Abstract
Candida albicans, the most prevalent fungal pathogen of humans, causes superficial mycoses, invasive mucosal infections, and disseminated systemic disease. Many studies have shown an intriguing association between C. albicans morphogenesis and the pathogenesis process. For example, hyphal cells have been observed to penetrate host epithelial cells at sites of wounds and between cell junctions. Ras- and Rho-type GTPases regulate many morphogenetic processes in eukaryotes, including polarity establishment, cell proliferation, and directed growth in response to extracellular stimuli. We found that the C. albicans Ras-like GTPase Rsr1p and its predicted GTPase-activating protein Bud2p localized to the cell cortex, at sites of incipient daughter cell growth, and provided landmarks for the positioning of daughter yeast cells and hyphal cell branches, similar to the paradigm in the model yeast Saccharomyces cerevisiae. However, in contrast to S. cerevisiae, CaRsr1p and CaBud2p were important for morphogenesis: C. albicans strains lacking Rsr1p or Bud2p had abnormal yeast and hyphal cell shapes and frequent bends and promiscuous branching along the hypha and were unable to invade agar. These defects were associated with abnormal actin patch polarization, unstable polarisome localization at hyphal tips, and mislocalized septin rings, consistent with the idea that GTP cycling of Rsr1p stabilizes the axis of polarity primarily to a single focus, thus ensuring normal cell shape and a focused direction of polarized growth. We conclude that the Rsr1p GTPase functions as a polarity landmark for hyphal guidance and may be an important mediator of extracellular signals during processes such as host invasion.
Candida albicans, an important fungal pathogen, causes infections in both healthy and ill persons ranging from superficial mucosal diseases (thrush and vaginitis) to life-threatening disseminated disease involving the bloodstream (sepsis), central nervous system (meningitis), and vital organs (renal, hepatic, or brain abscess, for example). C. albicans exists in three major morphologies: ellipsoid yeast-form (YF) cells, pseudohyphal cells (chains of elongated yeast cells, separated by constrictions), and extremely elongated filamentous hyphal-form (HF) cells. YF cells are thought to be the predominant morphology found during colonization at epithelial surfaces, whereas HF cells have been associated with invasion into and through epithelial and endothelial cell layers (40). Indeed, C. albicans strains with defects in hyphal morphogenesis appear to be less virulent in animal and tissue-based models of candidiasis (9, 36, 45, 48), leading to the idea that morphogenesis is required for virulence. On the one hand, this conclusion is complicated by the realization that many of these avirulent strains lack proteins (for example, transcription factors) that likely affect not only cell shape but also many other cellular processes. Thus, it remains unclear if cell shape per se is important for the pathogenesis process or if morphology is simply associated with other characteristics that are required for virulence (26). On the other hand, HF cells have been observed to penetrate through breaks in a membrane and wound sites in tissue specimens (18, 25, 50), supporting the hypothesis that the hyphal filament facilitates invasion, at least in some settings.
In unicellular as well as multicellular organisms, cell polarity is a fundamental process required for differentiation, proliferation, and morphogenesis. The polarized localization of proteins determines cell shape and cell fate and is regulated by internal cues or extracellular signals. Polarized growth mechanisms have been studied most extensively in the model yeast Saccharomyces cerevisiae (reviewed in references 12 and 14). During S. cerevisiae budding, morphogenesis is controlled by intracellular signals: cortically localized bud site selection proteins serve as a landmark for the next bud site by recruiting and activating the Ras-like GTPase Rsr1p. The proper, localized activation of the Rsr1p GTPase is required for the next step in the development of polarity, the recruitment, and the activation of the major polarity establishment factors, the Rho-type GTPase Cdc42p and its guanine nucleotide exchange factor, Cdc24p (35). Like other members of the GTPase superfamily, Rsr1p cycles between (active) GTP-bound and (inactive) GDP-bound states, and this cycling is required for the function of Rsr1p (7, 41, 42). S. cerevisiae cells lacking Rsr1p have defects in bud site selection but have normal morphology and growth (8), indicating that Cdc42p can be activated stochastically at alternative locations in the absence of Rsr1p. Localized activation of Cdc42p drives polarized morphogenesis by regulating the assembly and polarization of cytoskeletal components, including actin cables and patches, the septin ring (important for bud site selection and cytokinesis), and the polarisome complex, which anchors the actin cytoskeleton to the growing bud tip (reviewed in reference 46). In contrast, during S. cerevisiae mating, YF cells use pheromones as extrinsic signals, overriding the intrinsic cues provided by the bud site selection proteins and the Rsr1p GTPase, to effect reorganization of the actin cytoskeleton in the direction of the putative mating partner. Binding of pheromone to its receptor causes dissociation of the Gβγ subunit from the heterotrimeric G protein. The Gβγ effector Far1p is then responsible for the eventual activation of Cdc42p, by Cdc24p, and polarized growth of the mating projection (11, 37).
C. albicans YF and HF cells have different cell cycle and cell biological characteristics and thus are best described as two completely distinct morphogenesis programs rather than two extremes along one continuum (54). Pseudohyphal-form cells, best considered as a variant of the YF morphology (39, 54), are elongated YF cells that do not separate from each other. Consistent with the idea that they are more like yeast cells than hyphae, pseudohyphal cells have nuclear cell cycles and septin localization patterns that are more similar to that of YF cells (55). In general, C. albicans YF morphogenesis is similar to that described for YF growth in S. cerevisiae: YF buds grow in a polarized fashion until reaching approximately two-thirds of their eventual size and then grow in an isotropic manner in which growth is distributed evenly over the entire cell surface. C. albicans YF cells also exhibit discrete budding patterns (13), where Int1p (similar to the S. cerevisiae landmark protein Bud4p) and Rsr1p (similar to S. cerevisiae Rsr1p) are required for bud site selection patterns (19, 62). In contrast, during HF growth, hyphae grow in a completely polarized fashion that lacks isotropic growth. HF daughter cells lack constrictions but are compartmentalized by chitin-containing septa along their length. In contrast to the cell cycle-dependent localization of actin during S. cerevisiae and C. albicans YF growth, HF growth is associated with the continuous polarization of actin toward the hyphal tip (2, 3), a consequence of the activity of Cdc42p, which localizes to the tips of hyphal cells (29). Interestingly, C. albicans cells lacking the landmark proteins Int1p and Rsr1p cannot form hyphae in several types of hypha-inducing media (4, 20, 62). This is in contrast to the situation for S. cerevisiae, where ScBud4p and ScRsr1p, although required for the correct spatial regulation of budding, have no role in cell shape or the subsequent events of polarized growth. Thus, landmark proteins may have additional roles during C. albicans morphogenesis.
In this paper, we investigate the molecular basis for the morphogenesis defects in C. albicans cells lacking the Rsr1p GTPase or its predicted GTPase-activating protein, Bud2p. Phenotypic analysis, in parallel, of strains lacking either Rsr1p or Bud2p allowed us to gain insight into the relative roles of GTP-bound Rsr1p, GDP-bound Rsr1p, and cycling itself in the phenotypes observed. We found that cells lacking Rsr1p or Bud2p have molecular and cellular features more like pseudohyphae when grown under conditions that favor HF growth, implicating Rsr1p GTP-to-GDP cycling in specifying the morphogenesis program. In addition, Rsr1p and Bud2p were important for directing the axis of polarity in YF and HF cells, which during HF growth, was essential for hyphal guidance and agar invasion. An understanding of hyphal guidance in C. albicans will guide the design of models of tissue invasion mechanisms during C. albicans infections and of polarized growth mechanisms in other cell systems.
MATERIALS AND METHODS
Media and growth conditions.
For YF growth, strains (Table 1) were grown in synthetic dextrose complete (SDC) medium (49) at 28°C. Except where noted in the text, HF growth was achieved by growing strains at 37°C in SDC medium with 10% bovine serum added. Uridine (80 μg/ml) was added to all media, except when selecting for uridine prototrophs during strain constructions.
TABLE 1.
Yeast strains used in this study
| Species and strain | Relevant genotype | Source or reference |
|---|---|---|
| C. albicans | ||
| BWP17 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | 61 |
| JB6284 | BWP17 his1::hisG/hisG::his1::HIS1 arg4::hisG/ARG4-URA3::arg4::hisG | 9 |
| CA8815 | BWP17 RSR1/rsr1::HIS1 | This study |
| CA8876 | BWP17 RSR1/rsr1::HIS1 arg4::hisG/ARG4-URA3::arg4::hisG | This study |
| CA8832, 34 | BWP17 rsr1::ARG4/rsr1::HIS1 | This study |
| CA8880, 24 | BWP17 rsr1::ARG4/rsr1::HIS1 arg4::hisG/ARG4-URA3::arg4::hisG | This study |
| MG9215 | BWP17 rsr1::ARG4/pURA3-RSR1::rsr1::HIS1 | This study |
| DH7425 | BWP17 BUD2/bud2::HIS1 | This study |
| DH7451 | BWP17 BUD2/bud2::HIS1 arg4::hisG/ARG4-URA3::arg4::hisG | This study |
| DH7426 | BWP17 bud2::ARG4/bud2::HIS1 | This study |
| DH7453 | BWP17 bud2::ARG4/bud2::HIS1 arg4::hisG/ARG4-URA3::arg4::hisG | This study |
| MG9017 | BWP17 bud2::ARG4/pURA3-BUD2::bud2::HIS1 | This study |
| MG9061, 63 | BWP17 bud2::ARG4/pURA3-bud2::bud2::HIS1 | This study |
| MG8393 | BWP17 RSR1/GFPRSR1 | This study |
| MG9232 | BWP17 RSR1/URA3-PGAL1-GFPRSR1 | This study |
| MG8063 | BWP17 BUD2/bud2 pGFP-BUD2-URA3 | This study |
| MG5398 | BWP17 CDC3/CDC3GFP-URA3 | 22 |
| DH9237 | BWP17 rsr1::ARG4/rsr1::HIS1 CDC3/CDC3GFP-URA3 | This study |
| DH7849 | BWP17 bud2::ARG4/bud2::HIS1 CDC3/CDC3GFP-URA3 | This study |
| MG6748 | BWP17 SPA2/SPA2YFP-URA3 | This study |
| DH9146 | BWP17 rsr1::ARG4/rsr1::HIS1 SPA2/SPA2YFP-URA3 | This study |
| MG8869 | BWP17 bud2::ARG4/bud2::HIS1 SPA2/SPA2YFP-URA3 | This study |
| S. cerevisiae | ||
| JB1492 | MATaade2 ura3 lys2 trp1 his3 leu2 | I. Herskowitz |
| JB2762 | MATα ura3 trp1 his3 his4 leu2 can1 rsr1::URA3 | J. Pringle |
| JB2761 | MATaade2 ura3 lys2 trp1 his3 leu2 bud2::LEU2 | J. Pringle |
Construction of rsr1 and bud2 strains. (i) rsr1 and bud2 strains.
