Abstract
Animal enteroviruses shed in the feces of infected animals are likely environmental contaminants and thus can be used as indicators of animal fecal pollution. Previous work has demonstrated that bovine enterovirus (BEV) present in bovine feces contaminates waters adjacent to cattle herds and that BEV-like sequences are also present in shellfish and in deer feces from the same geographical area. However, little information is available about the prevalence, molecular epidemiology, and genomic sequence variation of BEV field isolates. Here we describe an optimized highly sensitive real-time reverse transcription-PCR method to detect BEV RNA in biological and environmental samples. A combination of the amplification procedure with a previously described filtration step with electropositive filters allowed us to detect up to 12 BEV RNA molecules per ml of water. The feasibility of using the method to detect BEV in surface waters at a high risk of fecal pollution was confirmed after analysis of water samples obtained from different sources. The method was also used to study the prevalence of BEV in different cattle herds around Spain, and the results revealed that 78% (78 of 100) of the fecal samples were BEV positive. BEV-like sequences were also detected in feces from sheep, goats, and horses. Nucleotide sequence analyses showed that BEV isolates are quite heterogeneous and suggested the presence of species-specific BEV-like variants. Detection of BEV-like sequences may help in the differentiation and characterization of animal sources of contamination.
The environment, including surface waters, is often contaminated with enteric viruses (4, 33), a heterogeneous group of viruses that comprises enteroviruses, noroviruses (Norwalk-like viruses), astroviruses, rotaviruses, enteric adenoviruses, and hepatitis A and hepatitis E viruses. Enteroviruses are characterized by their stability, both in the gastrointestinal tract and in the environment, and thus are excreted in feces in large amounts and persist in the environment for long times. As their main transmission route is the fecal-oral route, food and water contaminated with these viruses are major sources of infection. In addition, it is known that rain contributes greatly to dissemination of these viruses in the environment, and therefore, surface waters are believed to be important enteric virus reservoirs.
One of the best-studied groups of enteric viruses is the human enteroviruses, in the family Picornaviridae, which have long been recognized as indicators of fecal contamination of water (1, 4, 5, 23, 25, 26, 33). Although less is known about animal enteroviruses, it might be expected that these viruses are also good markers of environmental fecal contamination. Moreover, as most enteroviruses have a narrow host range, usually showing a strong preference for infection of one (or a few related) animal species, their presence in water can indicate that one or a few species are the source of contamination. In this regard, we have recently shown that viruses belonging to the genus Teschovirus, formerly included in the porcine enterovirus group in the Picornaviridae family (11) and specific for swine, are optimal indicators of porcine fecal contamination of water (9). Indeed, methods are now available for the use of animal enteric viruses as markers of fecal contamination (8, 9, 14, 15, 15a, 16).
Bovine enterovirus (BEV), a member of the Picornaviridae family (22), is endemic in some cattle herds and cattle environments around the world (2, 14, 17, 36). Currently, only two BEV serotypes are recognized, serotype 1 (BEV-1) and serotype 2 (BEV-2) (12, 18, 35). Infection with BEV does not cause serious animal health problems, and it is usually asymptomatic; however, viruses are excreted in feces in large amounts. The usefulness of BEV detection for tracking bovine fecal contamination of environmental waters after reverse transcription (RT)-PCR amplification of BEV RNA from feces and water samples has recently been demonstrated (8, 14). Further nucleotide sequencing of the amplified products allowed characterization of the detected strains circulating in the area studied, the Chesapeake Bay region of Maryland. The results indicated that BEV was endemic in the cattle population under study and that the virus contaminated waters (standing water, runoff, and animal watering tanks from the farm), as well as oysters collected from the adjacent waters. Moreover, a considerable number of feces from deer grazing in the same pastures were also BEV positive (14). Nucleotide sequence analysis showed the presence of at least three different groups of sequences, which allowed establishment of a relationship between BEV variants detected in cattle feces and BEV variants found in deer feces, oysters, and waters from the same area.
Even though these results confirmed that the detection of BEV RNA might be useful for tracking environmental pollution of animal origin, it should be noted that the low concentration at which viruses are usually found in water has greatly hampered their use as markers of fecal contamination. Consequently, successful virus detection often requires efficient concentration of water samples. Several procedures to do this have been described. These procedures are based essentially on filtration-elution though electropositive filters (6, 30), organic extraction (20), or ultracentrifugation (3) or a combination of two or more of these methods (10, 19). In addition, the detection methods themselves are of utmost importance for determining the sensitivity of the whole detection procedure. The recent advent of real-time PCR methodology with fluorogenic probes has provided a powerful tool that enables nucleic acid detection with high sensitivity and specificity.
In the present report we describe an optimized and highly sensitive procedure for detecting bovine enterovirus RNA in polluted water, environmental, and biological samples. The optimized method was used to determine the presence of BEV in cattle feces and water samples collected at various locations around Spain. In addition, fecal samples from other species (sheep, goats, horses, and donkeys) sharing or not sharing pastures with cattle were also analyzed. Nucleotide sequence analyses of representative samples from the different animal species and water supported the usefulness of the BEV detection method and suggested the presence of species-specific BEV-like variants.
MATERIALS AND METHODS
Viruses, cells, and virus propagation in cell cultures.
BEV strains PS87 (serotype 2) and G527 (serotype 1) were obtained from the American Type Culture Collection (Manassas, Va.). Virus isolation and propagation were performed with Mardin-Darby bovine kidney (MDBK) cells inoculated with BEV or filter-sterilized fecal extracts (see below), as previously described (14).
