Abstract
Citrinin produced by Aspergillus, Penicillium, and Monascus species is a polyketide compound that has nephrotoxic activity in mammals and is bactericidal toward gram-positive bacteria. To avoid the risk of citrinin contamination in other fermentation products produced by Monascus purpureus, knowledge of the citrinin biosynthetic genes is needed so that citrinin-nonproducing strains can be generated. We cloned a polyketide synthase (PKS) gene from M. purpureus with degenerate primers designed to amplify the conserved region of a ketosynthase domain of a fungal PKS. A 13-kb genomic DNA fragment was identified that contained a full-length PKS gene (pksCT) of 7,838 bp with a single 56-bp intron. pksCT encodes a 2,593-amino-acid protein that contains putative domains for ketosynthase, acyltransferase, acyl carrier protein (ACP), and a rare methyltransferase. There was no obvious thioesterase domain, which usually is downstream of the ACP domain in multi-aromatic-ring PKSs. pksCT transcription was correlated with citrinin production, suggesting that the pksCT gene product was involved in citrinin biosynthesis. Homologous recombination between the wild-type allele and a truncated disruption construct resulted in a pksCT-disrupted strain of M. purpureus. The disruptant did not produce citrinin, but a pksCT revertant generated by successive endogenous recombination events in the pksCT disruptant restored citrinin production, indicating that pksCT encoded the PKS responsible for citrinin biosynthesis in M. purpureus.
Fungi and actinomycetes are the primary producers of bioactive secondary metabolites (2), many of which are polyketides. Polyketides, such as pigments and mycotoxins, are structurally diverse and often complex compounds that are at least partially synthesized by multifunctional enzymes called polyketide synthases (PKSs).
There are three types of known PKSs—modular type I PKSs, iterative type I PKSs, and type II PKSs—with all described fungal PKSs belonging to the iterative type I PKS group. These enzymes contain single copies of several functional domains, a feature that distinguishes this class of PKSs from the modular type I PKS class (15). The functional domains typically found in the iterative type I PKSs are those for ketosynthase (KS), acyl transferase (AT), ketoreductase (KR), dehydratase (DH), enoyl reductase (ER), methyltransferase (MT), thioesterase (TE), and acyl carrier protein (ACP). Depending on the structural complexity of the polyketides produced, fungal PKSs have been divided into three groups: (i) single-aromatic-ring PKSs, (ii) multi-aromatic-ring PKSs, and (iii) reduced-complex-type PKSs (5). Single-aromatic-ring PKSs are the smallest and contain ∼1,800 amino acid residues. The characteristic positions of the KS and AT domains in multi-aromatic-ring PKSs are shifted closer to the C terminus than in the other two groups. The reduced-complex-type PKSs are the largest, with each protein containing >2,500 amino acid residues. The polyketides produced by this last PKS group contain more reduced and complex chemical structures than those produced by enzymes in the other two PKS classes.
The filamentous fungus Monascus is an ascomycete used to produce fermented foods, such as To-Fu-Yo and Benikouji, in East Asia. Monascus produces three kinds of described polyketides: citrinin (12), red pigments (7), and monacolin K (3). The red pigments are widely used as natural food colorants, and monacolin K is used as an antihypercholestrolemia agent. Citrinin is bactericidal against gram-positive bacteria, but its nephrotoxic activity (12) in mammalian systems prevents its use in medicine for humans. Nonproduction of citrinin is a very important character of M. purpureus strains used for the commercial production of pigments and monacolin K.
Our objective in this study was to identify and clone the PKS gene responsible for citrinin biosynthesis. Although some citrinin-nonproducing mutants of Monascus have been constructed by conventional mutagenesis, it has not been possible to determine the exact location of the mutation and thereby explain the nonproduction of citrinin, because the structural genes responsible for the enzymes in the citrinin biosynthestic pathway have not been cloned and sequenced. Although genetically modified strains are not allowed for use in the commercial production of food additives, UV/chemically mutagenized strains can be used for commercial production. Thus, by sequencing the genes in question we can determine the molecular nature of mutants with citrinin nonproduction phenotypes that are of commercial importance for producing other metabolites.
