Abstract
Homologous DNA recombination (HR) allows the deletion (knockout), repair (rescuing), and modification of a selected gene, thereby rendering a functional analysis of the gene product possible. However, targeting of nuclear genes has been an inefficient process in most eukaryotes, including algae, plants, and animals, due to the dominance of integration of the applied DNA into nonhomologous regions of the genome. We have shown for the green alga Chlamydomonas reinhardtii by repairing a previously introduced truncated aminoglycoside 3′-phosphotransferase gene, aphVIII, that single-stranded DNA can recombine with a homologous endogenous DNA region of interest. Nonhomologous DNA integration appeared to be more than 100-fold reduced compared with the use of double-stranded DNA, thus allowing isolation of the homologous recombinants. We propose that this method will be applicable to direct targeting of nuclear C. reinhardtii genes.
Green microalgae are of great value both as organisms for fundamental biological research and as a resource for biotechnological industry. Because of its well-defined genetics Chlamydomonas reinhardtii is an ideal system for studying photosynthesis, chloroplast biogenesis, flagellar function, phototaxis, etc. However, genetic and molecular analyses of nuclear transformants reveal that integration of the DNA occurs predominantly via nonhomologous recombination (NHR) resulting in the introduction of the marker DNA at apparently random loci (7). Further application of C. reinhardtii as a model system and for technical use urgently demands targeted gene disruption and gene replacement.
Use of targeted gene disruption.
Targeted gene disruption allows the introduction of in vitro-generated mutations, including null mutations, into the genome of an organism. Homologous gene replacement has significant advantages over random integration (see Fig. S1 in the supplemental material) because it does not have drawbacks such as gene dosage effects, position effects, and the necessity of detection against a background of endogenous gene activity. The successful application of targeted gene disruption and replacement is dependent on the ratio of homologous to illegitimate recombination events during integrative transformation. This ratio is extremely variable among different eukaryotes. In several lower eukaryotes such as yeasts (12), some filamentous fungi (8), Trypanosomatideae (6), and the moss Physcomitrella patents (a plant with the predominance of the haplophase in the life cycle) (28) have a ratio of homologous to random integration above 10%. In archaea, in many lower eukaryotes like algae, and especially in most higher eukaryotes, the ratio of homologous to nonhomologous recombination events is very low. It varies between 10−2 and 10−3 in animal cells (4) and between 10−3 and 10−6 in plant cells (20, 23). Several approaches for identifying, selecting, and enriching rare homologous recombination events have been developed for plants. They involve using two markers: one for positive selection and another outside a homologous region for negative selection and screening, (see Fig. S1 in the supplemental material) (20, 25, 40). The most promising negative selection marker in higher plants still is the diphtheria toxin A gene (39). However, the number of over all transformants generated in plants is reduced only by a factor of 10. Moreover, negative selection markers select for double-crossover events that do not always occur during homologous gene integration in yeast and apparently are extremely rare in higher plants or Chlamydomonas (22). This might explain why it was impossible to disrupt some plant genes despite the high quantity of analyzed transformants (40).
An alternative approach to overcome the problem of low frequency of homologous recombination in plants is to overexpress well-characterized heterologous genes recA and ruvC that encode proteins which are involved in homologous recombination (26, 30). For tobacco protoplasts it was found that expression of the nucleus-targeted Escherichia coli recA gene stimulated intrachromosomal recombination between rather short (only 325 bp) homologous regions 10-fold.
Orr-Weaver et al. (24) demonstrated that recombination in yeast can be stimulated by the introduction of double-stranded breaks into duplex DNA substrates. However, to find a restriction enzyme that cuts specifically enough in a large genome is difficult even if enzymes with 18-bp recognition sites are used (2).
Use of ssDNA.
Gene targeting using single-stranded DNA (ssDNA) has successfully been carried out on Saccharomyces cerevisiae (33). The authors have not only shown that gene targeting with ssDNA is possible, they also provided some evidence that ssDNA is more efficient than targeting with double-stranded DNA (dsDNA). Surprisingly, both variants, i.e., double crossover resulting in gene conversion and single crossover resulting in plasmid integration (reciprocal exchange), had occurred. Since the efficiency of both processes was quite the same, the authors concluded that the primary crossover event is rate limiting. However, these experiments in yeast do not monitor nonhomologous end joining (NHEJ) and do not allow determination of the ratio of homologous recombination (HR) to NHR, because NHEJ is under any condition a rare process in yeast.
Baur et al. (1) and Bilang et al. (3) studied extrachromosomal homologous recombination in tobacco protoplasts and found that ssDNA was an efficient substrate. However, in these and many later experiments efficiency of gene targeting was not evaluated because in general recombination between two overlapping truncated selection marker genes was tested. Neither is active by itself, and they can only provide resistance after homologous recombination. This finding did not stimulate application of ssDNA for gene targeting in plants, and the problem of the low ratio between HR and NHR has not been solved yet (5, 39).
A very popular method for introducing foreign DNA into a plant host is the application of the plant-infecting agrobacteria. The transfer of agrobacterial T-DNA to plant cells involves the induction of Ti plasmid virulence genes. This induction results in the generation of linear single-stranded copies of the T-DNA, which are thought to be transferred to the plant cell. A central requirement of this ssDNA transfer model is that the plant cell generates a second strand and integrates the resulting dsDNA into its genome, which may explain why integration normally occurs more or less randomly. Thus, the dsDNA is likely to be the active species. Furner et al. (11) incubated plant protoplasts with ssDNA and dsDNA and found that the transformation efficiency is similar. The authors also suggested that the introduced DNA becomes double stranded before integration.
Homologous recombination in Chlamydomonas.