Samples.
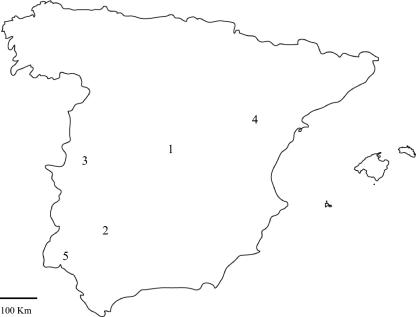
Fecal samples from cattle were collected in areas subjected to intensive farming (36 samples from 12 dairy farms) or extensive farming (64 samples from 12 sampling points, each corresponding to one herd). Most bovine samples (87 samples, including all samples from intensive farms) were collected in the center of Spain (area 1 in Fig. 1), a region with a high concentration of cattle farms. The remaining bovine samples were collected in two different areas (areas 2 and 3) that were far from area 1 (Fig. 1), where cattle are kept on extensive farms and share pastures with other ruminant species (goats and sheep). Hence, in addition to bovine samples, feces from goats were collected in areas 2 and 3 and feces from sheep were collected in areas 2 and 4. In the latter area, sheep was the only livestock species found. Finally, fecal samples from donkeys were obtained in area 3, and fecal samples from horses were obtained in areas 3 and 5. Additionally, 16 water samples were collected from different water sources that were under the influence of cattle farms, that were not near cattle farms, or that were under the influence of other livestock species. Samples were designated by their origin (fecal samples obtained from cattle [Bo], sheep [Ov], goats [Cp], or horses [Eq] or water samples [Wa]), followed by two digits; the first digit indicated the sampling area (areas 1 to 5 in Fig. 1), and the second digit indicated the sample number.
FIG. 1.
Map of the survey area. Feces from cattle, sheep, goats, donkeys, and horses and water samples were obtained from areas in the following provinces: Madrid (area 1), Seville (area 2), Cáceres (area 3), Teruel (area 4), and Huelva (area 5). See Table 3 for details.
Concentration of water samples.
Water samples were concentrated by a filtration-elution method using electropositive filters as described previously (6, 19). Briefly, 1.5- to 2-liter water samples were clarified by gravity filtration through Whatman no. 1 paper (Merck, Whatman 3MM, Darmstadt, Germany), followed by filtration through 47-mm prefilters (CUNO Inc., Meriden, Conn.) and electropositive filters (Virosorb 1-MDS; CUNO Inc.) at pH 6 to 7 and then eluted by soaking in 10 ml of elution buffer (0.1 M glycine, pH 9, with 3% beef extract) for 5 min. The pH of the eluted concentrate was immediately neutralized by addition of 1 N HCl to obtain a pH of 6 to 7. The concentrates were kept frozen at −20°C until they were analyzed for the presence of BEV. To check the efficiency of the concentration method used, 2 liters of distilled water was independently spiked in duplicate with three dilutions of the BEV PS87 prototype strain (4.6 × 103 to 4.6 × 104 50% tissue culture infective doses [TCID50]) and concentrated as described above, and then virus recovery was measured by RNA quantification using the quantitative fluorogenic RT-PCR protocol described below.
Virus extraction from fecal samples.
Fecal samples (1 to 2 g) were homogenized in phosphate-buffered saline containing antibiotics (100 U/ml penicillin, 100 μg/ml streptomycin) at a 1:2 ratio (wt/vol) and centrifuged at 14,000 × g for 15 min, and then the supernatants were filtered through 0.22-μm-pore-size membranes. Fecal extracts were conserved at −20°C until they were used.
Nucleic acid extraction.
Viral RNA was isolated from 140 μl of water concentrate, a fecal extract, or the supernatant of BEV-infected cells using a Qiamp viral RNA mini kit (QIAGEN, Valencia, Calif.) according to manufacturer's instructions. RNA was eluted in 60 μl of diethyl pyrocarbonate-treated water and kept at −20°C until analysis by RT-PCR.
Amplification procedures.
We optimized a quantitative real-time RT-PCR assay for BEV using a BEV-2 prototype strain (isolate PS87; American Type Culture Collection) and an ABI Prism 7700 sequence detection system (Perkin-Elmer Applied Biosystems, Branchburg, NJ). The virus was propagated in MDBK cell monolayers and concentrated as described above. Then the clarified supernatant from infected cells was purified by ultracentrifugation through a 2-ml 20% sucrose cushion for 2 h at 130,000 × g. Next, viral RNA was isolated as described above, except that an RNA carrier was excluded from the buffer to avoid interference with RNA quantification, as determined by spectrophotometry. Tenfold dilutions of this viral RNA stock were used as a standard, both to check the sensitivity and linearity of the method and to quantify BEV RNA in BEV-spiked water. To optimize the method for maximum sensitivity and specificity, several concentrations of primers and fluorogenic TaqMan probe, as well as different numbers of cycles, were tested.
Amplification was carried out in duplicate using a commercial RT-PCR amplification kit (TaqMan one-step RT-PCR master mixture reagents; Applied Biosystems) according to manufacturer's instructions. Briefly, real-time RT-PCR was carried out by mixing 13 μl of isolated RNA with 25 μl TaqMan 2× universal PCR master mixture, 1.25 μl of 40× Multiscribe and RNase inhibitor mixture, BEV-specific primers (BEV-5FL and BEV-3FL [Table 1]) at a final concentration of 0.5 μM, a fluorogenic TaqMan probe (BEV-SON) (Table 1) at a final concentration of 0.25 μM, and enough RNase-free water bring the volume to 40 μl. The amplification conditions were as follows: reverse transcription at 48°C for 30 min, followed by 10 min at 95°C (“hot start”) and 50 cycles of 15 s at 95°C and 1 min at 60°C.