(This paper is part of a Ph.D. dissertation by T. Shimizu.)
MATERIALS AND METHODS
Strain, growth conditions, and transformation.
Monascus purpureus IFO30873 is a wild-type strain. M. purpureus strains TNP13 and TNC21 are high producers of red pigments and citrinin, respectively. All three strains were maintained on plates (2% agar) of Monascus cultivation (MC) medium, consisting of 50 g/liter glucose, 7.5 g/liter polypepton (Nihon Seiyaku, Tokyo, Japan), 2.0 g/liter NH4H2PO4, 0.5 g/liter MgSO4 · 7H2O, 0.1 g/liter CaCl2 · 2H2O, and 2.0 g/liter KNO3, and incubated for 7 to 10 days at 28°C. For liquid cultivation, a mycelium mat (about 1 cm2 with agar) was taken from an agar plate, inoculated into 100 ml of MC medium in a 500-ml baffled Erlenmeyer flask, and incubated at 28°C with shaking at 120 strokes per min (spm).
For genetic manipulation in Escherichia coli, E. coli XL10-Gold Ultracompetent cells (Stratagene, La Jolla, California) were used as the cloning host.
Transformation protocol.
The transformation was done as described by Kubodera et al. (10) and according to the manufacturer's protocol of pPTR I DNA (Takara Bio, Inc., Otsu, Japan) with the following modifications. M. purpureus was grown for 40 h at 28°C and 120 spm in MC medium plus 30 g/liter N-acetylglucosamine. The protoplast buffer consists of 10 mM MES [2-(N-morpholino)ethanesulfonic acid] containing 0.6 M sucrose, adjusted to pH 6.0 with 1 M KOH, 20 mg/ml of Yatalase (Takara), and 5 mg/ml of Usukizyme (Kyowa Chemical Products Co., Ltd., Osaka, Japan). The collected protoplasts were washed with and suspended in Sol I (10 mM Tris-HCl [pH 8.0], 0.6 M sucrose, and 10 mM CaCl2), mixed with 4 ml of MC soft agar medium (0.5% Bacto agar [Difco, Detroit, Michigan]), and spread on MC medium containing 200 μg/ml of hygromycin B. Transformants were identified following incubation at 28°C for 7 to 10 days.
Analysis of citrinin production.
Mycelia were grown in 100 ml of MC medium in a 500-ml baffled Erlenmeyer flask, incubated at 28°C with shaking at 120 spm for 2 to 10 days, harvested by filtration, and dried in an oven at 80°C for 3 days. Dried mycelia (1.25 g) were frozen with liquid N2, ground to powder, and extracted with 50 ml of 70% ethanol (pH 8.0) by being stirred for 3 h at room temperature (from 15 to 25°C). The extracts, containing citrinin, were passed through a 0.20-μm filter (DISMIC-13 cp; Advantec, Tokyo, Japan) and analyzed by high-performance liquid chromatography (HPLC) on a C18 column (Inertsil ODS-2; GL Sciences, Inc., Tokyo, Japan) with acetonitrile/water/trifluoroacetic acid (1,000/1,000/1 [vol/vol]) as the mobile phase at a flow rate of 1.0 ml/min and detection by fluorescence (excitation at 330 nm and emission at 500 nm). Commercial citrinin (Wako Pure Chemical Industries, Ltd., Osaka, Japan) was used as the standard.
Isolation of genomic DNA and RNA.
Nucleic acids were extracted from M. purpureus as described by Feng et al. (4) with the following modifications. The ground mycelial powder was resuspended in TEN/SDS buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 150 mM NaCl, and 4% sodium dodecyl sulfate [SDS]) and mixed with 1 volume of TE-saturated phenol. After incubation at 120 spm for 15 min at room temperature, a water layer was obtained by centrifugation (900 × g; 10 min) at room temperature. A portion of the sample was digested with RNase A for 1 h at 37°C to degrade the RNA present. RNA was prepared from the remainder of the sample with the RNeasy Plant Mini-Kit (QIAGEN, Tokyo, Japan) according to the manufacturer's protocol.