RSR1 and BUD2 were sequentially disrupted in C. albicans strain BWP17 by PCR-mediated gene modification as previously described (61). An rsr1::HIS1 PCR product was amplified with primers 1460 (5′-AGGTATGTACATTCAACAAAAGCCCG TTACACTTGTATTTCAATAACCCTATATACTAACTTTTGTTTTGgttttcccagtcacgacgtt-3′; RSR1-specific sequence in uppercase letters) and 1265 (5′-TCACATAATTGTGCAGCACTTGCTTCCTGAGCTAGACTTTGGGTGATCATTAATTGATTGTTTCAATCTGtgtggaattgtgagcggata-3′) and with pGEM-HIS1 (61) as the DNA template. A bud2::HIS1 PCR product was amplified with primers 1161 (5′-ATGCCTCCAGCTTATCAATCTGCATTTCATAAAATCATTTCTCGTGATAAAGGAGTATTTGAAGGTCACAgttttcccagtcacgacgtt-3′; BUD2-specific sequence in uppercase letters) and 1040 (5′-TCAATTTTTCCGTAACCATTTTTTGAATGGATTCTTTGAAGTATTTGGTTTAATAGCACTACCAATACTAtgtggaattgtgagcggata-3′) and with pGEM-HIS1 (60) as the DNA template. The rsr1::HIS1 and bud2::HIS1 products were transformed individually into BWP17 to generate multiple independent heterozygous strains. Two such heterozygotes from each transformation were then transformed with either rsr1::ARG4 or bud2::ARG4 disruption cassettes (amplified by PCR with primers 1460 and 1265 or primers 1161 and 1040, respectively, and with pRS-ARGΔSpeI [61] as the DNA template) to generate two independent homozygous disruption strains for each gene. C. albicans rsr1/rsr1 and bud2/bud2 strains were made prototrophic for URA3 by transformation with pRS-ARG-URA-BN (17) linearized with NotI to target integration into ARG4.
(ii) RSR1 reintegrant strain (rsr1/rsr1::RSR1).
To reintroduce RSR1 into the rsr1-null strains, the RSR1 open reading frame, 825 bp of upstream promoter sequence, and 240 bp of sequence downstream of the stop codon were cloned from BWP17 using primers 1833 (5′-AAGGAAAAAAgcggccgcGGGAGAATTTTTGTTGGGCAACC-3′; NotI site in lowercase letters) and 1834 (5′-AACTGCAGAACCAatgcatGAAAACAAACAAAACAAGAAGTCG-3′; NsiI site in lowercase letters). The PCR product was purified, digested with the indicated restriction enzymes, and cloned into NotI- and NsiI-cut pGEM-URA3 (61) to generate pMG2128. pMG2128 was subsequently digested with MluI to target integration to the promoter of the disrupted rsr1 locus and used to transform rsr1-null strain MG8832 to uridine prototrophy and generate RSR1 reintegrant strain MG9215.
(iii) BUD2 reintegrant strain (bud2/bud2::BUD2).
To reintroduce BUD2 into the bud2-null strains, the BUD2 open reading frame, 1.4 kbp of upstream promoter sequence, and 160 bp of sequence downstream of the stop codon were cloned into pGEM-URA3 as two fragments: primers 1690 (5′-AAGGAAAAAAgcggccgcCAGTTGAGGGATTAAGTCTAAGTGG-3′, NotI site indicated in lowercase letters) and 1691 (5′-CTGACAGTATTTGTGGAGTTATTATATAGG-3′; generates a downstream, native XmaI site) were used to generate the 5′ half of the BUD2 sequence, and primers 1692 (5′-GGTTTGTCGATTTAAAATTTATGGACC-3′; generates a downstream, native XmaI site) and 1693 (5′-CGACGATGATGTTGATTctgcagCATATCAATCAAACACACATACAAGGG-3′; PstI site indicated in lowercase letters) were used to generate the 3′ half of the BUD2 sequence. Both PCRs used BWP17 genomic DNA as the template. The PCR products were digested with the indicated restriction enzymes and cloned into NotI/NsiI-digested pGEM-URA3 in two sequential steps to generate pMG2111. pMG2111 was subsequently digested with EcoRV to target integration to the promoter of the disrupted bud2 locus and used to transform bud2-null strain DH7426 to uridine prototrophy and generate BUD2 reintegrant strain MG9017. To control for inadvertent phenotypes due to differences in the genomic location of URA3, a disrupted BUD2 sequence was integrated to the disrupted bud2 locus (bud2Δ/bud2Δ::bud2) in the same way as full-length BUD2 was for strain MG9017, as follows. pMG2111 was digested with SmaI and SnaBI, and the large plasmid fragment was isolated and religated to generate pMG2122, which contains the BUD2 open reading frame lacking 1,500 bp of internal sequence (from bp 624 to 2040). pMG2122 was subsequently digested with EcoRV and used to transform strain DH7426 to uridine prototrophy and generate bud2 reintegrant strain MG9061. All plasmid constructions were verified by sequencing at the University of Minnesota Microchemical Facility. All strain constructions were verified by PCR using primer sequences outside of the integration site. In addition, Northern blot analyses were performed with all heterozygous, homozygous, and reintegrant strains to verify expression, or lack of expression, of RSR1 or BUD2.
Construction of FP fusions. (i) GFP-Rsr1p-expressing strain.
The gene for green fluorescent protein (GFP) was fused just after the ATG of RSR1 by transformation of an RSR1-specific GFP-URA3-GFP cassette into BWP17 and selection of uridine prototrophs, followed by selection, on 5-fluoroorotic acid medium (Sigma, St. Louis, MO), of transformants that had recombined the repetitive GFP sequences (and effectively lost URA3) to generate strain MG8393. Relative overexpression of GFP-Rsr1p was achieved by constructing a strain expressing GFP-RSR1 from the inducible C. albicans GAL1 promoter, using PCR-mediated gene modification as previously described (23). An RSR1-specific URA3-PGAL1-GFP cassette was amplified using primers 1844 (5′-CAATCTTCTGTCAGATTTCAATGAGAGGTATGTACATTCAACAAAAGCCCGTTACACTTGTATTTCAATAACCtctagaaggaccacctttgattg-3′; RSR1-specific sequence in uppercase letters) and 1845 (5′-CCTGGACAAATTGCACGGTGATTGAGGATTTACCTACCCCACCAGCACCCAATACTACGACTTTATAATCTCTtttgtacaattcatccatac-3′) and template plasmid pURA3-PGAL1-GFP (23). The PCR product was used for transformation into strain BWP17 to generate strain MG9232.
(ii) GFP-Bud2p-expressing strain.
GFP was fused immediately after the ATG of BUD2 by conventional cloning by the following strategy. A 1.4-kbp portion of the BUD2 promoter (immediately upstream of the BUD2 ATG) was generated by PCR using primers 1347 (5′-AAGGAAAAAAgcggccgcATCTTCTTTCTAGAGAATACTGAT-3′; NotI site in lowercase letters) and 1348 (5′-TTCCGCGGCCGCTATGGCCGACgtcgacCATTATTAGGGATGATAGATTATCTTTATAAGTAAT-3′; SalI site in lowercase letters) and with BWP17 genomic DNA as the template. This BUD2 5′ upstream activating sequence was ligated into NotI/SalI-digested pGEM-URA3 (61) to generate pMG2053. GFP was amplified from pURA3-GFP (22) using primers 1349 (5′-ACGCgtcgacTCTAAAGGTGAAGAATTATTC-3′) and 1350 (5′-cgcgacgcgtTTTGTACAATTCATCCATACCATGGG-3′) containing SalI and MluI sites (indicated by lowercase letters), respectively, followed by ligation of this GFP fragment into SalI/MluI-digested pMG2053 to generate p2065. A partial BUD2 sequence (bp 4 to 1200 from ATG) was generated by PCR of BWP17 genomic DNA using primers 1351 (5′-cgacgcgtCCTCCAGCTTATCAATCTGCATTTCATAAAATC-3′; MluI site in lowercase letters) and 1352 (5′-CGACGATGATGTTGATTctgcagCAGTATTTTCAAATTTGGCTTCAATAATG-3′; PstI site in lowercase letters) followed by ligation of this fragment into MluI/NsiI-digested pMG2065 to generate pMG2076, which contains C. albicans URA3 and the BUD2 promoter followed by a partial BUD2 open reading frame with GFP incorporated just after the ATG. pMG2076 was digested with XmaI (within the BUD2 promoter sequence) and used to transform BWP17 to uridine prototrophy, generating strain MG8063.
(iii) Septin Cdc3-GFP- and Spa2-YFP-expressing strains.
To tag septin Cdc3p and polarisome component Spa2p with GFP and yellow fluorescent protein (YFP), respectively, PCR-mediated gene modification using template plasmid pGFP-URA3 or pYFP-URA3 was performed as previously described (22). A CDC3-specific GFP-URA3 cassette was amplified using primers 620 (5′-ACAAAAATTATTACCACAAGACCCACCAGCACAACCAGCTCCACAAAAGAGTCGTAAAGGATTTTTACGTGGTGGTGGTtctaaaggtgaagaattatt-3′; gene-specific sequence in uppercase letters) and 621 (5′-AATTAAACAAACAGATTAACAAACAAATAAACTAAATTAAGTTACATACTATTTAGCTATACCTCGGCCCtctagaaggaccacctttgattg-3′). A SPA2-specific YFP-URA3 cassette was amplified using primers 881 (5′-ACGGTGGAAGAAGCTAGTCTTAAAGAAGATATTGCTTATCTTGATGCTAGAATAAGTCAAAATCTTGAAggtggtggttctaaaggtgaagaattatt-3′) and 882 (5′-ATAAATACATCGCAATTTATTTTCTAATTTCTTTTTTTTCATTTTATTTATTCTCTAGCTATTTTCTAGtctagaaggaccacctttgattg-3′). PCR products were used for transformation into C. albicans strains auxotrophic for ura3, and transformants were selected on SDC medium lacking uridine. Construction of all fluorescent protein (FP) fusions was verified by PCR, using primers outside of the region of integration, and expression of FP fusion proteins was confirmed by Western blot analyses as described previously (23). In addition, FP fusions to the carboxy termini of Cdc3p and Spa2p were functional when expressed over a null allele of the respective gene (data not shown; see reference 63).
Morphological analyses.