TABLE 1.
Oligonucleotide primers and probe used
| Primer or probe | Sequence | Target (positions)a |
|---|---|---|
| BEV-F | 5′-GGGGAGTAGTCCGACTCCGC-3′ | 126-145 |
| BEV-R | 5′-CAGAGCTACCACTGGGGTTGTGG-3′ | 375-397 |
| NBEV-F | 5′-ACGGAGTAGATGGTATTCCC-3′ | 189-208 |
| NBEV-R | 5′-CGAGCCCCATCTTCCAGAG-3′ | 393-411 |
| BEV-5FL | 5′-GCCGTGAATGCTGCTAATCC-3′ | 533-552 |
| BEV-3FL | 5′-GTAGTCTGTTCCGCCTCCACCT-3′ | 604-625 |
| BEV-SON | 5′FAM-CGCACAATCCAGTGTTGCTACGTCGTAAC-3′TAMRAb | 570-598 |
The nucleotide numbering in the target sequence corresponds to that of the PS87 isolate (18).
FAM, 6-carboxyfluorescein; TAMRA, 6-carboxy tetramethylrhodamine.
For sequencing purposes, a 286-bp fragment from the 5′ noncoding region (5′NCR) of the BEV genome was RT-PCR amplified with primers BEV-F and BEV-R (Table 1) using a SUPERSCRIPT One-Step RT-PCR kit (Invitrogen Inc., Carlsbad, Calif.). Reactions were carried out using 10 μl of extracted RNA and each primer at a concentration of 0.5 μM in a 25-μl (total volume) mixture by following the manufacturer's protocol. The thermal cycling conditions were as follows: 48°C for 30 min, 92°C for 2 min, and 35 cycles of 94°C for 30 s, 57°C for 45 s, and 72°C for 1 min followed by a final elongation step at 72°C. Next, 1.5 μl of the primary amplification product was used as the template in a nested PCR using the Expand High Fidelity PCR system (Roche Biochemicals, Mannheim, Germany) and primers NBEV-F and NBEV-R (Table 1) in a 50-μl (final volume) reaction mixture containing 1× reaction buffer, each deoxynucleoside triphosphate at a concentration of 0.4 mM, each primer at a concentration of 0.5 μM, and 0.875 U of Taq DNA polymerase. The thermal cycling procedure consisted of 92°C for 2 min, followed by 30 cycles of 94°C for 30 s, 57°C for 45 s, and 72°C for 1 min and then a final elongation step at 72°C for 10 min. Ten microliters of the 208-bp amplified product was visualized in 2% agarose gels after ethidium bromide staining. To avoid PCR contamination, general guidelines were strictly observed, and positive controls and specimens were never processed simultaneously. Amplified products were bidirectionally sequenced using inner primers described in Table 1 with a DyeDeoxy terminator cycle sequencing kit (Perkin-Elmer Applied Biosystems) and a 3100 DNA sequencer (Applied Biosystems). Electropherograms were analyzed using the Sequence Navigator software (Perkin-Elmer).
Sequence analysis.
Nucleotide sequences obtained in the present work, as well as some representative sequences previously obtained from cattle feces in the United States (14), all bovine enterovirus sequences from the 5′NCR deposited in the GenBank, either from BEV-1 isolates (SL305, BK-2577, VG-527, and BEV-1 prototype [accession numbers AF123432, AF123433, D00214, and AJ-250671, respectively]) or BEV-2 isolates (Bot-209, Bo261, RM2, and PS87M [accession numbers AJ250673, AJ250672, X79383, and X79368, respectively]), and a sheep sequence (accession number AJ250674), were aligned and compared with the CLUSTAL W 1.6 program (32). The levels of nucleotide identity and phylogenetic trees were analyzed with the PHYLIP (7) and MEGA (13) packages.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the GenBank database under accession numbers AY831684 to AY831718.
RESULTS
Sensitivity of the TaqMan assay.
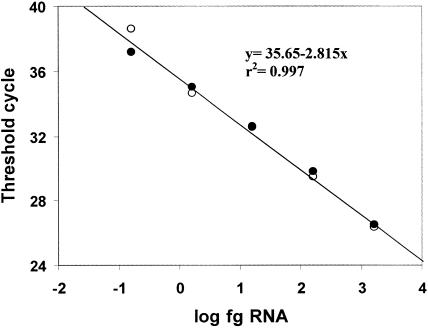
The sensitivity of the TaqMan assay was first evaluated by analyzing 10-fold dilutions of standard RNA obtained from a preparation of BEV-2 (isolate PS87; virus titer, 3.4 × 106 TCID50/ml) as described in Materials and Methods. As shown in Fig. 2, the assay was linear over a 5-log RNA concentration range. The method was able to detect as little as 0.156 fg of BEV RNA, which was equivalent to 11.6 RNA molecules per tube. Since this is the minimum amount of RNA that could be detected in 13 μl of a 60-μl RNA preparation obtained from a 0.14-ml sample, we concluded that this method can detect at least 5 fg (380 molecules) of RNA per ml of nonconcentrated sample. This figure corresponds to approximately 1.7 × 10−3 TCID50/ml, although it should be noted that enteroviruses can have up to 103 noninfectious particles per infectious particle.
FIG. 2.