An earlier study of recombination in C. reinhardtii indicated that the machinery for homologous recombination exists in vegetative cells and suggested that a targeted gene disruption technique could be developed (13, 35). Using the efficient endogenous marker genes NIT1 and ARG7 the authors have shown that homologous recombination between two cotransforming nonfunctional gene copies containing nonoverlapping mutations occurred to a high frequency to restore the repaired active gene. The transformation rate of such plasmid pairs reached 10 to 20% of those for intact genes and was dependent on the length of homologous regions. Moreover, homologous recombination and repair were found to occur between the introduced and endogenous mutated gene copies, but at a rate a few orders of magnitude lower than the rate of extrachromosomal recombination. For the NIT1 gene, the estimated ratio of homologous to nonhomologous recombination events ranges between 1:40 and 1:1,000, depending on transformation method used (35). Only rare but detectable gene-targeted insertion was revealed at the ARG7 locus, and the numbers reported by Sodeine and Kindle seemed to be by far overestimated (13). Consequently, attempts to generate nuclear mutations by targeted gene disruption have had very limited success. The only convincing evidence for a successful application of this technique was reported by Nelson and Lefebvre (22). For targeted disruption of the NIT8 locus, they used the bare NIT8 coding sequence interrupted by the CRY1-1 selectable marker gene that provides emetine resistance. One of 2,000 transformants selected for emetine resistance contained a homologous insertion of five copies of the disruption construct within the NIT8 gene. Gene targeting by using single-stranded DNA has not been reported for any alga yet. Up to now random double-stranded gene integration and the subsequent analysis of thousands of transformants is still the preferred method for deletion of a gene which one cannot directly select for (14, 15).
It was our intention to provide an uncomplicated, reliable, and inexpensive method to generate gene-targeted transformants of C. reinhardtii by HR and to suppress illegitimate gene integration into the genome by NHR events. This goal can be reached by using purified ssDNA with DNA segments that are homologous to the target sequence. The major finding is that the use of ssDNA and complete removal of any double-stranded DNA dramatically reduce NHR, strongly improving the HR/NHR ratio.
MATERIALS AND METHODS
Vector construction.
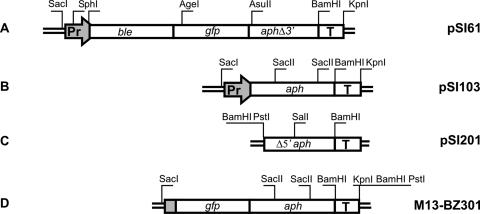
The full-length marker providing resistance to the antibiotic paromomycin is based on the aphVIII gene connected to the hemagglutinin (HA) promoter (HSP70/RBCS2) and an RBCS2 terminator (Fig. 1B, pSI103; 34).
FIG. 1.
Constructs that have been used for establishing directed gene targeting. Sequence accession numbers of the genes used are as follows: Pr = tandem promoter of HSP70/RBCS2, AY611535; ble, Z32751; gfp, AF188479; aphVIII, AF182845; and T, terminal RBCS2 3′, X04472. Sequences of the constructs A to D are specified below. Numbers in parentheses refer to the nucleotides listed under the respective accession numbers. Additional nucleotides are indicated as G, A, T, and C. (A) Plasmid pSI61: Pr(1-507), ble(1-370), tac, gfp(5-714), CTCGAGATTCGAAGC, aphVIII(1-629), CTCTACAACTAGT, T(2401-2633). (B) Plasmid pSI103: Pr(1-507), aphVIII(1-804), TGGTGGCTGGGTAGGGTTGCGTCGCGTGGGTGACAGCACAGTGTGGACGTTG, T(2401-2633). (C) Plasmid pSI201: aphVIII(121-804), TGGTGGCTGGGTAGGGTTGCGTCGCGTGGGTGACAGCACAGTGTGGACGTTG, T(2401-2633). (D) M13-BZ301: gfp(5-714), CTCGAGATTCGAAGC, aphVIII(1-804), TGGTGGCTGGGTAGGGTTGCGTCGCGTGGGTGACAGCACAGTGTGGACGTTG, T(2401-2633). Cleavage sites for restriction enzymes used for cloning and generation of transformation substrates are indicated.
Plasmids pKS-gfp-aphVIII and pKS-Δ3′aphVIII were constructed by cloning of the PCR-generated gfp-aphVIII (see the legend to Fig. 1) and Δ3′aphVIII (bp 1 to 634 of AF182845) into pBluscriptIIKS(−) (Stratagene). Plasmid pSI60 (HA promoter-ble-gfp-aph-3′rbcS2) was prepared from pMF59 (10) by the exchange of the AgeI-BamHI fragment for the AgeI-BamHI fragment containing gfp-aphVIII isolated from pKS-gfp-aphVIII. Plasmid pSI61 (HA promoter-ble-gfp-Δ3′aphVIII-3′rbcS2; accession no. DQ000653) was prepared from pSI60 by exchange of the AsuII-BamHI fragment, including aphVIII for the AsuII-BamHI fragment containing Δ3′aphVIII from pKS-Δ3′aphVIII. Plasmid pSI201 was constructed by cloning of the PCR-generated fragment containing Δ5′aphVIII-rbcS2 into the EcoRI + KpnI sites of pBluscriptKSII(−). Nucleotide sequences of all PCR-synthesized products were confirmed by sequencing. For detailed nucleotide sequences, see the GenBank entries given in the legend to Fig. 1.
Preparation of DNA.
Double-stranded DNA was used according to standard protocols and purified by gel electrophoresis. All extractions of dsDNA and ssDNA from the gel were done by using the Qiaquick gel extraction kit (QIAGEN, Hilden, Germany).
Linear single-stranded DNA.