To determine budding pattern, bud scars were stained with calcofluor white (Sigma, St. Louis, MO) as described previously (19) and visualized by fluorescence microscopy using a standard UV filter set. Viability of cells was determined by the ability to exclude the dye trypan blue. YF or HF cells, after 5 h of growth, were stained with 0.07% (final concentration) trypan blue (Sigma) and directly visualized by bright-field microscopy. To detect actin, C. albicans cells were fixed in 3.7% formaldehyde for 30 min, followed by incubation in PK buffer (50 mM potassium phosphate, pH 6.6) containing 3.7% formaldehyde for an additional 60 min. Formaldehyde-fixed cells were harvested by centrifugation, incubated in PK buffer containing 0.1% Triton X-100 for 30 min, washed twice in phosphate-buffered saline (PBS), and incubated overnight in PBS containing 2 U of Alexa Fluor 568 phalloidin (Molecular Probes, Eugene, OR). Cells were harvested by centrifugation and resuspended in PBS prior to fluorescence microscopy using a standard Texas red filter set. Bright-field, differential interference contrast (DIC), and epifluorescence microscopies were performed using a Nikon Eclipse E600 photomicroscope equipped with a 100-W mercury lamp, epifluorescence illumination with an Endow GFP or standard YFP filter set (both from Chroma Technology Corp., Brattleboro, VT), and a 60× objective fitted with an FCS2 objective heater (Bioptechs, Butler, PA). Immersion oil (type 37) optimized for GFP and YFP imaging at 37°C (Cargille, Cedar Grove, NJ) was used. Digital images were collected using a Photometrics CoolSNAP HQ camera (Tucson, AZ) and Metamorph software version 6.2r5 (Universal Imaging Corporation, Downingtown, PA). Microscopic analysis of colony morphology was performed using a Zeiss Stemi DRC stereomicroscope, and images were acquired with a Nikon CoolPix900 digital camera.
Time-lapse morphological analyses.
Yeast cells were cultured in SDC medium with uridine overnight at 28°C and then subcultured (1:20) into fresh medium with 10% serum, prewarmed to 37°C. After 5 min at 37°C, 200 μl of the subcultured cells was spread onto SDC agar containing uridine and 10% serum (prewarmed to 37°C for at least 2 h). The plate was incubated for about 15 min at 37°C before the time-lapse acquisition was begun, and the plate was then transferred immediately to the prewarmed (37°C) objective on the microscope. For DIC time-lapse microscopy, images were acquired every 2.5 min using a 2× camera bin and 5-ms exposure. For the Spa2-YFP time-lapse microscopy, fluorescence images were captured every 60 s using a 2× bin and 200-ms exposure, followed by a DIC image acquisition for 5 ms. Before measurements were made, the image stack was calibrated to adjust for the binning of the images. Stacks were translated into 8-bit images before Stack Arithmetic was done, in which the YFP stack was merged with the DIC stack using the Add function of Metamorph. All movies were compiled such that 5 min of real time equals 1 s of movie time. Stacks were assembled in Metamorph, and individual planes in the stack were aligned using the Align Stacks tool. Hyphal elongation rates were calculated by summing the distance traveled during each time point and dividing by the summed time intervals, using the Track Points feature of Metamorph to measure hyphal lengths. The summed distances were compared to a single Region Measurement of total hyphal length (using the segmented line tool) of the entire filament at the end of the experiment to ensure that the summed regions reflected the actual length (this controls for cell movements during measurements). To determine the extent of deviation of hyphal elongation from a straight line, the segmented line tool was used to define the two arms of the angle, along the length of the hypha. The angle was calculated using Region Measurements in the Angle Measure feature of Metamorph. Any curve that appeared to deviate from a straight line by more than 10° was measured; all such curves in each cell were included in the analysis. The total number of curves analyzed for each strain was at least 180 individual angles. Where indicated, differences in morphological measurements among the mutant strains and the wild-type strain were analyzed by Student's t test. Supplemental videos showing dynamics of Spa2-YFP during HF growth can be viewed at http://www.cbs.umn.edu/labs/gale/supplemental.html.
Agar invasion assay.
The ability of C. albicans strains to invade agar surfaces was determined as described by Warenda et al. (58) except SDC medium containing 10% serum plus 80 μg/ml uridine plus 1.5% agar was used. One microliter of cells (containing 104 cells) was spotted onto the surfaces of the plates, which were incubated for 5 days at 37°C. The resulting colonies were photographed and surface washed from the plate, and then cross-sectional slices of the underlying growth were also photographed.
RESULTS
C. albicans RSR1 and BUD2 are important for growth at high temperature.
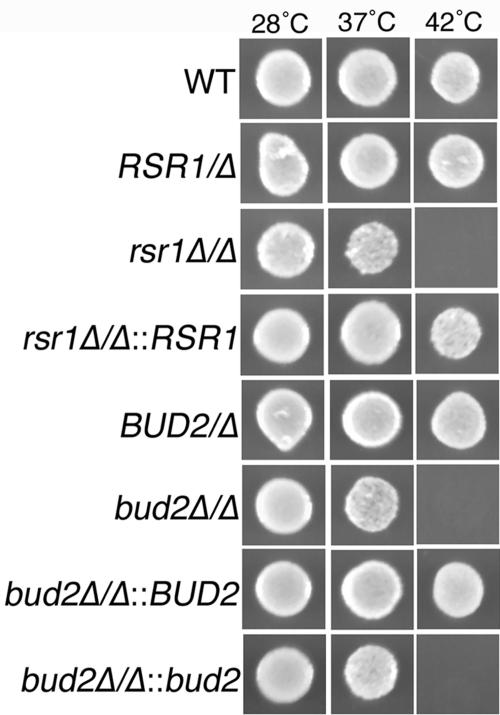
Open reading frame (ORF) 19.2614 is designated RSR1 (http://candida.bri.nrc.ca/candida), based on its similarity with S. cerevisiae RSR1/BUD1, and is identical to the sequence isolated as a suppressor of S. cerevisiae cdc24 and named CaRSR1 by Yaar et al. (62). C. albicans BUD2 was identified by a BLAST comparison to S. cerevisiae BUD2 (http://www.yeastgenome.org) to the C. albicans genome sequence (http://sequence-www.stanford.edu/group/candida). Subsequent annotation of the C. albicans genome defined this sequence as BUD2 and ORF 19.940 (http://candida.bri.nrc.ca/candida). To study the roles of RSR1 and BUD2 in C. albicans morphogenesis, we constructed strains lacking both copies of each gene by PCR-based gene disruption (61). YF growth of both disruption strains at 28°C and 37°C was similar to that of the isogenic parent strain (Fig. 1). However, in contrast to the analogous mutants of S. cerevisiae (15, 42) and to wild-type C. albicans, C. albicans strains lacking Rsr1p or Bud2p were sensitive to elevated temperature (42°C) (Fig. 1). Reintroduction of RSR1 or BUD2 into the respective null strains suppressed the temperature sensitivity. Furthermore, rsr1/rsr1 and bud2/bud2 strains were sensitive to growth on calcofluor white-containing medium (data not shown), indicating that a lack of Rsr1p or Bud2p causes defects in cell wall biogenesis.
FIG. 1.
Rsr1p and Bud2p are required for growth at high temperature. C. albicans strains JB6284 (wild type [WT]), CA8815 (RSR1/Δ), CA8880 (rsr1Δ/Δ), MG9215 (rsr1Δ/Δ::RSR1), DH7458 (BUD2/Δ), DH7453 (bud2Δ/Δ), MG9017 (bud2Δ/Δ::BUD2), and MG9061 (bud2Δ/Δ::bud2) were incubated on SDC medium at the indicated temperatures for 3 days and then photographed. An equal number of cells was inoculated in each case, as determined spectrophotometrically and verified by hemacytometer cell counts.
C. albicans Rsr1p and Bud2p direct the site of new daughter cell growth in yeast and hyphal cells.
In S. cerevisiae, Rsr1p and Bud2p are important for the maintenance of axial and bipolar budding patterns in haploid and diploid yeast cells, respectively. In C. albicans, an obligate diploid, YF cells exhibit a predominantly axial (adjacent) budding pattern when grown at 28°C, with more cells displaying nonadjacent budding patterns as growth temperatures approach 37°C (13). Yaar et al. previously reported that C. albicans YF cells lacking Rsr1p have a random budding pattern (62). To determine if C. albicans Bud2p also functions as a landmark protein, we analyzed the budding pattern of bud2/bud2 strains during YF growth. Like rsr1/rsr1 YF cells, bud2/bud2 YF cells have a completely random budding pattern (99% of cells), in comparison to the isogenic parent strain, which exhibits a predominantly axial pattern (77% of cells) at 28°C (Fig. 2, top).
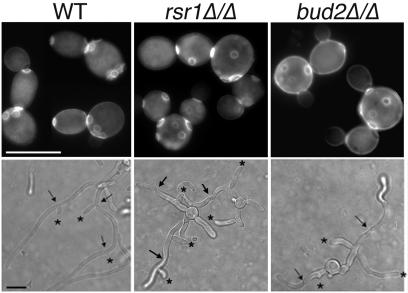
FIG. 2.
Bud2p provides a landmark for yeast and hyphal daughter cell site selection. Stationary-phase C. albicans strains JB6284 (WT), CA8880 (rsr1Δ/Δ), and DH4453 (bud2Δ/Δ) were subcultured under YF growth conditions for 4 h and stained with calcofluor white to visualize bud scars (top) or grown under HF conditions (bottom) and analyzed by DIC microscopy for branch position (asterisks) relative to hyphal septa (arrows). Scale bars, 5 and 10 μm (upper and lower panels, respectively).
In HF cells, daughter cell emergence patterns have distinct differences from YF bud site selection patterns. During HF growth, germ tubes emerge in predominantly nonaxial positions (bipolar or random [13]), whereas hyphal branches emerge adjacent to hyphal septa, on the mother cell (proximal) side (27, 28). To determine the role of Rsr1p and Bud2p as landmark proteins during HF growth, we analyzed the daughter cell emergence patterns (germ tubes and hyphal branches) of rsr1/rsr1 and bud2/bud2 HF cells. As expected, germ tubes emerged at nonaxial sites in the mutant strains as well as the parent strain. However, we observed a significant increase, during hyphal branching, in the number of hyphal daughters that emerged at random positions along the hypha in both rsr1/rsr1 (79% random, n = 42) and bud2/bud2 (69% random, n = 45) strains compared to that of the isogenic parent strain, which branched almost exclusively near septa (5% random, n = 50) (Fig. 2, bottom). Thus, Rsr1p and Bud2p function as landmarks that specify both bud emergence during YF growth and hyphal reinitiation during hyphal branching, but not during emergence of the first germ tube from the mother cell.
Rsr1p and Bud2p localize to the cell cortex.