Linearity and efficiency of the real-time RT-PCR assay for BEV detection. Serial 10-fold dilutions of BEV RNA were tested in duplicate. The data are the threshold cycle values calculated for each reaction and plotted against the quantity of RNA on a log scale. The open and solid circles represent data for duplicates. The threshold value was set at 0.03 fluorescence (delta) unit. The data are data from a representative example of one of the three experiments that produced essentially the same results.
Effectiveness of the concentration method.
To evaluate the effectiveness of the concentration method used for BEV detection in water samples, 2 liters of distilled water was independently spiked in duplicate with three concentrations of BEV-2 prototype strain PS87 as described above, and the viral RNA recovered after filtration and elution through electropositive filters was quantified by the real-time fluorogenic RT-PCR method. As shown in Table 2, the filtration-elution method resulted in a concentration factor of up to 32 in the most favorable conditions, which turned out to be the more diluted conditions (approximately 2 TCID50/ml). Higher concentrations, however, appeared to saturate the filter, since the amount of virus recovered remained almost constant, whereas the efficiency of virus recovery was drastically reduced. Bearing this in mind, the combination of concentration and fluorogenic RT-PCR described here allowed detection of viral concentrations equal to or greater than 12 BEV RNA molecules/ml of water sample.
TABLE 2.
Efficiency of the virus concentration method
| Virus input (ng)a | Virus recovered (ng/10 ml) | Efficiency of recovery (%) | Concn factor |
|---|---|---|---|
| 12.8 | 2.04 | 16 | 32 |
| 64 | 8.32 | 13 | 26 |
| 128 | 3.52 | 2.75 | 5.5 |
The RNA concentration was calculated, as described in Materials and Methods, for the three different amounts of BEV (4.6 × 103, 2.3 × 104, and 4.6 × 104 TCID50) that were individually added to 2 liters of water.
Survey of BEV in cattle herds.
Once the real-time RT-PCR procedure was optimized, we conducted a survey to determine the presence of BEV in bovine fecal samples (n = 100) collected in three different areas of Spain and obtained from different environments (Fig. 1 and Table 3). Area 1 is a region in which the population of cattle is high. In this area, two types of samples were analyzed, samples from intensive cattle farms (dairy cattle fed with fodder, with little or no grazing; obtained from 12 herds) and samples from extensive farms (meat cattle, with free grazing in mountain pastures; obtained from 10 herds). The remaining bovine samples studied were from areas where livestock is kept under extensive management (areas 2 and 3 in Fig. 1). A high prevalence (78 of 100 samples [78%]) of BEV was observed (Table 3), which supported the conclusion that BEV is highly endemic and widespread in the animals. However, the BEV prevalence was higher on dairy farms that were under intensive management, on which almost all samples were BEV positive (34 of 36 samples [94.4%]), than on extensive farms, on which the BEV prevalence was lower (44 of 64 samples [68.7%]). Nevertheless, for all cattle herds examined (n = 24) there was at least one BEV-positive sample, and in some herds all samples analyzed were BEV positive.
TABLE 3.
Results of the BEV survey of fecal samples using the real-time fluorogenic RT-PCR
| Area | Predominant species | No. of BEV-positive samples/total no. (%)
|
||||||
|---|---|---|---|---|---|---|---|---|
| Cattle
|
Sheep | Goats | Horses | Donkeys | ||||
| Intensive farming | Extensive farming | Total | ||||||
| 1 | Cattle | 34/36 (94) | 33/51 (65) | 67/87 (77) | 2/9 (22) | |||
| 2 | Cattle, sheep, and goats | 5/7 (71) | 5/7 (71) | 2/6 (33) | 0/4 (0) | |||
| 3 | Cattle and goats | 6/6 (100) | 6/6 (100) | 6/6 (100) | 3/5 (60) | 0/7 (0) | ||
| 4 | Sheep | 5/8 (63) | ||||||
| 5 | Horses | 3/5 (60) | ||||||
| Total | 34/36 (94) | 44/64 (69) | 78/100 (78) | 9/23 (39) | 6/10 (60) | 6/10 (60) | 0/7 (0) | |
Survey of BEV in water samples.
The usefulness of animal enteric viruses as markers of environmental pollution of surface waters has been described previously (8, 9, 14, 15, 15a, 16); therefore, in the present study water samples from the areas where animal fecal samples were collected were also examined for the presence of BEV sequences (Table 4). Our data indicated that 50% (8 of 16) of the water samples tested were BEV positive (7 of 10 samples from area 1, none of 5 samples from area 3, and the single sample from area 4). Four of the BEV-positive water samples from area 1 were collected from creeks, and one was collected from a swamp; all of the sampling sites were close to cattle farms on which the prevalence was high. The remaining two BEV-positive water samples were collected from a watering tank on a dairy farm. The only water sample collected from area 4 was also BEV positive. In this area, sheep were the only livestock observed, but it should be noted that this sample was obtained from a river that recently flooded due to heavy rain.
TABLE 4.
Real-time fluorogenic RT-PCR survey of BEV RNA in water samples collected in areas 1, 3, and 4a
| Sample | Source | BEVb |
|---|---|---|
| Wa1.1 | Watering tank | POS |
| Wa1.2 | Creek | POS |
| Wa1.3 | Creek | POS |
| Wa1.4 | Watering tank | POS |
| Wa1.5 | Swamp | POS |
| Wa1.6 | Watering tank | NEG |
| Wa1.7 | Creek | POS |
| Wa1.8 | Creek | POS |
| Wa1.9 | Creek | NEG |
| Wa1.10 | Creek | NEG |
| Wa3.1 | Pool | NEG |
| Wa3.2 | Pool | NEG |
| Wa3.3 | Pool | NEG |
| Wa3.4 | Pool | NEG |
| Wa3.5 | Reservoir | NEG |
| Wa4.1 | River | POS |
See Fig. 1.