The first method used for ssDNA preparation was linear PCR. It was performed by amplification of the SacI-KpnI fragment of pSI103 or the EcoRI-KpnI fragment of pSI201 only with the sense primer 5′ HSP (TGGAGCTCCACCGCGGTGG) and delta-5′-aph (GTCAAGGTGGCAGCTCTGG). Common PCR protocols were used: 95°C for 5 min, followed by 35 cycles of 95°C for 30 min, 60°C for 30 min, 72°C for 40 min, and finally 72°C for 5 min. Vent DNA polymerase (NEB) was used for all PCRs. Each PCR tube contained 50 μl reaction mixture with 300 to 500 ng DNA template. The total PCR product was precipitated by ethanol and cleaved with SacII for digestion of the double-stranded template. One to 1.5 μg of the final ssDNA was used for each transformation. Five independent experiments with at least 20 transformations each were carried out with all double-stranded constructs, and at least three experiments with 20 transformations were carried out with single-stranded DNA.
Circular single-stranded DNA.
The cloning vector pBluscriptKSII(−) is a phagemid and was used for the production of ssDNA by coinfection of E. coli cells with helper phage (VCSM13; Stratagene, Amsterdam, The Netherlands) according to the supplier's instruction. Briefly, 12 h after superinfection by the helper phage the cell culture was centrifuged, up to 3.5% polyethylene glycol 2000 was added to the supernatant, and the pellet was precipitated by centrifugation. The pellet was resuspended in 0.3 M NaOAc-1 mM EDTA followed by phenol-chloroform extraction. The ssDNA obtained was purified on 1% agarose gel in 4× TAE, digested with SacII to remove dsDNA contamination, and again purified by 1% agarose in 4× TAE (1× TAE is 40 mM Tris-acetate-1 mM EDTA, pH 8.0).
Cloning directly into M13mp18.
Plasmid pSI60 was cleaved with SphI and AgeI, blunted, and religated. This step removed the ble gene, the rbcS2 promoter, and about half of the hsp70 promoter. The obtained plasmid, pBZ62, was cleaved with SacI and KpnI. The resulting gfp-aphVIII-3′RBSC fragment was cloned between SacI and KpnI of the multiple cloning site of M13mp18 (GenBank accession no. M77815). Single-stranded DNA was prepared according to methods described in reference 27. ssDNA was purified on 1% agarose gels in 4× TAE. The DNA obtained was digested with SacII to remove residual dsDNA contaminations and run again through 1% agarose in 4× TAE.
Transformation.
The original recipient strain was C. reinhardtii cw15arg7 strain A (kindly provided by J. D. Rochaix). About 1 × 108 cells of the early exponential growth phase at an optical density at 800 nm of 0.2 to 0.3 were transformed with 2 to 3 μg of double-stranded plasmid DNA or 1 to 2 μg of ssDNA using glass beads (16). Zeocin-resistant Chlamydomonas clones were selected on 10 μg/ml Zeocin (Invitrogen, Karlsruhe, Germany) and analyzed by PCR and DNA blot hybridization. After transformation with pSI61, two independent transformants containing a single full-length ble-gfp-Δ3′aphVIII copy were isolated and named T61-5 and T61-9. The latter was used in all further experiments. After transformation of T61-9 with aphVIII or derivatives, transformants were selected on 15 μg/ml paromomycin (34). The resulting transformants were all still resistant to Zeocin.
PCR analysis and DNA blotting.
Total genomic DNA was prepared from C. reinhardtii with a DNeasy plant mini kit (QIAGEN; Hilden). For detection of clones with a repaired aphVIII gene and discrimination from transformants with nonhomologous gene integration (NHI), integration was tested by PCR with forward primer 2 (GAGATCGGCGAGCAGCCGTGG), according to Fig. 2, located inside the ble sequence and reverse primer 1 (ACCAGCGCGAGATCGGAGTGC) located at the very end of the aphVIII sequence, or reverse primer 3 (GAGCAGTATCTTCCATCCACC) belonging to the 3′-RBCS2 sequence (as in Fig. 2). Conditions for PCR analysis are similar to those described above. DNA blotting was carried out according to standard protocols. As a hybridization probe, we used a digoxigenin-labeled gfp-aphVIII-PCR fragment. Hybridizations were performed at 42°C in 50% formamide.
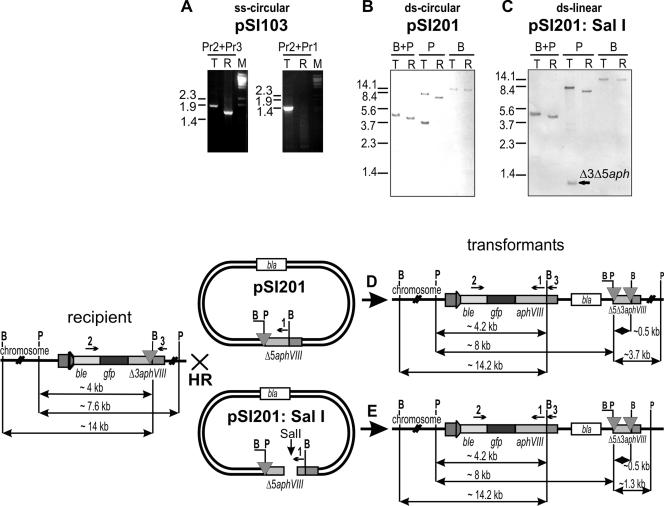
FIG. 2.