Genomic PCR.
Genomic PCR was used to amplify a portion of some of the PKS genes from Monascus with several primer sets, including the LC series primers of Bingle et al. (1) and the KS (5′-GGAATTCTGCAGGAYACNGCNTGYTC-3′) and AT (5′-GGAATTCTGCAGGCRATYTCNCCRGARGARTG-3′) primers that are based on the consensus sequences of the KS domain (DTACS) and the AT domain (HSSGEIA), respectively. The underlined letters indicate PstI and EcoRI sites inserted to enable the cloning of the resulting fragments. The amplification conditions were denaturation at 95°C for 10 min; then 30 cycles, each consisting of denaturation (94°C for 30 s), annealing (55°C for 1 min), and extension (72°C for 1 min); and finally a single extension at 72°C for 7 min. The PCR products were separated on a 1% agarose gel and subcloned into pUC19 (Takara). PCR products were sequenced with an ALFred DNA sequencer (Amersham Biosciences, Piscataway, N.J.) using the Thermo Sequenase Labeled Primer Cycle Sequencing kit with 7-deaza-dCTP (Amersham Biosciences).
RT-PCR.
Reverse transcription-PCR (RT-PCR) was used to detect the transcripts of pksCT and the actin gene from all of the transcripts in Monascus with the primer sets pksCT F (5′-GGAATTCTGCAGCCAGTGTGGCTATTCACC-3′) and pksCT R (5′-GGAATTCTGCAGAAGAGTAATGTCCTTAGG-3′) and actin F (5′-GGAATTCTGCAGATTCTACAACGAACTCCG-3′) and actin R (5′-GGAATTCTGCAGTCAGGGAGTTCATAGGAC-3′), which were designed to amplify a portion of pksCT and the actin gene, when the mRNA Selective PCR kit, version 1.1 (Takara), was used. The underlined letters indicate EcoRI and PstI sites inserted to enable the cloning of the resulting fragments. The amplification conditions were as follows: a reverse transcriptase reaction at 50°C for 30 min; then 25 cycles, each consisting of denaturation (85°C for 1 min), annealing (45°C for 1 min), and extension (72°C for 1 min); and finally a single extension at 72°C for 4 min. The PCR products were separated on a 2% agarose gel and subcloned into pUC19 (Takara). PCR products were sequenced with an ALFred DNA sequencer (Amersham Biosciences) using the Thermo Sequenase Labeled Primer Cycle Sequencing kit with 7-deaza-dCTP (Amersham Biosciences).
Sequence analysis.
Cloned DNA fragments were analyzed with the FASTA program in the DDBJ homology search system (http://www.ddbj.nig.ac.jp/E-mail/homology-j.html). The MT domains of PksCT and the lovastatin nonaketide PKS were compared by using the search homology program in GENETYX, version 6 (Genetyx Corp., Osaka, Japan).
GenBank database accession numbers of the PKS genes used in this paper are Aspergillus nidulans pksST, L39121; A. nidulans wA, X65866; Aspergillus parasiticus PksL1, L42766; Aspergillus terreus pksM, U31329; A. terreus lovB, AF151722; Bacillus subtilis ACP, Z14098; Botryotinia fuckeliana pks16, AY495621; B. fuckeliana pks17, AY495622; B. fuckeliana pks18, AY495623; B. fuckeliana pks19, AY495624; B. fuckeliana pks20, AY4956225; Cochliobolus heterostrophus pks1, U68040; C. heterostrophus Pks21, AY495662; C. heterostrophus pks22, AY495663; C. heterostrophus pks23, AY495664; Colletotrichum lagenarium pks1, D83643; Gibberella fujikuroi pks2, AJ421889; Microcystis aeruginosa mcyD, AB032549; Nodulisporium sp. ATCC 74245 pks1, AF151533; Penicillium patulum 6MSAS, X55776; Stigmatella aurantiaca stiH, AJ421825; S. aurantiaca stiJ, AJ421825; Streptomyces atroolivaceus lnmJ, AF484556; and Streptomyces avermitilis ACP, AB070946.