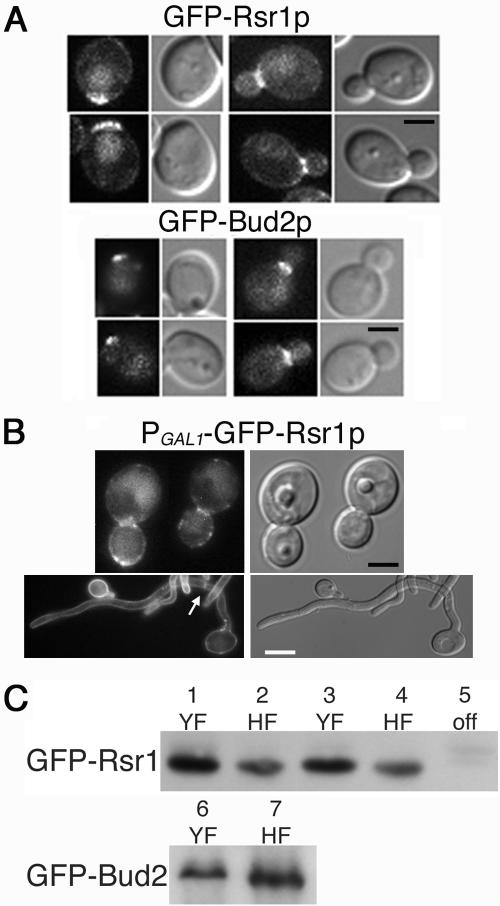
In order to function as landmarks for daughter cell site selection, Rsr1p and Bud2p should localize at the yeast cell cortex. To test this prediction, we fused GFP to Rsr1p and Bud2p and analyzed their localization in YF and HF cells. Rsr1p is predicted to be isoprenylated at its carboxy terminus, potentially facilitating membrane localization (8), and Bud2p is predicted to contain a coiled-coil domain near the carboxy terminus (http://smart.embl-heidelberg.de/). Therefore, to avoid potential functional defects from addition of GFP to the carboxy termini, we fused GFP to the amino termini of each protein. We observed that GFP-Rsr1p and GFP-Bud2p localized as a discrete cortical patch on a population of unbudded YF cells, presumably at the site of incipient bud emergence (Fig. 3A). In small and medium budded cells, both proteins localized to the neck region between mother and daughter yeast cells. Thus, like the similar proteins in S. cerevisiae (43, 44), C. albicans Rsr1p and Bud2p are cortical proteins that function as landmarks for future daughter cell emergence sites. Importantly, GFP-Bud2p was functional when expressed over a bud2-null allele: growth and morphogenesis phenotypes were identical to those of the parent strain (data not shown). In contrast, although GFP-Rsr1p localized as predicted from studies of ScRsr1p, it did not complement the morphogenesis defects of the rsr1/rsr1 strain. This is likely a result of the GFP tag interfering with the “small GTPase domain” predicted to be at the amino terminus of the protein (http://smart.embl-heidelberg.de/). Thus, although use of GFP-Rsr1p provided localization information for Rsr1p, alternative constructs will be needed for future studies of Rsr1p functional interactions.
FIG. 3.
GFP-Rsr1p and GFP-Bud2p localize to the cell cortex in yeast and hyphal cells. (A) C. albicans strains MG8393 (GFP-Rsr1p) and MG8063 (GFP-Bud2p) were grown under YF conditions and analyzed by GFP filter fluorescence and DIC microscopy. Scale bar, 2 μm. (B) C. albicans strain MG9232 (PGAL1-GFP-Rsr1p) was grown under YF (top; scale bar, 2 μm) or HF (bottom; scale bar, 5 μm) conditions and analyzed by GFP filter fluorescence and DIC microscopy. Localization of GFP-Rsr1p at a hyphal septum is indicated by an arrow. Adobe Photoshop v. 7 was used to process the acquired images in panels A and B. (C) Protein blots showing the relative levels of GFP-Rsr1p (top) and GFP-Bud2p (bottom) expressed in the strains shown in panels A and B. Strains MG8393 (lanes 1 and 2) and MG8063 (lanes 6 and 7) were grown in SDC medium at 28°C (YF) or in SDC medium plus 10% serum at 37°C (HF). Strain MG9232 (lanes 3 to 5) was grown in SDC medium lacking dextrose but containing 2% galactose at 28°C (YF), SC medium plus 2% galactose plus 10% serum at 37°C (HF), or in SDC medium at 28°C (off), as indicated.
Rsr1p and Bud2p clearly have a role in selecting daughter cell branch sites in HF cells (Fig. 2), yet we were unable to detect GFP-Rsr1p or GFP-Bud2p signal in HF cells by fluorescence microscopy. Western blot analysis, using antibody to GFP, did detect GFP-Rsr1p and GFP-Bud2p in hyphal cell extracts (Fig. 3C). To gain more information about the possible localization of Rsr1p during HF growth, we constructed a strain expressing GFP-Rsr1p from the galactose-inducible C. albicans GAL1 promoter. During YF growth in the presence of galactose, GFP-Rsr1p localized to the entire periphery of the cell, but the signal was most concentrated at the periphery of buds and at the mother bud necks (Fig. 3B). During HF growth in galactose-containing medium, GFP-Rsr1p localized to the entire periphery of germ tubes and hyphae and also to the septa of hyphal cells (Fig. 3B). Thus, GFP-Rsr1p can localize to the cell cortex of hyphal (and yeast) cells. Because GFP-Rsr1p is being constitutively overexpressed in this strain during growth in galactose-containing medium (Fig. 3B), these results suggest that, when expressed from its native promoter, Rsr1p localizes only very transiently to specific cortical sites in HF cells and/or that the level of GFP-Rsr1p signal at the cortex of HF cells is below the limit of detection of our microscope and camera system. Nevertheless, our results are consistent with a cortical location of Rsr1p and Bud2p that would facilitate their role as daughter cell landmark proteins.
C. albicans Rsr1p and Bud2p are important for the polarization of YF cells.
Similar to the cell shape abnormalities reported for C. albicans rsr1/rsr1 strains (62) (Fig. 2 and 4, DIC images), we observed that bud2/bud2 yeast cells were often larger and rounder than the yeast cells of the isogenic parent strain (Fig. 2 and 4, DIC images). Yeast cell shape abnormalities were rescued by reintroduction of RSR1 or BUD2 into the respective null strains (data not shown). Polarized growth in these strains is not completely defective, however, as rsr1/rsr1 and bud2/bud2 YF cells generate buds and form colonies of the same size as those of the isogenic parent strains. In addition, rsr1/rsr1 and bud2/bud2 YF cells are just as viable as the parent strain: in each strain, >99% of cells were viable after 5 h under YF growth conditions, as determined by trypan blue exclusion.
FIG. 4.
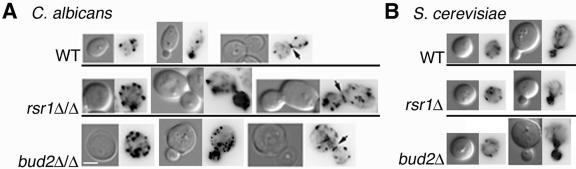
Rsr1p and Bud2p are important for the polarization of actin patches in C. albicans YF cells. (A) C. albicans strains JB6284 (WT), CA8880 (rsr1Δ/Δ), and DH7453 (bud2Δ/Δ) were grown under YF conditions for 4 h. (B) S. cerevisiae strains JB1492 (WT), JB2762 (rsr1Δ), and JB2761 (bud2Δ) were incubated under YF conditions for 4 h. All strains were fixed with formaldehyde, stained with Alexa phalloidin to visualize actin, and photographed. The left panel in each pair is a DIC image; the right panel of each pair is a Texas red filter fluorescent image. Scale bar, 3 μm (all panels). Adobe Photoshop v. 7 was used to invert the acquired fluorescent images in order to enhance visualization.
Polarization of the actin cytoskeleton is required for normal cell shape and for polarity establishment and maintenance. To investigate the cell biological features associated with the cell shape and polarity establishment abnormalities in strains lacking Rsr1p and Bud2p, we analyzed the organization of actin in rsr1/rsr1 and bud2/bud2 YF cells. Alexa-phalloidin staining of the parental strains showed that actin patches lacked polarization in unbudded cells, polarized to the small buds of budding cells, and localized in a ring at the mother-daughter neck prior to cytokinesis (Fig. 4A) similar to the patterns that have been reported for actin organization in S. cerevisiae (1, 34) and C. albicans (3) YF cells. Thus, Rsr1p and Bud2p are not absolutely required for the polarized localization of actin patches to the growing bud. However, in contrast to the parent strain, rsr1/rsr1 and bud2/bud2 YF cells exhibited an increased number of cortical actin patches during all phases of the yeast cell cycle (Fig. 4A), and some of the actin patches were larger than normal and/or aberrant in shape. Thus, a lack of either Rsr1p or Bud2p results in YF cells that are larger and rounder, which is associated with an increased number of cortical actin patches. Interestingly, in S. cerevisiae, Rsr1p and Bud2p are not important for yeast cell shape (15) (Fig. 4B) or for actin organization (Fig. 4B). This suggests that in C. albicans, Rsr1p and Bud2p have functions in morphogenesis in addition to their role in daughter cell site selection.