(A) PCR analysis of the recipient (R) and transformant (T); generated with single-stranded linear DNA (Pr-aphVIII-3′-rbcS2; Table 1, no. 5), which shows the repaired aphVIII gene. Primer positions are seen in the lower part of the figure: The product generated with Pr2+Pr3 indicates the increased size of the gfp-aphVIII region within the transformant. The product generated with Pr2+Pr1 indicates the repair of the 3′-aphVIII gene by insertion of the formerly missing 175-bp fragment. Pr1 does not anneal to the DNA of the recipient T61-9. (B and C) DNA blotting of transformants resulting from transformation of the T61-9 strain cells with circular double-stranded promoterless ds-Δ5′-aphVIII (ds-pSI201) (B) and linear double-stranded promoterless ds-Δ5′-aphVIII (ds-PSI201:SalI) (C). Total genomic DNA was cleaved with BamHI (B) and/or PstI (P). The bands are explained by the fragments defined for panels D and E. The blots were probed with an ss-DIG-labeled DNA covering the complete gfp-aphVIII coding sequence of 1,514 nucleotides. (C and D) Schematic representation of the integration loci in transformants analyzed in panels B and C. bla, ampicillin resistance gene.
RESULTS
Development of a detection system for determining the ratio of homologous recombination versus illegitimate gene integration, HR/NHR.
For comparing homologous recombination to nonhomologous gene integration (NHR), a system had to be created that evaluates the ratio and the efficiency of both processes. We transformed strain CW15 with the plasmid pSI61 (Fig. 1A) and generated the new C. reinhardtii recipient strain, T61-9. It contains an inserted genomic DNA element comprising the ble gene, the gfp gene, and a 3′-truncated aphVIII gene (Δ3′-aphVIII, 175 nucleotides missing at the 3′ end), all in frame. ble was used for selection of this strain on Zeocin (19, 36), Δ3′-aph for later selection of homologous recombinants on paromomycin, and gfp for potential monitoring of aphVIII expression as a fusion protein on protein blots or in living cells.
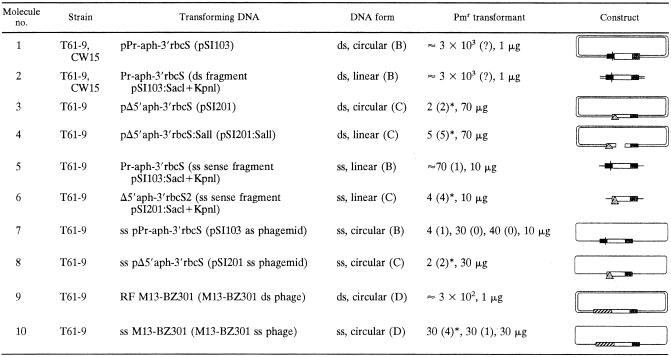
First, strain CW15 was transformed with a functional aphVIII gene (plasmid pSI103; Fig. 1B) (34). The expression was driven by a hybrid promoter, namely a combination of those of the HSP70 and RBCS2 genes (HA promoter) (29) and further enhanced by the RBCS2 3′ end. We obtained about 3,000 clones/1 μg of DNA, which is similar to the numbers obtained in earlier experiments (34). Similar numbers were also achieved with the recipient strain, T61-9 (Table 1, no. 1). The clones were not analyzed any further because from earlier experiments we could anticipate that most transformants were based on nonhomologous DNA integration into the genome. Almost the same number of clones was generated with 1 μg of a 1.8-kb SacI-KpnI fragment of pSI103 comprising the aphVIII coding region plus HA promoter and 3′ region of the RBCS2 gene (Table 1, no. 2). The molar amount of DNA was roughly four times higher than that of cyclic DNA used, but with respect to μg of DNA, there was no difference between transformations with linear or circular double-stranded DNA.
TABLE 1.
Transformation of CW15 and T61-9 with different DNA substratesa
DNA molecules 1, 2, 5, 7, 9, and 10 generate transformants after HR or NHR, whereas 3, 4, 6, and 8 generate transformants only after HR. In the DNA form cloning, letters in parentheses refer to constructs of Fig. 1. In the Pmr transformant column, the numbers of homologous recombinants are displayed in parentheses. If labeled with an asterisk, they have been analyzed by DNA blotting. The amount of DNA used for one experiment with 1 to 30 parallel transformations is given in μg; 1 to 3 μg was used per transformation.
Next, Chlamydomonas cells were transformed with a plasmid that contained two diphtheria toxin A fragment genes, dtA, plus HA promoter on both sites of the aphVIII in order to suppress illegitimate gene integration (negative selection). The dtA gene was resynthesized and adapted to the nuclear codon usage of strongly expressed C. reinhardtii genes according to reference 9 (accession no. AY611535). Similar to in rice (38), the total number of clones declined by a factor of about 8 in both strains CW15 and T61-9. Thus, the introduction of dtA did not sufficiently reduce the transformation rate so as to make it likely that the HR/NHR ratio was significantly increased. Therefore, transformants were not analyzed within this context and a detailed analysis might be reported elsewhere (B. Zorin, unpublished data).