Southern blot analyses.
Southern blots were made essentially as described by Sambrook et al. (13). Genomic DNA (20 μg) was digested with restriction enzymes overnight, separated on a 1% agarose gel, and transferred to a Hybond N+ membrane (Amersham Biosciences). The probe was labeled with [α-32P]dCTP using the Random Primer DNA Labeling kit, version 2 (Takara).
Colony hybridization.
Southern blot analysis detected a 4-kb BamHI-BamHI fragment and a 7-kb EcoRI-SalI fragment that could hybridize to the KS-LC5c PCR product as a probe and a 4.2-kb KpnI (2)-KpnI (3) fragment that could hybridize to the probe B which was amplified by PCR from the 3′ region of the EcoRI-SalI fragment with primers F (5′-ATTGCTTGGGCTGCCAGTG-3′) and R (5′-ATCAAAGTCCGGAAGCGC-3′). Monascus genomic DNA was digested with each restriction enzyme and electrophoresed on a 1% agarose gel, and corresponding regions were extracted and subcloned into the multicloning site of pUC19, resulting in construction of individual partial genomic libraries. From each library, 2,000 colonies were selected, and positive clones were identified by colony hybridization with each probe. The amino acid sequences inferred from these fragments were compared to amino acid sequences of the conserved active sites of known fungal PKS domains (11).
Construction of pCT1.
A 2.1-kb SphI-KpnI (1) fragment was treated with T4 DNA polymerase and inserted into the EcoRV site in pCSN44 (14), which contains a hygromycin B-resistant gene (Fungal Genetics Stock Center; http://www.fgsc.net/). DNA sequencing confirmed the orientation of the insert.
Analysis of phenotypes of the PKS disruptant and its revertant.
Citrinin content was measured either by HPLC or by enzyme-linked immunosorbent assay with the RIDASCREEN FAST citrinin kit (R-Biopharm AG, Darmstadt, Germany) according to the manufacturer's protocol. For the analysis of pigment production, dried mycelia (1.25 g) were frozen with liquid N2, ground to a powder, and extracted with 50 ml of 70% ethanol (pH 8.0) by being stirred for 1 h at room temperature. After filtration (Qualitative Filter Paper no. 2; Advantec, Tokyo, Japan), 1 ml of the filtrate was mixed with 50 ml of 70% ethanol (pH 4.0), and optical density at 500 nm was measured. The unit of measurement for the amount of citrinin and red pigment is micrograms per gram of dry mycelia and optical density at 500 nm per weight of dry mycelium, respectively.
RESULTS
Cloning of a PKS gene.
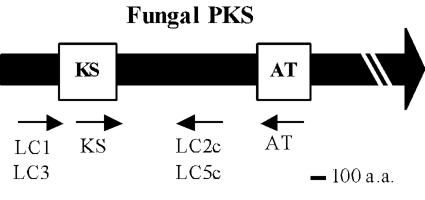
PCR with primer pair KS and LC5c (Fig. 1) yielded a single distinct product, and the PCR product (430 bp) was very close to the expected size, 420 bp, calculated from the sequences of other fungal PKSs. The deduced amino acid sequence was 49%, similar to the aflatoxin biosynthetic PKS from A. parasiticus and 43% similar to the bikaverin biosynthetic PKS from G. fujikuroi. These similarities suggested that the PCR fragment was part of a PKS gene responsible for the biosynthesis of multi-aromatic-ring polyketides.
FIG. 1.
Primer positions in the flanking regions of KS and AT domains of a consensus fungal PKS. The boldface arrow and open boxes show the fungal PKS gene and regions encoding the KS and AT domains, respectively. The small arrows indicate the primers' positions and directions.
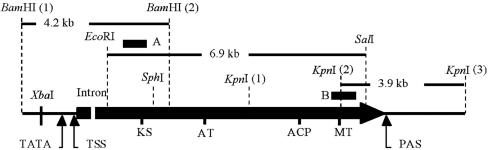
The complete PKS gene (7,838 bp; DDBJ accession no. AB167465) encoding a 2,593-amino-acid protein (Fig. 2) was obtained from the M. purpureus genome by three rounds of colony hybridization by using probe A to identify a BamHI (1)-BamHI (2) fragment and an EcoRI-SalI fragment and probe B to clone a KpnI (2)-KpnI (3) fragment (Fig. 2).
