. Bradley, and C. T. Walsh. 1997. Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry 36:8495-8503. [DOI] [PubMed] [Google Scholar]
- 14.Gornicki, P. 2003. Apicoplast fatty acid biosynthesis as a target for medical intervention in apicomplexan parasites. Int. J. Parasitol. 33:885-896. [DOI] [PubMed] [Google Scholar]
- 15.Henikoff, S., J. G. Henikoff, W. J. Alford, and S. Pietrokovski. 1995. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene 163:GC17-GC26. [DOI] [PubMed] [Google Scholar]
- 16.Hoover, D. M., and J. Lubkowski. 2002. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 30:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 18.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comp. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 19.Joshi, A. K., L. Zhang, V. S. Rangan, and S. Smith. 2003. Cloning, expression, and characterization of a human 4′-phosphopantetheinyl transferase with broad substrate specificity. J. Biol. Chem. 278:33142-33149. [DOI] [PubMed] [Google Scholar]
- 20.Kissinger, J. C., B. Gajria, L. Li, I. T. Paulsen, and D. S. Roos. 2003. ToxoDB: accessing the Toxoplasma gondii genome. Nucleic Acids Res. 31:234-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambalot, R. H., A. M. Gehring, R. S. Flugel, P. Zuber, M. LaCelle, M. A. Marahiel, R. Reid, C. Khosla, and C. T. Walsh. 1996. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem. Biol. 3:923-936. [DOI] [PubMed] [Google Scholar]
- 22.Lambalot, R. H., and C. T. Walsh. 1995. Cloning, overproduction, and characterization of the Escherichia coli holo-acyl carrier protein synthase. J. Biol. Chem. 270:24658-24661. [DOI] [PubMed] [Google Scholar]
- 23.Marrakchi, H., Y. M. Zhang, and C. O. Rock. 2002. Mechanistic diversity and regulation of type II fatty acid synthesis. Biochem. Soc. Trans. 30:1050-1055. [DOI] [PubMed] [Google Scholar]
- 24.Millership, J. J., P. Waghela, X. Cai, A. Cockerham, and G. Zhu. 2004. Differential expression and interaction of transcription co-activator MBF1 with TATA-binding protein (TBP) in the apicomplexan Cryptosporidium parvum. Microbiology 150:1207-1213. [DOI] [PubMed] [Google Scholar]
- 25.Mofid, M. R., R. Finking, and M. A. Marahiel. 2002. Recognition of hybrid peptidyl carrier proteins/acyl carrier proteins in nonribosomal peptide synthetase modules by the 4′-phosphopantetheinyl transferases AcpS and Sfp. J. Biol. Chem. 277:17023-17031. [DOI] [PubMed] [Google Scholar]
- 26.Mootz, H. D., R. Finking, and M. A. Marahiel. 2001. 4′-Phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. J. Biol. Chem. 276:37289-37298. [DOI] [PubMed] [Google Scholar]
- 27.Mootz, H. D., K. Schorgendorfer, and M. A. Marahiel. 2002. Functional characterization of 4′-phosphopantetheinyl transferase genes of bacterial and fungal origin by complementation of Saccharomyces cerevisiae lys5. FEMS Microbiol. Lett. 213:51-57. [DOI] [PubMed] [Google Scholar]
- 28.Praphanphoj, V., K. A. Sacksteder, S. J. Gould, G. H. Thomas, and M. T. Geraghty. 2001. Identification of the alpha-aminoadipic semialdehyde dehydrogenase-phosphopantetheinyl transferase gene, the human ortholog of the yeast LYS5 gene. Mol. Genet. Metab. 72:336-342. [DOI] [PubMed] [Google Scholar]
- 29.Puiu, D., S. Enomoto, G. A. Buck, M. S. Abrahamsen, and J. C. Kissinger. 2004. CryptoDB: the Cryptosporidium genome resource. Nucleic Acids Res. 32(Database issue):D329-D331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quadri, L. E., P. H. Weinreb, M. Lei, M. M. Nakano, P. Zuber, and C. T. Walsh. 1998. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37:1585-1595. [DOI] [PubMed] [Google Scholar]