To determine the frequency of homologous recombination, we transformed Chlamydomonas cells with an aphVIII gene that was truncated at the 5′ end by 120 nucleotides (plasmid pSI201). This Δ5′-aphVIII gene (Fig. 1C) could only generate paromomycin-resistant (Pmr) clones after recombination with the Δ3′-aphVIII of the recipient but not after integration elsewhere into the genome (I. Sizova, unpublished data). Only two T61-9 transformants were generated with 70 μg DNA (Table 1, no. 3). In yeast, integrative recombination is enhanced by introducing a double-stranded break in the region of homology shared between the transforming vector and the targeted gene (24). In line with these early findings, C. reinhardtii was transformed with a plasmid that has been linearized within the 5′Δ-aphVIII gene with SalI (named pSI201:Sal). The free double-stranded ends obviously promoted recombination only slightly, namely by a factor of 2.5 in five independent experiments, which was at the border of significance (Table 1, no. 4). PCR such as that in Fig. 2A has shown that, in all seven transformants tested (no. 3 and 4), the aphVIII gene has been successfully repaired. Primers from the ble part and the 175-bp fragment that is missing in the recipient strain T61-9 were used (see Fig. 2A). Two clones generated with pSI201 and five clones generated with pSI201 linearized with SalI were analyzed by DNA blot hybridization. One of each is shown in Fig. 2B and 2C. In both transformants, we observed one BamHI-PstI fragment that is enlarged compared to the recipient (from 4 to 4.2 kb) and consistent with the repair of the aphVIII gene. The larger shift of the PstI fragment (7.6 to 8 kb in Fig. 2B) again seen in all transformants is consistent with a single crossover leading to integration of plasmid DNA as outlined in Fig. 2D and E. However, the second PstI fragment including the Δ5′Δ3′aphVIII region is different in all five analyzed transformants, indicating an unclear and variable integration of the 3′end. In one transformant generated with circular plasmid, the Δ5′Δ3′aphVIII region is seen as 3.7-kb fragment (Fig. 2B), as expected from the PstI site position in the recipient. The interpretation was supported by probing the DNA blot with DIG-labeled pBluescriptIIKS− (vector only), which identified the 8-kb band but not the 3.7-kb PstI band (data not shown). In the five transformants resulting from linearized plasmid, the Δ5′Δ3′aphVIII region is located within short unexpected PstI fragments (1.3 kb in Fig. 2C and E) or is absent from all (data not shown). We suspect that the 0.5-kb BamHI fragment of Δ5′Δ3′-aphVIII has run off the gel. These observations show that even if a gene is effectively targeted through HR, DNA rearrangements can occur. But, under our experimental conditions, clones with 5′ rearrangements did not survive. Large rearrangements have been found after gene targeting and repair of NIT8 (22).
In summary, we have compared transformation rates with dsDNA carrying a similar length of homology, but either able or unable to transform Chlamydomonas via illegitimate recombination (experiments 1 versus 3 and 2 versus 4 of Table 1): we find that for linearized and circular DNA, the ratio HR/NHR is in the order of (4 × 104 to 105: [(3 × 103) × 70]/5 and [(3 × 103) × 70]/2).
Gene targeting using ssDNA.
By comparing natural homologous recombination processes with illegitimate gene integration processes as they occur in various systems like transposons, P-elements of Drosophila, retroviruses, retrotransposons, or T-DNA from agrobacteria, we have drawn the conclusion that all nonhomologous integration processes involve double-stranded DNA, whereas homologous recombination always occurs via single-stranded DNA or single-stranded reaction intermediates. We have hypothesized that the use of single-stranded DNA might prevent illegitimate integration processes.
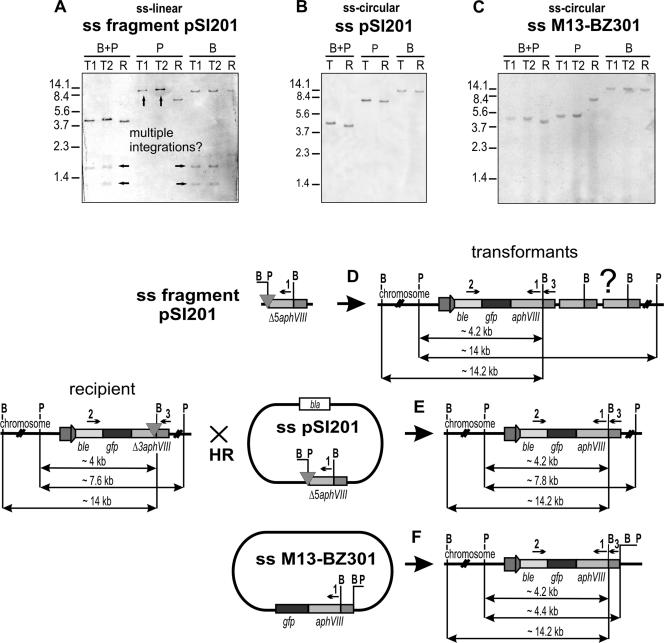
We transformed C. reinhardtii T61-9 cells with a fully functional single-stranded aphVIII gene (sense strand, SacI-KpnI fragment of pSI103) without plasmid extensions but including HA promoter and the 3′ end from the RBCS2 gene (Fig. 1B). Linear ssDNA was prepared by linear PCR from the ds-SacI-KpnI template and careful purification of the ss product. Ten micrograms of DNA generated only 70 transformants in T61-9 cells (Table 1, no. 5) instead of 3 × 104, which would appear after transformation with comparable amounts of the same double-stranded construct (Table 1, no. 2). From these numbers, it was not clear whether ssDNA is less efficient for transformation in general and to what extent HR and NHR had occurred. Moreover, all transformants could be based on nonhomologous gene integration caused by residual traces of dsDNA. All 70 transformants were tested by PCR. One transformant was a homologous recombinant. To test the frequency of homologous recombination with single-stranded DNA more directly, Chlamydomonas cells were transformed with an ss-aphVIII fragment that was deleted at the 5′ end (Table 1, no. 6). Four homologous recombinants were found. Two—not necessarily independent—were analyzed by DNA blotting (Fig. 3A). Both were based on double-crossover events but showed multiple integrations of the aphVIII genes into the aphVIII site of the recipient according to Fig. 3A and D.
FIG. 3.
(A to C) DNA blotting of transformants resulting from transformation of T61-9 strain cells with linear single-stranded promoterless ss-Δ5′-aphVIII (ss-SacI-KpnI fragment of PSI201) (A), circular single-stranded promoterless ds-Δ5′-aphVIII (ss-pSI201) (B), and circular single-stranded promoterless gfp-aphVIII (ss-M13-BZ301) (C). Total genomic DNA was cleaved with BamHI (B) and/or PstI (P). The blots were probed with the same ss-DIG-labeled DNA as used in Fig. 2. (D, E, F) Schematic representation of the integration loci of the transformants analyzed in panels A to C.