FIG. 2.
Restriction map of the cloned 12.9-kb BamHI (1)-KpnI (3) fragment and structure of the cloned PKS. The black boxes indicate the locations of probes A and B used to obtain the BamHI (1)-BamHI (2) and EcoRI-SalI fragments and the KpnI (2)-KpnI (3) fragment, respectively, by colony hybridization. The thick black arrow indicates the deduced open reading frame for pksCT in the cloned DNA sequence. The small arrows indicate the TATA box, the transcriptional start site (TSS), and polyadenylation site (PAS), respectively.
The C residue 974 bp downstream from the XbaI site in the BamHI (1)-BamHI (2) fragment was identified as the transcriptional start site by primer extension (data not shown). The putative start codon and TATA box were localized 223 bp downstream and 112 bp upstream of the transcription start site, respectively. A TTA codon 390 bp downstream of the SalI site served as the stop codon and was followed by a polyadenylation site 49 bp downstream of the TTA codon. An intron was present at bp 1836 to 1891, flanked by a typical splice site (5′-GT-AG-3′).
The product of pksCT gene contained domains for KS, AT, and ACP based on their similarities to the corresponding domains in fungal PKSs and the presence of typical active-site sequences DXACXS, GHSXG, and GXDS (X refers to amino acids that are not strictly conserved), respectively (11). There was also a region with 32% similarity to the MT domain of lovastatin nonaketide PKS from Aspergillus terreus (8) downstream from the ACP domain, which suggests that PksCT may include a MT domain.
In addition to pksCT, another putative PKS gene fragment was obtained by genomic PCR with the primer sets of LC1 and LC2c. Cloning and sequencing of this putative PKS gene revealed that it was a part of pks1 (accession no. AJ414729 in GenBank) in Monascus purpureus, reported only in a database. Although the putative PKS gene was disrupted in this study, it did not affect citrinin production (data not shown). PCR with other combinations of primers was unsuccessful, probably due to the low similarity of primers to other PKSs of M. purpureus.
Transcriptional analysis of the putative PKS gene.
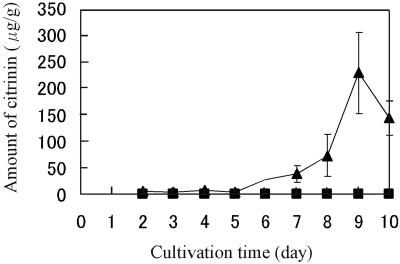
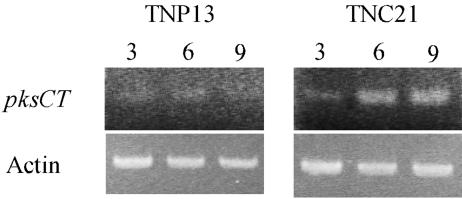
pksCT transcription was examined to investigate the relationship between citrinin production and pksCT transcription in a strain that produces high levels of citrinin (TNC21) and one that produces little or no citrinin (TNP13). Citrinin production by strain TNC21 began on day 2 of cultivation and continued until day 9 (Fig. 3), while very little production (<1 μg/g) was detected at any time for strain TNP13 throughout cultivation. mRNA levels analyzed by RT-PCR clarified that the pksCT gene was more highly transcribed in TNC21 than in TNP13 throughout the cultivation and that the amount of the transcript in TNC21 increased from day 3 to 6, accompanying the increase in citrinin production (Fig. 4). This result suggests that pksCT may be part of the citrinin biosynthetic pathway.
FIG. 3.
Comparison of citrinin production between the red-pigment high-producer strain TNP13 and the citrinin high-producer strain TNC21. A profile of citrinin production of strains TNP13 (▪) and TNC21 (▴) during cultivation for 10 days is shown. Citrinin content was determined by reverse-phase HPLC. Error bars indicate standard deviations.