POS, positive; NEG, negative.
Survey of BEV in other animal species.
While sampling bovine feces for the BEV survey, since we previously described the presence of BEV-like sequences in deer sharing cattle pastures (14), we also collected fecal samples from other animal species grazing or not grazing in the areas with cattle in order to evaluate the possible presence of BEV-like sequences in these animal feces. A total of 50 fecal samples from sheep, goats, horses, and donkeys collected in the five areas studied were analyzed (Fig. 1 and Table 3). Nine of the 23 (39.1%) sheep samples tested were positive as determined by the fluorogenic real-time RT-PCR assay, and the values were similar to those obtained with cattle feces. These samples were obtained from two areas (areas 1 and 2) where cattle were the major livestock species and from one area (area 4) where sheep were the only ruminants grazing. Additional analysis of 10 fecal samples from goats resulted in detection of BEV-like sequences in 6 of 10 (60%) samples tested, but while all samples from a herd in area 3 were positive, none of the four samples collected from a herd in area 2 gave positive results. Finally, the presence of BEV-like sequences was assessed in 17 equine fecal samples. Our results showed that while none of the seven donkey samples tested from area 3 were positive, BEV-like sequences were detected in 60% of the horse samples collected in areas 3 (three of five samples) and 5 (three of five samples).
Sequence and phylogenetic analysis.
Representative samples of BEV-positive feces (from 13 cattle, eight sheep, three goats, and five horses) and water (n = 6) from the five areas studied were amplified by a nested RT-PCR of the 5′NCR as described in Materials and Methods. Nucleotide sequence comparison with previously sequenced isolates (14) and all available GenBank BEV sequences indicated that, with few exceptions (see below), the sequences analyzed were genetically more closely related to BEV-2 sequences than to BEV-1 sequences. The mean level of nucleotide sequence identity with BEV-2 sequences retrieved from the GenBank database was 85.3%, while it was 76.2% with BEV-1 sequences, a value equal to that obtained when previously published BEV-1 and BEV-2 sequences were compared (76.2%). The relatively high genetic heterogeneity observed among the 5′NCR of BEV isolates from Spain (86% nucleotide identity) was similar to that recorded for BEV-2 GenBank isolates (86.7%). The only exceptions were the three BEV-like sequences amplified from goat feces and one of the ovine isolates (Ov2.3) that exhibited similar levels of homology with the remaining Spanish isolates and the BEV-2 or BEV-1 GenBank sequences (79.1%, 78.7%, and 76.2%, respectively).
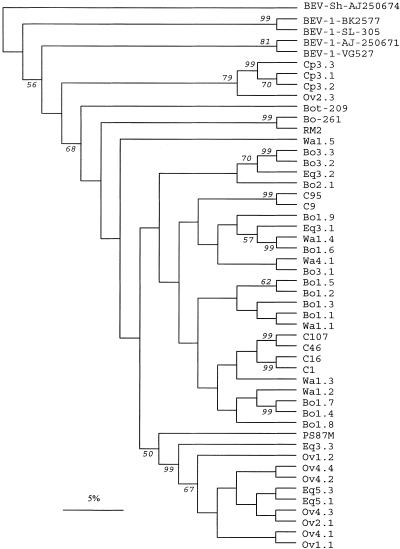
Phylogenetic analysis (Fig. 3) confirmed the sequence comparison data. All samples grouped with BEV-2 variants, except the cluster of goat sequences that seemed to segregate from both BEV-1 and BEV-2 strains. The heterogeneity found among bovine sequences was quite high, and so was the heterogeneity of the equine samples analyzed. On the other hand, with one exception, all ovine fecal samples had almost the same sequence and clustered together. Notably, this cluster included most of the amplified sequences from sheep and horse feces but none of the sequences amplified from bovine samples.
FIG. 3.
Phylogenetic tree of the amplified BEV 5′NCR of isolates used in the present study. Previously published BEV sequences from cattle (C) feces obtained in the United States (14), BEV-1 isolates SL305 (GenBank accession number AF123433), BK-2577 (AF123433), and VG-527 (D00214), a BEV-1 prototype (AJ250671), and BEV-2 isolates Bot-209 (AJ250673), Bo261 (AJ250672), RM2 (X79383), and PS87 (X79368), as well as the only available sheep sequence (AJ250674), were included in the tree for comparison. The designations for cattle (beginning with Bo), sheep (Ov), goat (Cp), horse (Eq), and water (Wa) samples used in the present study include two digits; the first digit indicates the sampling area (areas 1 to 5 in Fig. 1), and the second digit indicates the sample number. The numbers at the nodes indicate the bootstrap values (expressed as percentages) that support the adjacent nodes based on 100 resampling iterations. Only bootstrap values higher than 50% are shown. Scale bar = 5% nucleotide sequence divergence.