C. albicans Rsr1p and Bud2p are important for the polarization of HF cells.
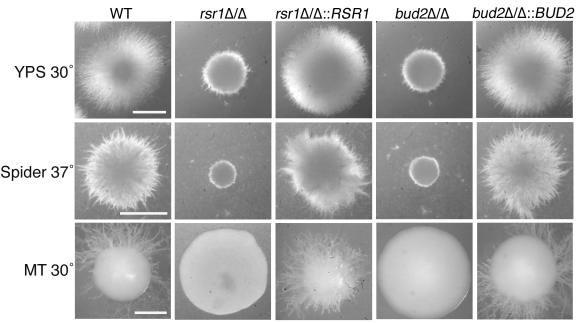
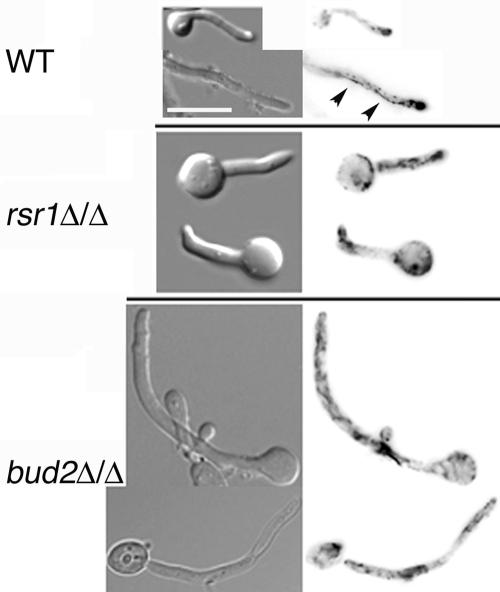
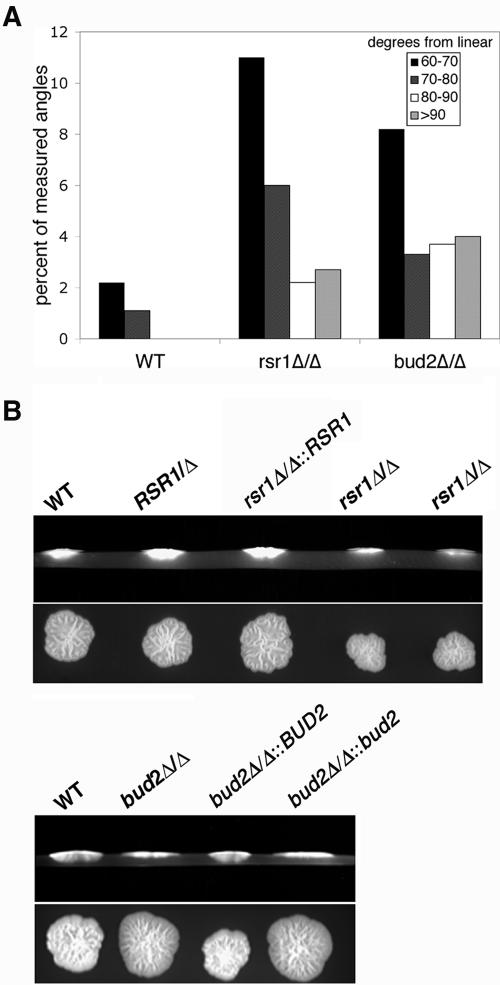
While previous studies reported that C. albicans rsr1/rsr1 strains form hyphae less well in serum-containing liquid media than do nonisogenic wild-type strains (4, 62), these studies did not provide molecular or cellular details to aid in understanding how the Rsr1 GTPase affects hyphal morphogenesis. To further investigate the role of Rsr1p and Bud2p during HF growth, we analyzed the ability of two independently generated rsr1/rsr1 strains and two independently generated bud2/bud2 strains to form hyphae during growth on milk-Tween agar (32), in embedded Spider agar at 30°C and 37°C, in embedded YPS agar at 30°C and 37°C (10), in serum-containing liquid medium, or on serum-containing agar medium. All four mutant isolates were very defective in their ability to form hyphae on milk-Tween agar and under all of the embedded conditions tested, at both 30°C and 37°C (Fig. 5 A and data not shown). This defect in HF growth was rescued by reintroduction of a single RSR1 or BUD2 sequence into the respective null strains (rsr1Δ/rsr1Δ::RSR1 or bud2Δ/bud2Δ::BUD2) (Fig. 5A). Microscopic analysis of cells harvested from colonies revealed that rsr1/rsr1 and bud2/bud2 HF cells were larger, wider, and contained more constrictions throughout their length (Fig. 2; see Fig. 7), characteristics of pseudohyphal rather than HF cells. Thus, the elongated rsr1/rsr1 and bud2/bud2 daughter cells that form in response to hyphal induction conditions will henceforth be referred to as “filaments” rather than hyphae. In addition, rsr1/rsr1 and bud2/bud2 cells were more bent and curved than the cells of the isogenic parent strains (Fig. 5B; see Fig. 7). Together, these data show that both Rsr1p and Bud2p are necessary for a similar subset of morphogenetic characteristics.
FIG. 5.
Rsr1p and Bud2p are important for hyphal morphogenesis. C. albicans strains JB6284 (WT), CA8880 (rsr1Δ/Δ), MG9215 (rsr1Δ/Δ::RSR1), DH7453 (bud2Δ/Δ), and MG9017 (bud2Δ/Δ::BUD2) were grown to stationary phase under YF conditions. Equal numbers of cells were then inoculated (embedded) into yeast-peptone-sucrose agar (YPS) and Spider agar, both at 37°C for 3 days, and on milk-Tween agar (MT) at 30°C for 10 days and photographed. Scale bars, 1 mm (for each row).
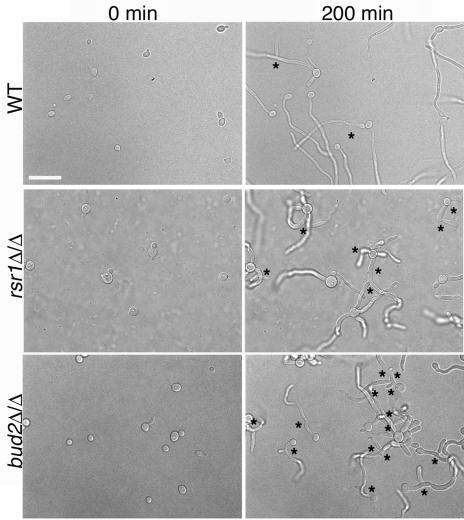
FIG. 7.
Rsr1p and Bud2p regulate branching events in HF cells. Stationary-phase C. albicans strains JB6284 (WT), CA8880 (rsr1Δ/Δ), and DH7453 (bud2Δ/Δ) were inoculated onto serum-containing agar and photographed at the indicated times after inoculation. Asterisks indicate hyphal branches. Scale bar, 20 μm.
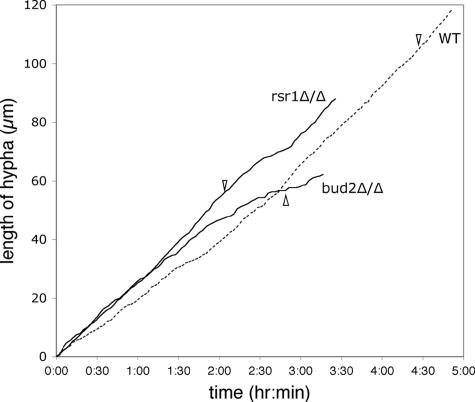
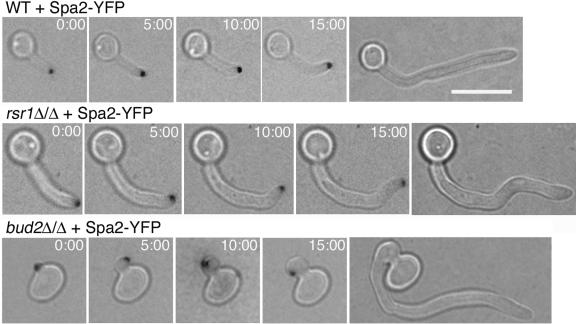
To explore the defects in HF growth in rsr1 and bud2 strains in more detail, we used time-lapse microscopy to monitor filament growth rates and morphogenetic characteristics during growth on serum-containing agar. During the first hour after hyphal induction, wild-type hyphae elongated at an average rate of 18 μm/h (n = 14) (Fig. 6), identical to wild-type hyphal elongation rates reported over 20 years ago (27). In contrast, rsr1/rsr1 and bud2/bud2 filaments both elongated faster than wild-type HF cells (Fig. 6), with average rates of 29 μm/h (n = 12; P = 0.002) and 25 μm/h (n = 11; P = 0.0001), respectively. During the first hour of growth in serum, the filament elongation rates of the rsr1/rsr1 and bud2/bud2 strains were not statistically different from each other (P = 0.3). At later time points, however, the filament elongation rates of the rsr1/rsr1 and bud2/bud2 strains were more variable, with individual hyphae exhibiting periods of faster and slower growth. In contrast, the wild-type strain exhibited stable, linear growth kinetics (Fig. 6) (27, 28). These observations support the idea that Rsr1p and Bud2p function in the maintenance of hyphal growth.
FIG. 6.
Rsr1p and Bud2p influence hyphal growth rates. Time-lapse images were obtained from C. albicans strains JB6284 (WT), CA8880 (rsr1Δ/Δ), and DH7453 (bud2Δ/Δ) during hyphal induction on serum-containing agar and filament elongation rates (filament length in μm/time) were plotted using the Track Points function of Metamorph, as described in Materials and Methods. For clarity, only one representative example for each strain is shown. The emergence of the first branch is indicated by an arrowhead.
Interestingly, the rsr1/rsr1 and bud2/bud2 strains exhibited an increased frequency of branching events from the primary filament (Fig. 7). For example, within 4 h after emergence of the primary filament, rsr1/rsr1 and bud2/bud2 filaments typically underwent more than two branching events, in comparison to a wild-type hypha which formed, at most, one branch during the same period of time. The average time between the emergence of the primary hypha and the first hyphal branch was 252 min for the parent strain (n = 8), 113 min for the rsr1/rsr1 strain (n = 12; P < 0.0001), and 140 min for the bud2/bud2 strain (n = 16; P < 0.0001) (Fig. 6). The branch emergence times of rsr1/rsr1 and bud2/bud2 filaments were not statistically different from each other (P = 0.05). Thus, Rsr1p and Bud2p appear to suppress branching during HF growth, which may promote the sustained elongation of the primary hypha.
To investigate the role of Rsr1p and Bud2p in directing actin polarization during HF growth, we stained rsr1/rsr1 and bud2/bud2 HF cells with Alexa-phalloidin. During C. albicans HF growth, cortical actin patches localize almost exclusively to the hyphal apex, leaving the remainder of the hyphal cell relatively devoid of actin patches (3). This pattern of actin localization correlates with the observation that hyphal cell walls expand primarily from the apical one-third of the cell (51, 53). Similarly, we observed that in the isogenic parent strain, actin patches were polarized to the majority (73%; n = 30) of hyphal tips after 4 h of growth in serum-containing medium (Fig. 8, top). In cells lacking Rsr1p, actin patches polarized to the tips almost as well as in the parent strain (58% of cells; n = 30), but an increase in the number of actin patches was observed in mother cells and at sites within the filament distant from the tip (Fig. 8, middle). Interestingly, in cells lacking Bud2p, even fewer tips contained polarized actin (18%; n = 50), and similar to the rsr1/rsr1 strain, actin was observed in nonapical locations. In addition, whereas wild-type strains contain actin cables directed toward the hyphal apex (Fig. 8) (3), rsr1/rsr1 and bud2/bud2 strains exhibited more randomly directed actin cables, consistent with the pattern observed in nonbudding yeast cells (3). These observations indicate that both Rsr1p and Bud2p are important for polarizing actin cables and patches during hyphal growth and, furthermore, that Bud2p has an additional role in the maintenance of polarized actin patches at the tips of primary hyphae. Thus, the overall lower rates of filament elongation at later time points in the mutant strains and the wider appearance of rsr1/rsr1 and bud2/bud2 filaments may be due to a loss of focused growth at the tip and increased growth along the entire filament.
FIG. 8.
Rsr1p and Bud2p are important for actin patch polarization during HF growth. Stationary-phase C. albicans strains JB6284 (WT), CA8880 (rsr1Δ/Δ), and DH7453 (bud2Δ/Δ) were induced to form hyphae by growth in serum-containing agar for 4 h, fixed with formaldehyde, stained with Alexa-phalloidin, and photographed. Left panels, DIC images; right panels, Texas red filter fluorescent images. Scale bar, 10 μm. Adobe Photoshop v. 7 was used to invert the acquired fluorescent images in order to enhance visualization. Arrowheads, actin cables.
Rsr1p and Bud2p are important for hyphal guidance and invasion.