FIG. 4.
RT-PCR on the pksCT transcriptions in strains TNP13 and TNC21 with the primer set of pksCT F and R. The number above each lane indicates the days of cultivation when the RNA sample used for template of RT-PCR was extracted. Actin gene was used as a control.
Phenotype of pksCT disruptant and its revertant.
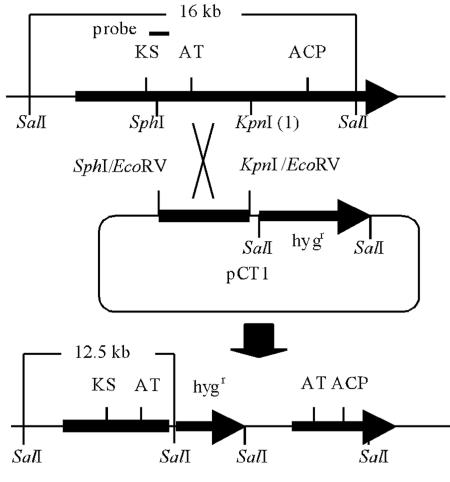
We disrupted the pksCT gene in M. purpureus by homologous recombination between an SphI-KpnI (1) fragment on a disruption plasmid and on the chromosome (Fig. 5). The disruptant has a truncated pksCT at either the 5′ region lacking the KS domain or at the 3′ region lacking the ACP domain, and neither protein should have catalytic activity, if either is synthesized at all. Integration of the disruption plasmid in the correct position was confirmed by Southern blot analysis with the KS-LC5 fragment as a probe (Fig. 6).
FIG. 5.
Disruption of the pksCT gene in M. purpureus. The strategy for disrupting the pksCT gene by a single crossover. A pCNS44 derivative containing the internal 2.1-kb SphI-KpnI (1) fragment (pCT1) was transformed into M. purpureus IFO30873. The single-crossover event between the Monascus genome and the SphI-KpnI (1) fragment on pCT1 results in truncated open reading frames that lack either the ACP domain or the KS domain.
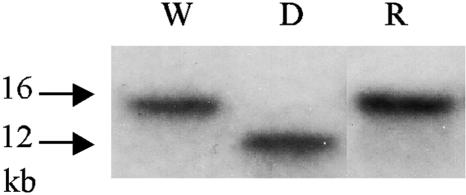
FIG. 6.
Southern blot analysis of the wild-type strain and the pksCT disruptant. Southern blotting was carried out against the SalI-digested genomic DNA using the KS-LC5 PCR product as a probe. The 16-kb and 12.5-kb bands indicate the existence of the intact pksCT and truncated pksCT, respectively. W, wild type; D, pksCT disruptant; R, pksCT revertant.
The pksCT disruptants were similar to the wild-type strain in red pigmentation: wild type, 1.66 × 103; and pksCT disruptant, 0.82 × 103 optical density/g of dry mycelia after 10 days of cultivation. These values are within the usual range of variation for red pigment production, suggesting that the pksCT gene is not involved in the biosynthesis of the red pigments. A tiny peak was found by reverse-phase HPLC at the same elution position as that of citrinin in the pksCT disruptant: wild type, 1.4 × 102; and pksCT disruptant, 0.56 μg/g-dry mycelia after 10 days of cultivation. However, an enzyme-linked immunosorbent assay with the RIDASCREEN FAST citrinin kit (R-Biopharm AG) detected no citrinin until day 10 of cultivation: wild type, 3.0 × 102 μg/g of dry mycelia; pksCT-disruptant, not detectable (detection limit, 15 ng/g) after 10 days of cultivation. The HPLC result is probably attributable to a contaminant that eluted at the same position as that of citrinin.