It was conceivable that ssDNA degradation contributed to the reduction of the total number of transformants. Especially, degradation of the promoter would cause a reduced number of nonhomologous integrations in the case of construct no. 5 of Table 1. To test this possibility, transformation was repeated with cyclic single-stranded DNA (Table 1, no. 7). ss-aphVIII DNA was produced in pBluescriptIIKS(−) phagemid. Ten micrograms of ss phage DNA resulted in four transformants. In one of them, the 3′ deletion of the aphVIII had been repaired as verified by PCR and sequencing of the aphVIII PCR product. In the other three transformants, aphVIII had integrated somewhere else, possibly into homologous plasmid sequences of the T61-9 recipient outside the aphVIII or into the endogenous rbcS2 5′ or 3′ sequences. This has not been experimentally verified. Nevertheless, the result looked highly promising. But, unfortunately, we were unable to reproduce this finding. In several subsequent repeats, the number of clones was higher and no recombinant was found (see Table 1, no. 7). To get more reliable numbers for the frequency of HR after transformation with cyclic ssDNA, experiments were repeated with 5′-deleted single-stranded aphVIII in plasmid (Table 1, no. 8) and with 3 times more DNA (30 μg ss-Δ5′-aphVIII). Two clones, both homologous recombinants, were found. DNA blotting revealed that the two transformants generated with circular ss-Δ5′-aphVIII gives rise to only one repaired aphVIII gene in each transformant with no double-deleted copy (Fig. 3B and E). For the clone analyzed in Fig. 3B, the double-truncated Δ3′Δ5′aphVIII copy (small PstI fragment), as created by a single “crossover” event using ds-circular DNA (Fig. 2B and C), did not appear. In addition, the size of the large PstI fragment increased only slightly, from 7.6 to 7.8 kb, in agreement with repair only. Both observations are consistent with a double-crossover event as outlined in Fig. 3E (further discussed below).
Single-stranded plasmid linearized within the aphVIII gene comparable to the double-stranded plasmid in Table 1, no. 4, was not used for two reasons. First, linearization is difficult with ssDNA and only possible by using adapter primers. Second, it is thought that linearized double-stranded plasmid integrates into the host genome by the two 3′ ends of the opposite plasmid strands (18). ssDNA with only one 3′ end cannot integrate via such mechanism (further discussed below).
By comparing transformation efficiencies, we draw the conclusion that the efficiency of homologous recombination is identical with ssDNA and dsDNA within the experimental errors. In contrast, the frequency of nonhomologous integration was dramatically reduced when ssDNA was applied.
It was not unlikely that the small absolute number of recombinants was correlated with the shortness of homology 5′ of the aphVIII deletion that had to be repaired. In a final set of experiments, we transformed with a promoterless full-length aphVIII connected to 720 nucleotides of gfp (ss-M13-BZ301), resulting in a 1.4-kb sequence of homology 5′ contiguous to the recipient deletion. In former experiments, promoter deletion from double-stranded aphVIII caused a 5- to 140-fold reduction of transformants compared to homologues that were linked to promoters of different strength (34). Promoterless aphVIII is able to jump in frame into any other gene, the transcription of which is driven by a moderate promoter. However, the production of the circular ss-gfp-aphVIII plasmid proved to be difficult in our hands using the phagemid system. With increasing plasmid size, the yield of ssDNA of interest drastically dropped. Therefore, gfp-aphVIII was directly cloned into M13mp18 (New England BioLabs) phage (plasmid M13-BZ301). After two independent transformations with 30 μg DNA, in every case 30 transformants appeared; 4 and 1 were homologous recombinants (Table 1, no. 10). Two were analyzed by DNA blotting (Fig. 3C). The blots are similar, and the transformants obviously did not originate from independent events. Both showed single integration and repair of the aphVIII gene. As outlined in Fig. 3F, the 5′ crossover was homologous, whereas the 3′ crossover is less obvious. It must have occurred within the 8-kb Pst fragment of the recipient but outside the Pst-fragment of the transforming plasmid. Thus, the resulting Pst fragment was smaller than that of the recipient. We do not know to what respect the DNA regions outside the RBCS2 site are homologous since the sequence of the recipient is not known. But, other than in the case of no. 8, a rearrangement of the 3′ end cannot be excluded. By comparing the number of clones that had appeared after transformation with the single-stranded M13-BZ301 vector (Table 1, no. 10) and double-stranded replicative form (Table 1, no. 9), the number of nonhomologous recombinants is reduced about 300 times with promoterless constructs. One should keep in mind that with promoterless constructs, only recombinations that occurred in frame into an active exon become visible as a clone. This reduced the number of nonhomologous recombinants and the NHR/HR ratio.
DISCUSSION
The experiments presented above demonstrate successful targeted recombination of single-stranded DNA with homologous nuclear DNA segments in Chlamydomonas. Our data allow the conclusion that the tendency of ssDNA to integrate at nonhomologous locations of the genome is much lower than that of dsDNA. For a linear gene fragment linked to a strong promoter, the reduction was several hundred to a few thousand times (lines 2 and 5 of Table 1). The reduction for circular DNA was less clear because the number of transformants was quite variable, but the reduction was of the same order of magnitude (lines 1 and 7). For genes with a weak promoter or no promoter, the reduction of the number of transformants was smaller since nonhomologous integration resulted in a functional resistance marker only after insertion into selected places of the genome like exons of expressed genes (lines 9 and 10).