Our data also indicated that the heterogeneity observed was not directly related to the geographic origin of the BEV strains circulating in Spain (Fig. 3). Some contemporary bovine isolates from the same herd had relatively divergent sequences (e.g., the Bo1.7 and Bo1.9 sequences, with 85.4% nucleotide similarity), while bovine isolates collected in areas separated by hundreds of kilometers were genetically more closely related (e.g., the Bo1.3 and Bo2.1 sequences, with 92.7% nucleotide similarity). Therefore, our data suggest that many BEV variants cocirculate in cattle around Spain. Likewise, while the two sequences obtained from horse feces in area 5 (Eq5.1 and Eq5.3) were very similar to each other, sequences obtained from area 3 (Eq 3.1, Eq3.2, and Eq3.3) were more divergent (Fig. 3). In contrast, all of the sequences except one (Ov2.3) from sheep feces were almost identical, independent of the area where the samples were collected (area 1, 2, or 4), as were the BEV-like sequences obtained from goat feces, although in this case it should be noted that all samples came from a single herd.
Analysis of water samples showed that most of the amplified sequences were unique. Although these sequences were genetically closely related to BEV sequences from feces of cattle pasturing in the same area, they did not exactly match any of them. Water samples were collected mainly from creeks and swamps shared by different herds, and thus, the lack of sequence identity probably reflected the heterogeneity found among BEV sequences amplified from cattle feces from the same areas. The only exception was a water sample collected from a watering tank (Wa1.4) that had a sequence identical to that found in a bovine fecal sample from the same farm (Bo1.6).
DISCUSSION
In the present study, we optimized a real-time RT-PCR assay able to detect 5 fg (380 molecules, equivalent to approximately 1.7 × 10−3 TCID50/ml) of BEV RNA per ml of nonconcentrated sample. A combination of this methodology with a previous concentration step with electropositive filters, which concentrated the sample up to 32-fold, allowed detection of as little as 12 BEV RNA molecules/ml of water tested.
Once the BEV detection procedure was optimized, it was applied to a survey study of BEV in biological and environmental samples collected around Spain in areas with intensive or extensive farming of different animal species. Our results indicated that, as in other regions of the world (2, 14, 17, 36), BEV is widespread in cattle herds around Spain. Up to 78% (78 of 100) of the bovine fecal samples tested were BEV positive as determined by the fluorogenic real-time RT-PCR assay. Nevertheless, as expected, BEV was less prevalent in cattle from extensive farms (69%) than in cattle from intensive farms (94%), even though all 24 herds tested had at least one BEV-positive sample, stressing the endemicity of BEV infection in Spain.
BEV is very stable in the environment, and thus, it has been proposed that this virus could be a useful indicator of water and environmental contamination (14), as well as a potential surrogate for other highly infectious picornaviruses, such as foot-and-mouth disease virus, in order to optimize extraction and/or detection procedures for contamination of the environment (8, 34). The first description of the genetic relationship between simultaneously collected water and fecal samples from a single closed herd showed that in most cases, the sequences detected in water samples were quite similar to those present in feces (14). In the present study, the BEV sequences detected in water samples were also quite heterogeneous and closely related to those amplified from bovine samples. In only one case did the BEV sequence amplified from a water sample (Wa1.4) obtained from a watering tank on a farm have the same sequence as one of the fecal bovine samples (Bo1.6) simultaneously collected on the same farm. Since most water samples were obtained from sources shared by different herds, the lack of sequence homology between water and bovine samples may be explained by the relatively high heterogeneity found in the bovine samples. Recent data (8) describing the detection of BEV RNA in water samples at a time when no BEV RNA was amplified from cattle feces obtained in the same area suggest that the virus may persist for long time in the environment and/or may be shed by a few animals of the herd. Actually, while we detected BEV RNA in 70% of the water samples from area 1, in which the population of cattle is high and where intensive farming is common, none of the water samples collected in area 3, a predominant extensive farming region, gave positive results. Thus, as expected, our data support the finding that the greater the concentration of animals that share pastures and drinking sources, the higher the risk of BEV environmental contamination and, consequently, the greater the possible usefulness of BEV RNA detection for tracing animal viral contamination.
Enteroviruses are shed in feces, and consequently, they are spread through contaminated soil and water. Therefore, different animal species grazing in the same pastures and/or drinking from the same water sources are likely to be infected by similar virus variants. However, little information is available regarding the presence of BEV-like viruses in other ruminants. Recently, amplification of BEV-like sequences from deer feces collected in an area where there was cattle pasturing was described (14). In the present work, analysis of feces from animals other than cattle that were grazing in the areas under study revealed that the prevalence of BEV-like sequences in feces from horses, goats, and sheep was relatively high (39 to 60%). At this point, it cannot be clearly established whether these animals are susceptible to BEV-like virus infection or just mechanical carriers of the virus. The fact that the amplification values obtained when these animal feces were tested by real-time RT-PCR were similar to those obtained with cattle feces and the fact that some samples were obtained in areas where no cattle farming was observed suggest that the animals are not just mechanical carriers. In any case, experiments to fully characterize these BEV-like isolates are currently under way.
To gain further insight into the genetic relationship among the different BEV-positive samples detected, nucleotide and phylogenetic analyses of the viral 5′NCR were performed. The 5′NCR of all picornaviruses is directly implicated in viral translation and genome replication, having conserved regions that maintain the secondary structures, as well as variable regions that allow genetic classification (24, 29, 31, 35). In fact, nucleotide changes in this region that interfere with RNA folding and/or RNA-protein interactions may have drastic consequences in cell tropism (27, 35) and pathogenicity (21). Our data indicate that with a few exceptions, the sequences in most of the samples analyzed were more closely related to BEV-2 sequences than to BEV-1 sequences. Differences affecting the folding pattern of the two BEV types may be the result of viral evolution (35), and thus, they may have important biological consequences.