- 31.Rawlings, B. J. 1998. Biosynthesis of fatty acids and related metabolites. Nat. Prod. Rep. 15:275-308. [DOI] [PubMed] [Google Scholar]
- 32.Roos, D. S., M. J. Crawford, R. G. Donald, M. Fraunholz, O. S. Harb, C. Y. He, J. C. Kissinger, M. K. Shaw, and B. Striepen. 2002. Mining the Plasmodium genome database to define organellar function: what does the apicoplast do? Phil. Trans. R. Soc. LondonB Biol. Sci. 357:35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato, O., and Y. Ogawa. 2001. Myosin assembly critical for the enzyme activity of smooth muscle myosin phosphatase: effects of MgATP, ionic strength, and Mg(2+). J. Biochem. (Tokyo) 129:881-889. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 35.Scott, D. J., B. M. da Costa, S. C. Espy, J. D. Keasling, and K. Cornish. 2003. Activation and inhibition of rubber transferases by metal cofactors and pyrophosphate substrates. Phytochemistry 64:123-134. [DOI] [PubMed] [Google Scholar]
- 36.Smith, S., A. Witkowski, and A. K. Joshi. 2003. Structural and functional organization of the animal fatty acid synthase. Prog. Lipid Res. 42:289-317. [DOI] [PubMed] [Google Scholar]
- 37.Staunton, J., and K. J. Weissman. 2001. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 18:380-416. [DOI] [PubMed] [Google Scholar]
- 38.Thelen, J. J., and J. B. Ohlrogge. 2002. Metabolic engineering of fatty acid biosynthesis in plants. Metab. Eng. 4:12-21. [DOI] [PubMed] [Google Scholar]
- 39.van Roermund, C. W., H. R. Waterham, L. Ijlst, and R. J. Wanders. 2003. Fatty acid metabolism in Saccharomyces cerevisiae. Cell Mol. Life. Sci. 60:1838-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waller, R. F., P. J. Keeling, R. G. K. Donald, B. Striepen, E. Handman, N. Lang-Unnasch, A. F. Cowman, G. S. Besra, D. S. Roos, and G. I. McFadden. 1998. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 95:12352-12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waters, N. C., K. M. Kopydlowski, T. Guszczynski, L. Wei, P. Sellers, J. T. Ferlan, P. J. Lee, Z. Li, C. L. Woodard, S. Shallom, M. J. Gardner, and S. T. Prigge. 2002. Functional characterization of the acyl carrier protein (PfACP) and beta-ketoacyl ACP synthase III (PfKASIII) from Plasmodium falciparum. Mol. Biochem. Parasitol. 123:85-94. [DOI] [PubMed] [Google Scholar]
- 42.Zhu, G. 2004. Current progress in the fatty acid metabolism in Cryptosporidium parvum. J. Eukaryot. Microbiol. 51:381-388. [DOI] [PubMed] [Google Scholar]
- 43.Zhu, G., M. J. LaGier, F. Stejskal, J. J. Millership, X. Cai, and J. S. Keithly. 2002. Cryptosporidium parvum: the first protist known to encode a putative polyketide synthase. Gene 298:79-89. [DOI] [PubMed] [Google Scholar]
- 44.Zhu, G., Y. Li, X. Cai, J. J. Millership, M. J. Marchewka, and J. S. Keithly. 2004. Expression and functional characterization of a giant type I fatty acid synthase (CpFAS1) gene from Cryptosporidium parvum. Mol. Biochem. Parasitol. 134:127-135. [DOI] [PubMed] [Google Scholar]
- 45.Zhu, G., M. J. Marchewka, K. M. Woods, S. J. Upton, and J. S. Keithly. 2000. Molecular analysis of a type I fatty acid synthase in Cryptosporidium parvum. Mol. Biochem. Parasitol. 105:253-260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.