The enhancement of recombination observed with dsDNA after double strand breaks in E.coli and yeast indicated that the created free ends are used as starting points for generation of partially single stranded 3′-OH fragments. Such overhangs are generated in E. coli by the RecBCD complex and in yeast by analogous proteins during double-stranded break repair (32), or as originally postulated for P-element-induced recombination in Drosophila during synthesis-dependent strand repair (synthesis-dependent strand annealing) (21).
At homologous sites the ssDNA invades the dsDNA molecule, forming a D-loop with annealing of the ssDNA with one of the target strands. In case of single crossover the second 3′ end anneals with the opposite strand at the same location. The two 5′ ends of the invading DNA are connected with the target DNA, implying two strand transfer reactions via Holliday-like rearrangement integrating the whole plasmid into the host DNA (18). Such DNA integration at one location is according to our understanding not possible with single-stranded DNA because the second free 3′ end does not exist. Thus, it appears surprising that HR was almost equally efficient with dsDNA and ssDNA. The only explanation available at the moment for single-stranded integration is that homologous single-stranded sense DNA annealed with the host antisense strand at two locations at opposite sites of the deletion. Next, both free ends are covalently connected to the same sense strand by DNA break and religation (double crossover), forming a heteroduplex with a mismatched region. This gene conversion event occurs preferentially without integration of additional plasmid DNA. Replication of the strand with the corrected aphVIII (including the missing 175 bp) results in resistant progenies.
Simon and Moore (33) found integration of single-stranded plasmids into the yeast genome. They were likewise surprised and explained their finding by replication of the single-stranded plasmid into double-stranded plasmid during the integration process, initiated by the nick in the host DNA.
Invasion of ssDNA into the double-stranded host DNA is mediated by RecA or its eukaryotic analog, Rad51. They form right-handed helical nucleoprotein filaments and carry out a homology search followed by strand exchange (17). The computer analysis of Chlamydomonas databases revealed homologues of RAD51 (ChlamyRAD51; accession no. AAS75433) and genes related to RAD51 paralogs: ChlamyRAD51B (accession no. AAS75434), and ChlamyRAD51C (accession no. AAL27842), which were likely to participate in gene targeting in the alga. Recently, we have shown that ChlamyRAD51C in vitro promotes DNA strand exchange (31).
The mechanism of DNA integration for both cases, namely single and double crossovers for ssDNA and dsDNA, is not clear yet, and more experiments need to be done. For most described cases like “double-stranded break repair,” “synthesis-dependent strand annealing,” and the “postreplication repair model,” double-stranded breaks and single-stranded overhangs initiate the recombination process (32). The finding that linearized dsDNA did not significantly facilitate recombination was therefore unexpected. In yeast linear transforming DNA is more recombinogenic than circular DNA and mating-type switching in yeast is initiated by a double-stranded break (37). This apparent discrepancy may be explained in such a way that in Chlamydomonas the break itself is not the rate-limiting step.
The strongly improved ratio of HR/NHR in C. reinhardtii provides promising expectations for ssDNA as substrates for gene targeting of endogenous nuclear genes. However, we do not underestimate the difficulties that may be encountered with real genes. Enlarged nonhomologous segments caused by the introduction of a selection marker may prevent the second crossover. Positional effects may also reduce the targeting efficiency, especially when genes are targeted that are expressed weakly or only under special conditions. For gene repair or introduction of point mutations, the marker must be included outside the coding region, which might raise additional complications. But nevertheless the results achieved with ss-aphVIII at least let us conclude that ssDNA is a more efficient targeting tool than dsDNA. In the future, the absolute number of homologous recombinants will be the limiting factor.
Acknowledgments
Many thanks go to Markus Fuhrmann, Wolfgang Oertel, and Paul Lefebvre for stimulating discussion and suggestions.
The work was supported by the DFG (P.H.) and the INTAS program.
Footnotes
This publication is dedicated to Karen Kindle, who convinced P.H. in 1994 that nuclear-gene targeting is possible in Chlamydomonas.
REFERENCES
- 1.Baur, M., I. Potrykus, and J. Paszkowski. 1990. Intermolecular homologous recombination in plants. Mol. Cell. Biol. 10:492-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibikova, M., K. Beumer, J. K. Trautman, and D. Caroll. 2003. Enhanced gene targeting with designed zinc finger nucleases. Science 300:764. [DOI] [PubMed] [Google Scholar]
- 3.Bilang, R., A. Peterhans, A. Bogucki, and J. Paszkowski. 1992. Single-stranded DNA as a recombination substrate in plants as assessed by stable and transient recombination assays. Mol. Cell. Biol. 12:329-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bollag, R. J., A. S. Waldmann, and R. M. Liskay. 1989. Homologous recombination in mammalian cells. Annu. Rev. Genet. 23:199-225. [DOI] [PubMed] [Google Scholar]
- 5.Bouche, N., and D. Bouchez. 2001. Arabidopsis gene knock-out: phenotype wanted. Curr. Opin. Plant Biol. 4:111-117. [DOI] [PubMed] [Google Scholar]
- 6.Cruz, A., and S. M. Beverley. 1990. Gene replacement in parasitic protozoa. Nature 348:171-173. [DOI] [PubMed] [Google Scholar]
- 7.Debuchy, R., S. Purton, and J.-D. Rochaix. 1989. The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 8:2803-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fotheringham, S., and W. K. Holloman. 1989. Cloning and disruption of Ustilago maydis genes. Mol. Cell. Biol. 9:4052-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuhrmann, M., W. Oertel, and P. Hegemann. 1999. A synthetic gene coding for the green fluorescent protein GFP is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 19:353-361. [DOI] [PubMed] [Google Scholar]
- 10.Fuhrmann, M. 2001. Grün fluoreszierendes Protein und gezielte Genstilllegung in Chlamydomonas reinhardtii. Dissertation. Universität Regensburg, Regensburg, Germany.