The pksCT revertant was obtained from the pksCT disruptant by endogenous homologous recombination between the duplicated regions of SphI-KpnI (1) fragment (Fig. 5), which generated original gene organization with the deletion of the vector region. After several rounds of cultivation from the spore of the pksCT disruptant in medium without hygromycin, hygromycin-sensitive colonies were obtained and confirmed to be the pksCT-revertant strains (Fig. 6). Citrinin levels measured by HPLC clarified that citrinin production was restored in the revertant: wild type, 8.6 × 10 μg/g of dry mycelia; pksCT revertant, 7.4 × 10 μg/g of dry mycelia after 7 days of cultivation. These results confirmed that pksCT encodes the PKS gene responsible for citrinin biosynthesis.
DISCUSSION
Citrinin is nephrotoxic in mammalian systems (12), but other Monascus metabolites, e.g., red pigments and monacolins, are widely used as natural food colorants or antihypercholesterolemia agents. Thus, there are strong economic and safety demands for suppressing citrinin production in commercially useful Monascus strains. In this report, we identified and characterized a gene encoding a PKS that participates in citrinin biosynthesis by the transcriptional analysis and gene disruption. Based on the DNA sequence, PksCT has a domain organization that is unique thus far among characterized fungal PKSs.
Fungal PKSs have been divided into three groups based on the chemical structure of the polyketide produced and the domain organization of the polypeptide (5). More recently, Kroken et al. (9) classified fungal PKSs into 10 small clades in three primary groups based on conserved sequences in the KS domain. Each group in the earlier grouping corresponds to one of three main groups in the phylogenetic grouping. Single-aromatic-ring PKSs are found in two fungal PKS clades, multi-aromatic-ring PKSs are found in fungal nonreducing PKSs, and reduced-complex-type PKSs are equivalent to fungal reducing PKSs.
We assumed that the citrinin biosynthetic PKS would belong to the multi-aromatic-ring PKS group, based on the structure of citrinin. This type of PKS should have a KS-AT-ACP-TE domain organization. However, no region encoding the TE domain consensus sequence (GPYXLXGWSXXG) was detected in PksCT, and the amino acid sequence contains a putative MT region, typical of clade III nonreducing PKSs (Table 1). Thus, the domain organization of PksCT does not match that expected for a multi-aromatic-ring PKS but is consistent with that expected for a clade III nonreducing PKS (KS-AT-ACP-MT). The putative MT domain is consistent with the need for a methylation step in the citrinin biosynthetic pathway (6).
TABLE 1.
Similarity of MT domains in the PksCT protein of M. purpureus
| Organism | Protein | %b | Amino acid sequencec |
|---|---|---|---|
| Monascus purpureus | PksCT | 100 | ILEMGAGTGG-TDLSSS-VHAT-LLLLEMT |
| Botryotinia fuckeliana | Pks17 | 80 | ILEMGAGTGG-TDLSSS-VHAT-LLLIEMT |
| Cochliobolus heterostrophus | Pks21 | 61 | ILEMGAGTGA-TDLAPS-VHAT-LMLLEMT |
| B. fuckeliana | Pks20 | 54 | ILEVGAGTGG-TDLSRS-VHAT-VVLLEGT |
| B. fuckeliana | Pks18 | 43 | ILEIGGGTGG-SDISPM-IHAT-CSLVEFT |
| C. heterostrophus | Pks23 | 44 | ILEVGAGTGG-TDISPT-VHAT-VVLSEVT |
| Microcystis aeruginosa | McyDa | 46 | ILEIGGGTGA-TDISSS-LHAT-LILLEST |
A functionally identified PKS.
Amino acid identities for the 100-amino-acid sequences centered on the conserved active site.
-, a space among the conserved regions. Boldface letters are amino acids identical to PksCT.
In addition to the presence of MT domain, the overall deduced amino acid sequence from pksCT, excluding the intron, had the highest similarity to five members of the clade III nonreducing PKS class (61% to Pks17 in Botryotinia fuckeliana, 43% to Pks 21 in Cochliobolus heterostrophus, 32% to Pks16 and Pks18 in B. fuckeliana, and 28% to Pks22 in C. heterostrophus), followed by other known PKSs, e.g., 1,3,6,8-tetrahydroxynaphthalene (melanin) synthase of Nodulisporium sp. ATCC 74245, which belongs to clade II of the nonreducing PKSs. Thus, we classified PksCT as a member of clade III of the fungal nonreducing PKSs. This report is the first of a characterized PKS that belongs to this clade, although nonreducing PKSs have been identified in both clade I (e.g., the aflatoxin PKS and bikaverin PKS) and clade II (e.g., the melanin PKS).