The hyphae of filamentous fungi generally grow in stable directions for long distances, and the process by which the direction of hyphal growth is controlled has recently been termed “hyphal growth guidance” (6). The observation that rsr1/rsr1 and bud2/bud2 HF cells are more bent and curved (Fig. 2 and 7) indicated that hyphal guidance was defective in these cells. Using time-lapse microscopy, we confirmed that wild-type cells maintained a relatively stable, focused direction of growth on serum-containing agar (Fig. 9A; see supplemental movie 1 at http://www.cbs.umn.edu/labs/gale/supplemental.html). In contrast, rsr1/rsr1 and bud2/bud2 cells had more filaments deviate from a straight line of growth by more than 60° (Fig. 9A; see supplemental movies 2 and 3 at http://www.cbs.umn.edu/labs/gale/supplemental.html).
FIG. 9.
Rsr1p and Bud2p are important for hyphal guidance. (A) C. albicans strains JB6284 (WT), CA8880 (rsr1Δ/Δ), and DH7453 (bud2Δ/Δ) were grown on serum-containing agar for 4 h and photographed. The angles of hyphal curvature were determined for each strain using the Angle Measure function of Metamorph and plotted as degrees from linear, with linear being 0°. (B) C. albicans strains JB6284 (WT), CA8878 (RSR1/Δ), CA8880 (rsr1Δ/Δ), MG9215 (rsr1Δ/Δ::RSR1), DH7453 (bud2Δ/Δ), MG9017 (bud2Δ/Δ::BUD2), and MG9061 (bud2Δ/Δ::bud2) were inoculated onto serum-containing agar and incubated at 37°C for 5 days, and the extent of invasion was determined by microscopic examination as described in Materials and Methods. The experiment was repeated three times with the same results obtained each time. The top panel of each pair is a cross-sectional view of yeast colonies and agar; the lower panel of each pair is a view from the top of the agar plate prior to colony washing.
Invasion into agar surfaces is a property of filamentous fungi that, in part, requires directed polarized growth and the ability to control hyphal guidance. To further investigate the role of C. albicans Rsr1p and Bud2p in invasion, we analyzed the ability of rsr1/rsr1 and bud2/bud2 strains to penetrate serum-containing agar. In three replica experiments, we observed that rsr1/rsr1 and bud2/bud2 strains were defective in the ability to invade agar in comparison to that of the isogenic parent, heterozygous, and reintegrant strains (Fig. 9B). Thus, C. albicans Rsr1p and Bud2p are required for effective agar invasion, most likely because they maintain the fidelity of hyphal guidance.
Rsr1p and Bud2p stabilize the polarisome at hyphal tips.
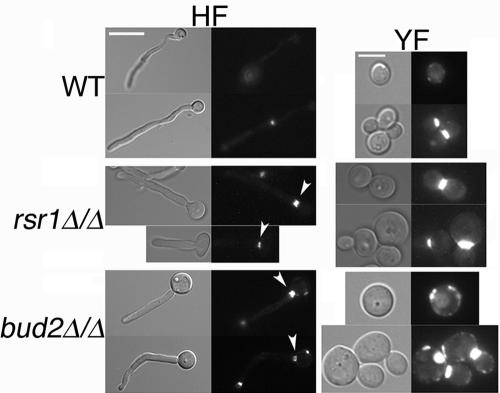
S. cerevisiae Spa2p is a component of the polarisome that controls cell polarity. In C. albicans, Spa2p localizes constitutively to the tips of hyphae, and in cells lacking Spa2p, hyphal growth is defective (63). We used time-lapse microscopy of Spa2 fused to YFP to determine whether rsr1/rsr1 and bud2/bud2 strains had defects in the localization of this important polarity determinant. In wild-type cells, Spa2p-YFP consistently localized to the central portion of the hyphal tip, which predicted the direction of hyphal extension (Fig. 10; see supplemental movie 1 at http://www.cbs.umn.edu/labs/gale/supplemental.html). In cells lacking Rsr1p or Bud2p, Spa2-YFP localized within the tip region of daughter cells (primary filament and filament branches) as well. However, the Spa2-YFP signal position was not consistently at the apex of the primary hypha at all time points; it often moved from the extreme tip, to either side, and back again (Fig. 10; see supplemental movies 2 and 3 at http://www.cbs.umn.edu/labs/gale/supplemental.html). During these fluctuations in Spa2-YFP localization, the daughter cell often increased in size, not only in length but also in width, resulting in isotropic-like, rather than polarized, growth. The times at which oscillations in Spa2-YFP localization occurred correlated with the periodic slowing in growth that we had observed previously (Fig. 6). In addition, for both mutant strains, the Spa2p signal sometimes completely disappeared from the tip. Disappearance of Spa2 from the tip usually coincided with the initiation of a filament branch (with Spa2-YFP relocalizing to the tip of the incipient branch; 4/17 events) or with the appearance of a septum by DIC microscopy (with Spa2-YFP relocalizing to the septum; 13/17 events) (see supplemental movie 4 at http://www.cbs.umn.edu/labs/gale/supplemental.html) within the primary filament. Changes in Spa2-YFP position (from the tip to the side of the tip) were observed prior to a change in the direction of polarized growth that resulted in a bend in the primary filament (Fig. 10; see supplemental movies 2 and 3 at http://www.cbs.umn.edu/labs/gale/supplemental.html). Even during the rare occasions when a hypha of the parent strain exhibited a bend, the Spa2-YFP signal remained at the apex of the hypha and did not exhibit the dramatic changes in position that it did in either of the mutant strains (Fig. 10). Upon resumption of polarized growth, in both mutant strains, Spa2-YFP maintained a more stable position, similar to the parent strain, at the tip of the extending daughter cell. We conclude that Rsr1p and Bud2p are important for stabilizing Spa2p at hyphal tips, thus limiting polarized growth to a confined area and regulating the direction of hyphal growth. Furthermore, Rsr1p and Bud2p appear to have an additional role in the maintenance of Spa2p at the tips of HF cells.
FIG. 10.
Rsr1p and Bud2p stabilize Spa2p localization at hyphal tips. C. albicans strains MG6748 (WT), DH9146 (rsr1Δ/Δ), and MG8868 (bud2Δ/Δ), all expressing Spa2-YFP, were induced to form hyphae on serum-containing agar, and time-lapse images were acquired. DIC and YFP filter fluorescence images were merged to obtain the montages shown. The final DIC image on the right of each row shows the hyphal shape at the end of the experiment. Scale bar, 10 μm (all panels). Adobe Photoshop v. 7 was used to process the acquired images. Supplemental videos showing dynamics of Spa2-YFP during HF growth can be viewed at http://www.cbs.umn.edu/labs/gale/supplemental.html.
C. albicans Rsr1p and Bud2p regulate septin localization during morphogenesis.
Strains lacking either Rsr1p or Bud2p produce daughter cells that look more like pseudohyphae (wider and less elongated) and contain more constrictions when grown under conditions that induce HF growth in wild-type cells. Since the timing of septin ring formation is a critical difference between yeast and hyphal cells (55), we investigated the role of Rsr1p and Bud2p in septation. Using calcofluor white to visualize chitin within septa, no defects in septum formation were evident in the mutant strains in comparison to the isogenic parent strain (data not shown). Next, we analyzed septation in more detail by comparing the localization of septin Cdc3p in rsr1/rsr1 and bud2/bud2 strains to that in the isogenic parent strain. In wild-type HF cells, septins Cdc10p and Cdc11p localize to a “basal septin band” structure at the neck between the basal mother cell and HF daughter cells as well as in the true septin ring structure within the HF daughter cell, which prescribes the position of the septum at cytokinesis (55, 59). Septin Cdc3p also localizes to the true septin ring within hyphae, but unlike Cdc10p and Cdc11p, Cdc3p is generally absent from the basal septin band (7% of cells with Cdc3p at the neck; n > 100) (Fig. 11) (54). Interestingly, in rsr1/rsr1 and bud2/bud2 filaments, Cdc3p localized as a ring or band at the necks of the majority of cells (53% and 78% of cells, respectively; n ≥ 50), consistent with a pseudohyphal, rather than HF, localization pattern for this septin (Fig. 11). These results suggest that Rsr1p and Bud2p affect the regulation of Cdc3p localization during morphogenesis.
FIG. 11.
Rsr1p and Bud2p regulate septin Cdc3p localization during YF and HF growth. C. albicans strains MG5398 (WT), DH9237 (rsr1Δ/Δ), and MG7849 (bud2Δ/Δ), all expressing Cdc3-GFP, were grown under HF or YF conditions, and DIC and GFP filter fluorescence microscopies were used to analyze Cdc3-GFP localization. Arrowheads indicate Cdc3-GFP at the necks of the filaments. Scale bars, 10 and 5 μm (HF and YF, respectively). Adobe Photoshop v. 7 was used to process the acquired images.
Interestingly, during YF growth, strains lacking Bud2p had excessive ectopic aggregates of cortical Cdc3p (Fig. 11), whereas in strains lacking Rsr1p, Cdc3p localized solely to the mother-daughter neck (Fig. 11) as it did in the isogenic parent strain. This result was not due to differences in the expression levels of Cdc3-GFP in the strains, as equal amounts of Cdc3-GFP were detected in the cell lysates of all strains by Western blot analysis (data not shown). Thus, in contrast to Rsr1p, Bud2p has an additional function in septin dynamics during YF growth.
DISCUSSION
Based on the high degree of sequence similarity between ScBUD2 and C. albicans ORF 19.940, we predicted that this sequence encoded the GTPase-activating protein (GAP) for CaRsr1p. In functional analyses, we found that both Rsr1p and Bud2p are cortically localized proteins and that, like Rsr1p (62), Bud2p functions as a landmark for daughter cell site selection during YF growth and HF branching in C. albicans. Importantly, unlike in S. cerevisiae, C. albicans Rsr1p and Bud2p are both important for YF and HF morphogenesis. Strains lacking either Rsr1p or Bud2p have less-polarized YF and HF cell shapes, are unable to invade agar surfaces, and fail to orient hyphal growth in a stable direction. Thus, Bud2p is likely the GAP for Rsr1p. Because loss of Bud2p is predicted to result in a constitutively active (GTP-bound) Rsr1p GTPase, the almost identical phenotypes of the rsr1/rsr1 and bud2/bud2 strains suggest that cycling of Rsr1p between GDP- and GTP-bound states is required for its function in polarity establishment, cell shape, and hyphal guidance. In mammalian cells, Ras GTPase proteins control cell proliferation and cell cycle progression, and deregulation of GTPase activity results in oncogenic transformation (16). The observation that rsr1/rsr1 and bud2/bud2 strains have promiscuous branching during HF growth (Fig. 7) suggests that deregulated CaRsr1p interacts with the cell cycle to drive cell proliferation, as deregulated ras proteins do in mammalian cells.