Even though there is little information regarding BEV sequence homology, our data confirmed the previously described significant sequence variation in the 5′NCR found among BEV isolates (8, 14). This variation is similar to that described for other enteroviruses that exhibit levels of identity between isolates ranging from 70 to 96% (28). In our study, none of the sequences were identical to any of the previously described sequences, including the three main groups of sequences reported for BEV isolates from the United States (14). Therefore, our data support the hypothesis that many different BEV populations coexist. Moreover, different BEV sequences were detected in cattle feces from the same herds, while genetically more closely related variants were amplified from feces collected in herds from very distant farms.
The BEV-like variants found in feces from deer were very similar to those detected in cattle feces from the same closed area (14). In the present study, nucleotide and phylogenetic analyses of fecal samples from animals other than cattle suggested the presence of species-specific BEV-like sequences, so that sequences obtained from goat feces were very similar to each other but clearly different from the rest of the sequences analyzed. In any case, additional analysis of BEV-like variants found in bovine and nonbovine feces are needed to clarify whether there are species-specific variants and what the actual degree of genetic variation found among different isolates around the world is.
In conclusion, we optimized a highly sensitive BEV RNA detection procedure that, by combining a simple water concentration step involving electropositive filters with a real-time RT-PCR, allows detection of as little as 12 BEV RNA molecules/ml of water tested. This method may be useful for the detection of environmental pollution by animal viruses, and it allowed a clear demonstration that BEV infection in cattle is widespread in Spain. In addition, BEV-like sequences were amplified from feces of sheep, goats, and horses, which in some instances were collected in areas where these animals were the only livestock pasturing. Thus, the specificity of the method described here, when applied to the detection of animal fecal contamination of environmental water, should be extended to animals other than cattle. Finally, sequencing and phylogenetic analysis highlighted the relatively high nucleotide variation found in the 5′NCR of BEV isolates and suggested that there are species-specific BEV-like variants.
Acknowledgments
This work was supported in part by grants from CICYT (grant RTA02-35) and Comunidad Autónoma de Madrid (grant 07 M/0023/2002) to J.C.S. N.G was support by the Comunidad Autónoma de Madrid, and L.C. was supported by a scholarship from INIA.
We are indebted to S. Goens and M. Purdue for sharing their results with us before publication.
REFERENCES
- 1.Abbaszadegan, M., P. Stewart, and M. Le Chevallier. 1999. A strategy for detection of viruses in ground water by PCR. Appl. Environ. Microbiol. 65:444-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, A. A. 1978. Cross reaction between enterovirus and South African Territories 15 foot-and-mouth disease virus. Am. J. Vet. Res. 39:59-63. [PubMed] [Google Scholar]
- 3.Bergh, O., K. Y. Borsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in the aquatic environment. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 4.Bosch, A. 1998. Human enteric viruses in the water environment: a minireview. Int. Microbiol. 1:191-196. [PubMed] [Google Scholar]
- 5.Chapron, C. D., N. A. Ballester, J. H. Fontaine, C. N. Frades, and A. B. Margolin. 2000. Detection of astroviruses, enteroviruses, and adenovirus types 40 and 41 in surface waters collected and evaluated by the information collection rule and an integrated cell culture-nested PCR procedure. Appl. Environ. Microbiol. 66:2520-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clesceri, L. S., A. E. Greenberg, and A. D. Eaton (ed.). 1998. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, D.C.
- 7.Felsenstein, J. 1993. PHYLIP (phylogeny inference package version 3.5c). Department of Genetics, University of Washington, Seattle. (Distributed by the author.)
- 8.Goens, S. D., S. Botero, A. Zemla, E. Zhou, and M. L. Perdue. 2004. Bovine enterovirus type 2. Complete genomic sequence and molecular modeling of the reference strain and wild type isolate from endemically infected US cattle. J. Gen. Virol. 85:3195-3203. [DOI] [PubMed] [Google Scholar]
- 9.Jimenez-Clavero, M. A., C. Fernandez, J. A. Ortiz, J. Pro, G. Carbonell, J. V. Tarazona, N. Roblas, and V. Ley. 2003. Teschoviruses as indicators of fecal contamination of water. Appl. Environ. Microbiol. 69:6311-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katzenelson, E., B. Fattal, and T. Hostovestky. 1976. Organic flocculation: an efficient second step concentration method for the detection of viruses in tap water. Appl. Environ. Microbiol. 32:638-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King, A. M. Q., F. Brown, P Christian, T. Hovi, T. Hyypiä, N. J. Knowles, S. M. Lemon, P. D. Minor, A. C. Palmemberg, T. Skern, and G. Stanway. 2000. Picornaviridae, p. 657-673. In M. H. V. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, C. H. Calisher, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, New York, N.Y.
- 12.Knowles, N. J., and I. T. Barnett. 1985. A serological classification of bovine enteroviruses. Arch. Virol. 83:141-155. [DOI] [PubMed] [Google Scholar]
- 13.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 14.Ley, V., J. Higgins, and R. Fayer. 2002. Bovine enteroviruses as indicators of fecal contamination. Appl. Environ. Microbiol. 68:3455-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ley, V., and M. A. Jimenez-Clavero. March. 2004. Method to identify environmental contamination by detecting enteric viruses. U.S. patent application 2004/0058312 A1.
- 15a.Ley, V., and M. A. Jimenez-Clavero. February. 2004. Method to identify environmental contamination by detecting enteric viruses. European patent publication EP 336 663 A3 (application 03380032.7).