In vitro enzymatic characterization of PksCT, e.g., to determine the number of malonyl-coenzyme As to be condensed, and the ability to catalyze cyclization and methylation will clarify the individual function of each domain in PksCT. Additional knowledge of the citrinin biosynthetic pathway and the enzymes that participate in it should more readily enable the construction of citrinin-nonproducing strains of Monascus.
REFERENCES
- 1.Bingle, L. E. H., T. J. Simpson, and C. M. Lazarus. 1999. Ketosynthase domain probes identify two subclasses of fungal polyketide synthase genes. Fungal Genet. Biol. 26:209-223. [DOI] [PubMed] [Google Scholar]
- 2.Demain, A. L. 1999. Pharmaceutically active secondary metabolites of microorganisms. Appl. Microbiol. Biotechnol. 52:455-463. [DOI] [PubMed] [Google Scholar]
- 3.Endo, A. 1979. Monacolin K, a new hypocholesterolemic agent produced by a Monascus species. J. Antibiot. 32:852-854. [DOI] [PubMed] [Google Scholar]
- 4.Feng, G. H., F. S. Chu, and T. J. Leonard. 1992. Molecular cloning of genes related to aflatoxin biosynthesis by differential screening. Appl. Environ. Microbiol. 58:455-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujii, I., A. Watanabe, Y. Mori, and Y. Ebizuka. 1998. Structures and functional analysis of fungal polyketide synthase genes. Actinomycetologica 12:1-14. [Google Scholar]
- 6.Hajjaj, H., A. Klaébé, M. O. Loret, G. Goma, P. J. Blanc, and J. François. 1999. Biosynthetic pathway of citrinin in the filamentous fungus Monascus ruber as revealed by 13C nuclear magnetic resonance. Appl. Environ. Microbiol. 65:311-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hajjaj, H., A. Klaébé, G. Goma, P. J. Blanc, E. Barbier, and J. François. 2000. Medium-chain fatty acids affect citrinin production in the filamentous fungus Monascus ruber. Appl. Environ. Microbiol. 66:1120-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendrickson, L., C. R. Davis, C. Roach, D. K. Nguyen, T. Aldrich, P. C. McAda, and C. D. Reeves. 1999. Lovastatin biosynthesis in Aspergillus terreus: characterization of blocked mutants, enzyme activities and a multifunctional polyketide synthase gene. Chem. Biol. 6:429-439. [DOI] [PubMed] [Google Scholar]
- 9.Kroken, S., N. L. Glass, J. W. Taylor, O. C. Yoder, and B. G. Turgeon. 2003. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. USA 100:15670-15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubodera, T., N. Yamashita, and A. Nishimura. 2000. Pyrithiamine resistance gene (ptrA) of Aspergillus oryzae: cloning, characterization and application as a dominant selectable marker for transformation. Biosci. Biotechnol. Biochem. 64:1416-1421. [DOI] [PubMed] [Google Scholar]
- 11.Pažoutová, S., M. Linka, Š. Štorková, and H. Schwab. 1997. Polyketide synthase gene pksM from Aspergillus terreus expressed during growth phase. Folia Microbiol. 42:419-430. [DOI] [PubMed] [Google Scholar]
- 12.Sabater-Vilar, M., R. F. M. Maas, and J. Fink-Gremmels. 1999. Mutagenicity of commercial Monascus fermentation products and the role of citrinin contamination. Mutat. Res. 444:7-16. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Staben, C., B. Jensen, M. Singer, J. Pollock, M. Schechtman, J. Kinsey, and E. Selker. 1989. Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal. Genet. Newsl. 36:79-81. [Google Scholar]
- 15.Staunton, J., and K. J. Weissman. 2001. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 18:380-416. [DOI] [PubMed] [Google Scholar]