Rsr1p and Bud2p influence cell shape and hyphal guidance via polarized components of the cytoskeleton.
In the generation of YF or HF daughter cells, the Rho-type GTPase Cdc42p, cortical actin patches, and the polarisome protein Spa2p all polarize to the site of active growth (29, 63). In S. cerevisiae, polarity establishment (polarization of the actin cytoskeleton and the localization and assembly of other polarized components, such as the polarisome and septins) is achieved through the activity of Cdc42p (14). The S. cerevisiae Ras-like GTPase Rsr1p is important for determining the site of polarity establishment but not in the subsequent steps of polarized growth and morphogenesis (47). Interestingly, we found that C. albicans strains lacking components of the Rsr1p GTPase module had defects in morphogenesis that were associated with abnormal or unstable localization of cytoskeletal proteins. Strains lacking Rsr1p and Bud2p contained multiple, ectopically localized actin patches in YF and HF cells and mislocalized septin Cdc3p and had unstable Spa2p localization at hyphal tips. The finding that these phenotypes were similar in both mutant strains is consistent, again, with the idea that aspects of actin, septin, and polarisome localization dynamics require the cycling of Rsr1p between GDP- and GTP-bound states. In S. cerevisiae, actin, septin, and polarisome functions are controlled by Cdc42p. Recently, it was shown that polarisome components, along with the Cdc42p effector kinase Cla4p and Cdc42p GAPs, are required for the actin cable-dependent assembly of the septin ring (33). Thus, functional interactions exist among the polarisome, septin ring, actin, and Cdc42p. In addition, C. albicans strains lacking Wal1p, a protein predicted to mediate interaction of Cdc42p with actin, have phenotypes similar to those of strains lacking Rsr1p and Bud2p: wal1/wal1 strains have abnormal organization of actin, random YF bud site selection, and a pseudohypha-like morphology during growth in hypha-inducing media (57). Our findings raise the possibility that the Rsr1p GTPase positions Wal1p to the proper daughter cell site and, in addition, that rsr1 and bud2 phenotypes, like wal1 phenotypes, may be mediated through altered Cdc42p activity and/or interactions.
We found that in filamentous cells lacking Rsr1p or Bud2p, morphology and cytoskeletal features were more like those of pseudohyphal than hyphal cells. Filaments were wider and contained more constrictions, and actin was less polarized. rsr1/rsr1 and bud2/bud2 filaments had pauses in growth associated with loss of Spa2p signal from the tips, supporting the idea that the Rsr1-GTPase promotes the maintenance of Spa2p at the tips of primary hyphae. That these pausing events usually occurred at the time of lateral branch or septum formation suggests that Rsr1p and Bud2p are important for the normally cell cycle-independent localization of Spa2p during HF growth. Thus, in the absence of Rsr1p and Bud2p, Spa2p localization becomes more cell cycle dependent, similar to the regulation observed during pseudohyphal (and YF) growth (63). Finally, in rsr1Δ/rsr1Δ and bud2Δ/bud2Δ strains, septin Cdc3p localizes to the neck between mother cell and filament, identical to the localization seen in wild-type pseudohyphae but not in wild-type hyphae. Together, these results indicate that Rsr1p may indirectly influence polarization of cytoskeletal proteins by regulation of the overall morphogenesis program (pseudohyphal versus hyphal).
During budding in S. cerevisiae, yeast cells select a site for growth in response to internal signals mediated by Rsr1p and Cdc42p. In contrast, during mating, the direction of shmoo formation occurs in response to external signals that are mediated by Cdc42p but that are independent of Rsr1p. However, in the absence of pheromone receptor signaling (i.e., in cells lacking Cdc24p, the guanine nucleotide exchange factor for Cdc42p), Rsr1p is required to stabilize the axis of polarized growth during shmoo formation. S. cerevisiae rsr1 cdc24 double mutant strains can initiate polarized growth but cannot stabilize the direction of growth: the site of polarized growth and the localization of Spa2p wander, causing cell shape defects (38). Similarly, we observed that C. albicans rsr1/rsr1 and bud2/bud2 strains are able to initiate polarized growth but are unable to maintain a stable axis of polarity. The similar genetic relationships exhibited by S. cerevisiae shmoos and C. albicans hyphae suggest that S. cerevisiae mating projection formation may provide a more relevant paradigm for C. albicans hyphal growth than either the S. cerevisiae pseudohyphal or yeast morphogenesis programs. This idea is supported by the observation that Cdc42p localization to the tips of hyphae, as in S. cerevisiae shmoos but not YF cells, depends on actin (29). It is likely that growth orientation during mating in S. cerevisiae (38) and during HF growth in C. albicans requires continuous orientation of growth direction, whereas during budding in either organism, a single positioning event is sufficient for polarity establishment.
A ras-like GTPase similar to S. cerevisiae Rsr1p also has been reported to be important for hyphal guidance in the filamentous fungus Ashbya gossypii (6). Cells lacking AgRsr1p have slowed hyphal elongation due to frequent pauses of growth associated with asymmetric distribution of actin patches as well as loss of Spa2p at the tip. Resumption of polarized growth was associated with relocalization of Spa2p to the tips, but usually with a different axis of polarity, resulting in a distorted hyphal morphology. The findings that the Rsr1p GTPase is important for shmoo formation in S. cerevisiae cdc24 strains and for hyphal morphology and growth guidance in A. gossypii and C. albicans demonstrate that stabilization of the direction of cell surface growth is critical for cell shape determination in many fungal organisms.
Insights from subtle differences in the phenotypes of rsr1/rsr1 and bud2/bud2 strains.
In addition to the very similar phenotypes described above, rsr1/rsr1 and bud2/bud2 strains exhibited some differences in actin and septin localization characteristics. First, only in bud2/bud2 cells did we observe a significant population of cells that lacked polarized actin patches at the tips of hyphae. This finding suggests that constitutively active Rsr1p either interferes with maintenance or promotes disassembly/depolarization of polarized actin at hyphal tips. Alternatively, the GDP-bound form of Rsr1p may be required for actin maintenance at the tips. Second, in bud2/bud2 YF cells, but not in rsr1/rsr1 YF cells, septin Cdc3p localized to multiple ectopic sites, suggesting that constitutive Rsr1p GTP drives either unregulated septin assembly or improper septin ring disassembly during the yeast cell cycle. The different phenotypes of rsr1/rsr1 and bud2/bud2 cells with respect to actin and Cdc3p localization imply that some functions of Rsr1p are mediated through guanine nucleotide-specific species whereas others require GTP/GDP cycling of Rsr1p. Interestingly, the ectopic cortical Cdc3p was observed only in bud2/bud2 YF cells and not in filaments. This observation is consistent with previous results that demonstrated that septin ring dynamics are fundamentally different in YF and HF cells (55).
Hyphal guidance is important for invasion.
C. albicans HF cells lacking Rsr1p or Bud2p were unable to maintain a stable axis of polarity. Hyphae had frequent bends and curves, during growth on hypha-inducing agar media, which were associated with unstable Spa2p localization at the hyphal tips. In addition, these mutant strains were unable to invade agar surfaces. C. albicans HF cells lacking septins (Cdc10p or Cdc11p) or Spa2 are also curvier than the parent strains and, in the case of the septin mutant strains, also defective in agar invasion (58, 59, 63). Our study, along with these additional studies, supports the idea that hyphal guidance is required for effective agar invasion and that daughter cell site selection proteins and polarized components of the cytoskeleton are important for this process. Alternatively, site selection proteins such as Rsr1p and Bud2p may have a more global role in coupling morphogenesis to gene expression, potentially by regulation of Cdc42p. For example, C. albicans strains with altered Cdc42p activity have reduced expression of, or are unable to maintain transcription of, hypha-specific genes during growth in serum (5, 56).
Previously, it was reported that C. albicans rsr1/rsr1 strains were less virulent in a mouse model of systemic candidiasis, but histological analyses of the effectiveness of tissue invasion were not discussed. How might hyphal guidance influence the pathogenesis of C. albicans infections? There are abundant reports in the literature supporting the idea that C. albicans hyphae exhibit thigmotropism, i.e., regulation of the direction of growth along grooves, into breaks on a surface, or into pores on a membrane (25, 50). In addition, hyphae have been observed to penetrate the cell membrane of epithelial cells (18) and between cell junctions and at sites of local wounds (39). Finally, hyphal cells, in contrast to yeast cells, are capable of apical secretion of proteases and lipases (31). Together, these studies implicate the existence of a positional control pathway, influenced by the site of fungus-host interaction, and support the hypothesis that directional growth of a hypha facilitates the invasion of host cells. Hyphal guidance responses involve reorientation of the cytoskeleton and thus may require landmark proteins, such as Rsr1p, to do this. Indeed, Ras- and Rho-type GTPases are likely to be components of a general mechanism used for growth guidance in many eukaryotic cell systems. For example, Ras- and Rho-type GTPases mediate extracellular signals that guide the migration of axons and dendrites to specific nervous system locations (21, 24, 30, 52). We propose that C. albicans Rsr1p also mediates extracellular information to guide hyphal growth during host tissue invasion.
Acknowledgments
We thank Nicole Dauer for technical assistance and Aaron Mitchell, Ira Herskowitz, and John Pringle for providing strains and/or plasmids. We are very grateful to Judith Berman, Ken Finley, Peter Sudbery, Dana Davis, and Ben Distel for helpful discussions during the preparation of the manuscript. We acknowledge the Stanford Candida Genome sequencing project and the Canadian Bioinformatics Resource at the National Research Council of Canada for providing sequence and sequence comparison information.
This work was supported by a Viking Children's Fund Award and a University of Minnesota Grant-in-Aid of Research and Artistry to C.A.G., NIH AI/DE R01 14666 to Judith Berman, and an Undergraduate Research Opportunites Program award to C.K.-A.