- 11.Furner, I. J., E. S. Higgins, and A. W. Berrington. 1989. Single stranded DNA transforms protoplasts. Mol. Gen. Genet. 220:65-68. [Google Scholar]
- 12.Grimm, C., and J. Kohli. 1988. Observation on integrative transformation in Schizosaccharomyces pombe. Mol. Gen. Genet. 154:97-100. [DOI] [PubMed] [Google Scholar]
- 13.Gumpel, N. J., J.-D. Rochaix, and S. Purton. 1994. Studies on homologous recombination in the green alga Chlamydomonas reinhardtii. Curr. Genet. 26:438-442. [DOI] [PubMed] [Google Scholar]
- 14.Harris, E. H. 2001. Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52:363-406. [DOI] [PubMed] [Google Scholar]
- 15.Kathir, P., M. LaVoie, J. Brazelton, N. A. Haas, P. A. Lefebvre, and C. D. Slflow. 2003. Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot. Cell 2:363-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kindle, K. L. 1990. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87:1228-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156-165. [DOI] [PubMed] [Google Scholar]
- 18.Li, J., and M. D. Baker. 2000. Formation and repair of heteroduplex DNA on both sites of the double-strand break during mammalian gene targeting. J. Mol. Biol. 295:505-516. [DOI] [PubMed] [Google Scholar]
- 19.Lumbreras, V., D. R. Stevens, and S. Purton. 1998. Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 14:441-447. [Google Scholar]
- 20.Miao, Z.-H. and E. Lam. 1995. Targeted disruption of the TGA3 locus in Arabidopsis thaliana. Plant J. 7:359-365. [DOI] [PubMed] [Google Scholar]
- 21.Nassif, N., J. Penney, S. Pal, W. R. Engels, and G. B. Gloor. 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14:1613-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson, J. A. E., and P. A. Lefebvre. 1995. Targeted disruption of the NIT8 gene in Chlamydomonas reinhardtii. Mol. Cell. Biol. 15:5762-5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohi, S., R. Offringa, P. J. M. van den Elzen, and P. J. J. Hooykas. 1994. Gene replacement in plants, p. 191-217. In J. Paszkowski (ed.), Homologous recombination and gene silencing in plants. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 24.Orr-Weaver, T. L., J. W. Szostak, and R. J. Rothstein. 1981. Yeast transformation: a model system for the study of recombination. Proc. Natl. Acad. Sci. USA 78:358-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puchta, H., B. Dujon, and B. Hohn. 1996. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc. Natl. Acad. Sci. USA 93:5055-5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reiss, B., M. Klemm, H. Kosak, and J. Schell. 1996. RecA protein stimulates homologous recombination in plants. Proc. Natl. Acad. Sci. USA 93:3094-3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russel. 2001. Molecular cloning, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Schaefer, D. G., and J.-P. Zryd. 1997. Efficient gene targeting in the moss Physcomitrella patents. Plant J. 11:1195-1206. [DOI] [PubMed] [Google Scholar]
- 29.Schroda, M., D. Blöcker, and C. F. Beck. 2000. The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 21:121-131. [DOI] [PubMed] [Google Scholar]
- 30.Shalev, G., Y. Sitrit, N. Avivi-Ragolski, C. Lichtenstein, and A. Levy. 1999. Stimulation of homologous recombination in plants by expression of the bacterial resolvase RuvC. Proc. Natl. Acad. Sci. USA 96:7398-7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shalguev, V. I., O. K. Kaboev, I. A. Sizova, P. Hegemann, and V. A. Lanzov.2005. Identification of Chlamydomonas reinhardtii Rad51C: recombinational characteristics. Mol. Biol. 39:98-104. [PubMed] [Google Scholar]
- 32.Shinohara, A., and T. Ogava. 1995. Homologous recombination and the role of single stranded breaks. Trends Biol. Sci. 20:387-391. [DOI] [PubMed] [Google Scholar]
- 33.Simon, J. R., and P. D. Moore. 1987. Homologous recombination between single stranded chromosomal genes in Sacharomyces cerevisiae. Mol. Cell. Biol. 7:2329-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sizova, I., M. Fuhrmann, and P. Hegemann. 2001. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to the green alga C.reinhardtii. Gene 277:221-229. [DOI] [PubMed] [Google Scholar]
- 35.Sodeinde, O. A., and K. L. Kindle. 1993. Homologous recombination in the nuclear genome of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 90:9199-9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens, D. R., J.-D. Rochaix, and S. Purton. 1996. The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol. Gen. Genet. 251:23-30. [DOI] [PubMed] [Google Scholar]
- 37.Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein, and F. W. Stahl. 1983. The double-strand-break repair model for recombination. Cell 33:25-35. [DOI] [PubMed] [Google Scholar]
- 38.Terada, R., H. Urava, Y. Inagaki, K. Tsugane, and S. Iida. 2002. Efficient gene targeting by homologous recombination in rice. Nature Biotechnol. 20:1030-1034. [DOI] [PubMed] [Google Scholar]
- 39.Terada, R., H. Asa, and S. Iida. 2004. A large scale Agrobacterium-mediated transformation procedure with a strong positive-negative selection for gene targeting in rice (Oryza sativa L.). Plant Cell Rep. 22:653-659. [DOI] [PubMed] [Google Scholar]
- 40.Thykjar, T., J. Finnemann, L. Schauser, and L. Christensen. 1997. Gene targeting approaches using positive-negative selection and large flanking regions. J. Mol. Biol. 35:523-530. [DOI] [PubMed] [Google Scholar]