- 16.Maluquer de Motes, C., P. Clemente-Casares, A. Hundesa, M. Martin, and R. Girones. 2004. Detection of bovine and porcine adenoviruses for tracing the source of fecal contamination. Appl. Environ. Microbiol. 70:1448-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarthy, F. M., G. A. Smith, and J. S. Mattick. 1999. Molecular characterization of Australian bovine enteroviruses. Vet. Microbiol. 68:71-81. [DOI] [PubMed] [Google Scholar]
- 18.McNally, R. M., J. A. Earle, M. McIlhatton, E. M. Hoey, and S. J. Martin. 1994. The nucleotide sequence of the 5′ non-coding and capsid coding regions of two bovine enterovirus strains. Arch. Virol. 139:287-299. [DOI] [PubMed] [Google Scholar]
- 19.Mehnert, D. U., K. E. Stewien, C. M. Harsi, A. P. Queiroz, J. M. Candeias, and J. A. Candeias. 1997. Detection of rotavirus in sewage and creek water: efficiency of the concentration method. Mem. Inst. Oswaldo Cruz 92:97-100. [DOI] [PubMed] [Google Scholar]
- 20.Mendez, I. I., I. I. Hermann, P. R. Hazelton, and K. M. Coombs. 2000. A comparative analysis of Freon substitutes in the purification of reovirus and calicivirus. J. Virol. Methods 90:59-67. [DOI] [PubMed] [Google Scholar]
- 21.Minor, P. D. 1992. The molecular biology of polio vaccines. J. Gen. Virol. 73:3065-3077. [DOI] [PubMed] [Google Scholar]
- 22.Pallanch, M. A., and R. P. Ross. 2001. Enteroviruses: polioviruses, coxsackie, echoviruses and newer enteroviruses, p. 723-775. In D. Knipe and P. Howley (ed.), Fields virology, 4th ed. Williams and Wilkins Publishers, Baltimore, Md.
- 23.Pianetti, A., W. Baffone, B. Citterio, A. Casaroli, F. Bruscolini, and L. Salvaggio. 2000. Presence of enteroviruses and reoviruses in the waters of the Italian coast of the Adriatic Sea. Epidemiol. Infect. 125:455-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilipenko, E. V., V. M. Blinov, L. I. Romanova, A. N. Sinyakov, S. V. Maslova, and V. I. Agol. 1989. Conserved structural domains in the 5′-untranslated region of picornaviral genomes: an analysis of the segment controlling translation and neurovirulence. Virology 168:201-209. [DOI] [PubMed] [Google Scholar]
- 25.Schvoerer, E., F. Bonnet, V. Dubois, G. Cazaux, R. Serceau, H. J. Fleury, and M. E. Lafon. 2000. PCR detection of human enteric viruses in bathing areas, waste waters and human stools in southwestern France. Res. Microbiol. 151:693-701. [DOI] [PubMed] [Google Scholar]
- 26.Schvoerer, E., M. Ventura, O. Dubos, G. Cazaux, R. Serceau, N. Gournier, V. Dubois, P. Caminade, H. J. Fleury, and M. E. Lafon. 2001. Qualitative and quantitative molecular detection of enteroviruses in water from bathing areas and from a sewage treatment plant. Res. Microbiol. 152:179-186. [DOI] [PubMed] [Google Scholar]
- 27.Shiroki, K., T. Ishii, T. Aoki, Y. Ota, W.-X. Yang, T. Komatsu, Y. Ami, M. Arita, S. Abe, S. Hashizume, and A. Nomoto. 1997. Host range phenotype induced by mutations in the internal ribosomal entry site of poliovirus RNA. J. Virol. 71:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siafakas, N., P. Markoulatos, and G. Stanway. 2002. Molecular classification of coxsackie A viruses on the basis of the 5′-UTR: structural and evolutionary aspects. J. Mol. Evol. 55:638-652. [DOI] [PubMed] [Google Scholar]
- 29.Skinner, M. A., V. R. Racaniello, G. Dunn, J. Cooper, P. D. Minor, and J. W. Almond. 1989. New model for the secondary structure of the 5′ non-coding RNA of poliovirus is supported by biochemical and genetic data that alsoshow that RNA secondary structure is important in neurovirulence. J. Mol. Biol. 207:379-392. [DOI] [PubMed] [Google Scholar]
- 30.Sobsey, M. D., and B. L. Jones. 1979. Concentration of poliovirus from tap water using positively charged microporous filters. Appl. Environ. Microbiol. 37:588-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanway, G., T. Hovi, N. J. Knowles, and T. Hyypiä. 2002. Molecular and biological basis of picornavirus taxonomy, p. 17-24. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 32.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyn-Jones, A. P., and J. Sellwood. 2001. Enteric viruses in the aquatic environment. J. Appl. Microbiol. 91:945-962. [DOI] [PubMed] [Google Scholar]
- 34.Yilmaz, A., and E. F. Kaleta. 2003. Evaluation of virucidal activity of three commercial disinfectants and formic acid using bovine enterovirus type 1 (ECBO virus), mammalian orthoreovirus type 1 and bovine adenovirus type 1. Vet. J. 166:67-78. [DOI] [PubMed] [Google Scholar]
- 35.Zell, R., and A. Stelzner. 1997. Application of genome sequence information to the classification of bovine enteroviruses: the importance of the 5′- and 3′-nontranslated regions. Virus Res. 51:213-229. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, A. Q., J. R. Smith, and G. W. Burgess. 1990. Blocking enzyme-linked immunosorbent assay for detection of antibodies against bovine enterovirus. Vet. Microbiol. 21:275-281. [DOI] [PubMed] [Google Scholar]