REFERENCES
- 1.Adams, A. E., and J. R. Pringle. 1984. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J. Cell Biol. 98:934-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akashi, T., T. Kanbe, and K. Tanaka. 1994. The role of the cytoskeleton in the polarized growth of the germ tube in Candida albicans. Microbiology 140:271-280. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, J. M., and D. R. Soll. 1986. Differences in actin localization during bud and hypha formation in the yeast Candida albicans. J. Gen. Microbiol. 132:2035-2047. [DOI] [PubMed] [Google Scholar]
- 4.Bassilana, M., J. Blyth, and R. A. Arkowitz. 2003. Cdc24, the GDP-GTP exchange factor for Cdc42, is required for invasive hyphal growth of Candida albicans. Eukaryot. Cell 2:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassilana, M., J. Hopkins, and R. A. Arkowitz. 2005. Regulation of the Cdc42/Cdc24 GTPase module during Candida albicans hyphal growth. Eukaryot. Cell 4:588-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer, Y., P. Knechtle, J. Wendland, H. Helfer, and P. Philippsen. 2004. A Ras-like GTPase is involved in hyphal growth guidance in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell 15:4622-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bender, A. 1993. Genetic evidence for the roles of the bud-site-selection genes BUD5 and BUD2 in control of the Rsr1p (Bud1p) GTPase in yeast. Proc. Natl. Acad. Sci. USA 90:9926-9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bender, A., and J. R. Pringle. 1989. Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene. RSR1. Proc. Natl. Acad. Sci USA 86:9976-9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bensen, E. S., S. G. Filler, and J. Berman. 2002. A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot. Cell 1:787-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, D. H., Jr., A. D. Giusani, X. Chen, and C. A. Kumamoto. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34:651-662. [DOI] [PubMed] [Google Scholar]
- 11.Butty, A. C., P. M. Pryciak, L. S. Huang, I. Herskowitz, and M. Peter. 1998. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 282:1511-1516. [DOI] [PubMed] [Google Scholar]
- 12.Casamayor, A., and M. Snyder. 2002. Bud-site selection and cell polarity in budding yeast. Curr. Opin. Microbiol. 5:179-186. [DOI] [PubMed] [Google Scholar]
- 13.Chaffin, W. L. 1983. Site selection for bud and germ tube emergence in Candida albicans. J. Gen. Microbiol. 130:431-440. [Google Scholar]
- 14.Chang, F., and M. Peter. 2003. Yeasts make their mark. Nat. Cell Biol. 5:294-299. [DOI] [PubMed] [Google Scholar]
- 15.Chant, J., and I. Herskowitz. 1991. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell 65:1203-1212. [DOI] [PubMed] [Google Scholar]
- 16.Coleman, M. L., C. J. Marshall, and M. F. Olson. 2004. RAS and RHO GTPases in G1-phase cell-cycle regulation. Nat. Rev. Mol. Cell Biol. 5:355-366. [DOI] [PubMed] [Google Scholar]
- 17.Davis, D., R. B. Wilson, and A. P. Mitchell. 2000. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrell, S. M., D. F. Hawkins, and T. A. Ryder. 1983. Scanning electron microscope study of Candida albicans invasion of cultured human cervical epithelial cells. Sabouraudia 21:251-254. [PubMed] [Google Scholar]
- 19.Gale, C., M. Gerami-Nejad, M. McClellan, S. Vandoninck, M. S. Longtine, and J. Berman. 2001. Candida albicans Int1p interacts with the septin ring in yeast and hyphal cells. Mol. Biol. Cell. 12:3538-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gale, C. A., C. M. Bendel, M. McClellan, M. Hauser, J. M. Becker, J. Berman, and M. K. Hostetter. 1998. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science 279:1355-1358. [DOI] [PubMed] [Google Scholar]
- 21.Gallo, G., and P. C. Letourneau. 1998. Axon guidance: GTPases help axons reach their targets. Curr. Biol. 8:R80-R82. [DOI] [PubMed] [Google Scholar]
- 22.Gerami-Nejad, M., J. Berman, and C. A. Gale. 2001. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18:859-864. [DOI] [PubMed] [Google Scholar]
- 23.Gerami-Nejad, M., D. Hausauer, M. McClellan, J. Berman, and C. Gale. 2004. Cassettes for the PCR-mediated construction of regulatable alleles in Candida albicans. Yeast 21:429-436. [DOI] [PubMed] [Google Scholar]
- 24.Goi, T., G. Rusanescu, T. Urano, and L. A. Feig. 1999. Ral-specific guanine nucleotide exchange factor activity opposes other Ras effectors in PC12 cells by inhibiting neurite outgrowth. Mol. Cell. Biol. 19:1731-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gow, N. A. 1997. Germ tube growth of Candida albicans. Curr. Top Med. Mycol. 8:43-55. [PubMed] [Google Scholar]
- 26.Gow, N. A., A. J. Brown, and F. C. Odds. 2002. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5:366-371. [DOI] [PubMed] [Google Scholar]
- 27.Gow, N. A., and G. W. Gooday. 1982. Growth kinetics and morphology of colonies of the filamentous form of Candida albicans. J. Gen. Microbiol. 128:2187-2194. [DOI] [PubMed] [Google Scholar]
- 28.Gow, N. A., and G. W. Gooday. 1982. Vacuolation, branch production and linear growth of germ tubes in Candida albicans. J. Gen. Microbiol. 128:2195-2198. [DOI] [PubMed] [Google Scholar]
- 29.Hazan, I., and H. Liu. 2002. Hyphal tip-associated localization of Cdc42 is F-actin dependent in Candida albicans. Eukaryot. Cell 1:856-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoshino, M., and S. Nakamura. 2002. The Ras-like small GTP-binding protein Rin is activated by growth factor stimulation. Biochem. Biophys. Res. Commun. 295:651-656. [DOI] [PubMed] [Google Scholar]
- 31.Hube, B., and J. Naglik. 2001. Candida albicans proteinases: resolving the mystery of a gene family. Microbiology 147:1997-2005. [DOI] [PubMed] [Google Scholar]
- 32.Jitsurong, S., S. Kiamsiri, and N. Pattararangrong. 1993. New milk medium for germ tube and chlamydoconidia production by Candida albicans. Mycopathologia 123:95-98. [DOI] [PubMed] [Google Scholar]
- 33.Kadota, J., T. Yamamoto, S. Yoshiuchi, E. Bi, and K. Tanaka. 2004. Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell 15:5329-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kilmartin, J. V., and A. E. Adams. 1984. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J. Cell Biol. 98:922-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozminski, K. G., L. Beven, E. Angerman, I. Herskowitz, D. G. Drubin, and H. Park. 2003. Interaction between a Ras and a Rho GTPase couples selection of a growth site to the development of cell polarity in yeast. Mol. Biol. Cell 14:4958-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 37.Nern, A., and R. A. Arkowitz. 1999. A Cdc24p-Far1p-Gβγ protein complex required for yeast orientation during mating. J. Cell Biol. 144:1187-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nern, A., and R. A. Arkowitz. 2000. G proteins mediate changes in cell shape by stabilizing the axis of polarity. Mol. Cell 5:853-864. [DOI] [PubMed] [Google Scholar]
- 39.Odds, F. C. 1988. Candida and candidosis. Balliere Tindall, London, United Kingdom.
- 40.Odds, F. C. 1994. Pathogenesis of Candida infections. J. Am. Acad. Dermatol. 31:S2-S5. [DOI] [PubMed] [Google Scholar]
- 41.Park, H. O., E. Bi, J. R. Pringle, and I. Herskowitz. 1997. Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc. Natl. Acad. Sci. USA 94:4463-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park, H. O., J. Chant, and I. Herskowitz. 1993. BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature 365:269-274. [DOI] [PubMed] [Google Scholar]
- 43.Park, H. O., P. J. Kang, and A. W. Rachfal. 2002. Localization of the Rsr1/Bud1 GTPase involved in selection of a proper growth site in yeast. J. Biol. Chem. 277:26721-26724. [DOI] [PubMed] [Google Scholar]
- 44.Park, H. O., A. Sanson, and I. Herskowitz. 1999. Localization of Bud2p, a GTPase-activating protein necessary for programming cell polarity in yeast to the presumptive bud site. Genes Dev. 13:1912-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phan, Q. T., P. H. Belanger, and S. G. Filler. 2000. Role of hyphal formation in interactions of Candida albicans with endothelial cells. Infect. Immun. 68:3485-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pruyne, D., A. Legesse-Miller, L. Gao, Y. Dong, and A. Bretscher. 2004. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 20:559-591. [DOI] [PubMed] [Google Scholar]
- 47.Ruggieri, R., A. Bender, Y. Matsui, S. Powers, Y. Takai, J. R. Pringle, and K. Matsumoto. 1992. RSR1, a ras-like gene homologous to Krev-1 (smg21A/rap1A): role in the development of cell polarity and interactions with the Ras pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:758-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saville, S. P., A. L. Lazzell, C. Monteagudo, and J. L. Lopez-Ribot. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2:1053-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-20. [DOI] [PubMed] [Google Scholar]
- 50.Sherwood, J., N. A. Gow, G. W. Gooday, D. W. Gregory, and D. Marshall. 1992. Contact sensing in Candida albicans: a possible aid to epithelial penetration. J. Med. Vet. Mycol. 30:461-469. [DOI] [PubMed] [Google Scholar]
- 51.Soll, D. R., M. A. Herman, and M. A. Staebell. 1985. The involvement of cell wall expansion in the two modes of mycelium formation of Candida albicans. J. Gen. Microbiol. 131:2367-2375. [DOI] [PubMed] [Google Scholar]
- 52.Spencer, M. L., H. Shao, and D. A. Andres. 2002. Induction of neurite extension and survival in pheochromocytoma cells by the Rit GTPase. J. Biol. Chem. 277:20160-20168. [DOI] [PubMed] [Google Scholar]
- 53.Staebell, M., and D. R. Soll. 1985. Temporal and spatial differences in cell wall expansion during bud and mycelium formation in Candida albicans. J. Gen. Microbiol. 131:1467-1480. [DOI] [PubMed] [Google Scholar]
- 54.Sudbery, P., N. Gow, and J. Berman. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317-324. [DOI] [PubMed] [Google Scholar]
- 55.Sudbery, P. E. 2001. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localisation. Mol. Microbiol. 41:14-31. [DOI] [PubMed] [Google Scholar]
- 56.vandenBerg, A. L., A. S. Ibrahim, J. E. Edwards, Jr., K. A. Toenjes, and D. I. Johnson. 2004. Cdc42p GTPase regulates the budded-to-hyphal-form transition and expression of hypha-specific transcripts in Candida albicans. Eukaryot. Cell 3:724-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walther, A., and J. Wendland. 2004. Polarized hyphal growth in Candida albicans requires the Wiskott-Aldrich Syndrome protein homolog Wal1p. Eukaryot. Cell. 3:471-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warenda, A. J., S. Kauffman, T. P. Sherrill, J. M. Becker, and J. B. Konopka. 2003. Candida albicans septin mutants are defective for invasive growth and virulence. Infect. Immun. 71:4045-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warenda, A. J., and J. B. Konopka. 2002. Septin function in Candida albicans morphogenesis. Mol. Biol. Cell 13:2732-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson, R. B., D. Davis, B. M. Enloe, and A. P. Mitchell. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65-70. [DOI] [PubMed] [Google Scholar]
- 61.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yaar, L., M. Mevarech, and Y. Koltin. 1997. A Candida albicans RAS-related gene (CaRSR1) is involved in budding, cell morphogenesis and hypha development. Microbiology 143:3033-3044. [DOI] [PubMed] [Google Scholar]
- 63.Zheng, X. D., Y. M. Wang, and Y. Wang. 2003. CaSPA2 is important for polarity establishment and maintenance in Candida albicans. Mol. Microbiol. 49:1391-1405. [DOI] [PubMed] [Google Scholar]