Abstract
An in situ mesocosm system was designed to monitor the in situ dynamics of the microbial community in polluted aquifers. The mesocosm system consists of a permeable membrane pocket filled with aquifer material and placed within a polypropylene holder, which is inserted below groundwater level in a monitoring well. After a specific time period, the microcosm is recovered from the well and its bacterial community is analyzed. Using this system, we examined the effect of benzene, toluene, ethylbenzene, and xylene (BTEX) contamination on the response of an aquifer bacterial community by denaturing gradient gel electrophoresis analysis of PCR-amplified 16S rRNA genes and PCR detection of BTEX degradation genes. Mesocosms were filled with nonsterile or sterile aquifer material derived from an uncontaminated area and positioned in a well located in either the uncontaminated area or a nearby contaminated area. In the contaminated area, the bacterial community in the microcosms rapidly evolved into a stable community identical to that in the adjacent aquifer but different from that in the uncontaminated area. At the contaminated location, bacteria with tmoA- and xylM/xylE1-like BTEX catabolic genotypes colonized the aquifer, while at the uncontaminated location only tmoA-like genotypes were detected. The communities in the mesocosms and in the aquifer adjacent to the wells in the contaminated area consisted mainly of Proteobacteria. At the uncontaminated location, Actinobacteria and Proteobacteria were found. Our results indicate that communities with long-term stability in their structures follow the contamination plume and rapidly colonize downstream areas upon contamination.
Benzene, toluene, ethylbenzene, and the xylenes (BTEX) form one of the main groundwater contaminant groups. In situ bioremediation, either active or passive, is increasingly being applied as a technology for the treatment of BTEX-contaminated aquifers. BTEX are biodegradable under aerobic and anaerobic conditions, and the technologies involved are becoming more reliable and accepted by the public (12, 28, 35). Passive in situ bioremediation consists of monitoring natural bioattenuation, i.e., the biological processes that act without human intervention to reduce the mass and toxicity of the contaminants in the aquifer. Active in situ bioremediation consists of stimulation of the local microbial activity by the addition of nutrients and/or electron acceptors (12). In both cases, an understanding of the response of the indigenous microbial community to naturally occurring or human-implemented changes in environmental conditions is of interest. Laboratory microcosm studies provide insight into how a microbial community in the aquifer responds to specific treatments and help in the development of hypotheses about the behavior and functioning of the community under these treatments, but ultimately, the field relevance of a laboratory study must be compared to field data (20). However, studies of aquifer microbiology in the field are hampered by (i) the reproducibility of soil/aquifer sampling under in situ conditions due to soil heterogeneity and (ii) the labor-intensive nature and high cost of core drilling. Therefore, the use of partially enclosed, easily sampled, outdoor experimental model ecosystems or in situ mesocosms, which allow the exchange of matter and energy, including nutrients, pollutants, and biota, with the surrounding environment, is recommended. Recently, different in situ mesocosm systems have been reported for aquifer studies, but they were mainly designed to monitor in situ physicochemical parameters (for a review, see reference 29). Recent advances such as nucleic acid-based fingerprinting techniques allow for rapid assessments of changes in the structure of a complex microbial community (24). However, the development and use of in situ mesocosms for the study of microbial community dynamics in aquifers by means of molecular techniques have not been reported, and most data have been obtained from arduous full-scale field studies.
For this study, an in situ mesocosm system was designed to monitor the response of a bacterial aquifer community upon contact with a BTEX-contaminated groundwater plume. This is a typical situation found in the field when a migrating BTEX plume reaches the uncontaminated area downstream of the aquifer. The mesocosm system consisted of a membrane pocket filled with aquifer material and contained within a polypropylene holder. Identically treated mesocosms were installed into a monitoring well, and after each specified incubation period, one microcosm was recovered, DNAs were extracted from its contents, and the microbial community structure was studied by the use of molecular tools. Mesocosms filled with aquifer material from an uncontaminated area were inserted into wells in the contaminated area of the same site, and the in situ dynamics of the oligotrophic aquifer community and the presence of specific BTEX-catabolic genotypes within this community were monitored.
MATERIALS AND METHODS
Membrane materials for mesocosm manufacturing.
Membrane materials were purchased from Solana N.V., Schoten, Belgium, and included polypropylene (PP), stainless steel (RVS), ethylene-tetrafluorethylene (FK1), polyester, and polyamide (PA) with different mesh widths and open surfaces (Table 1).
TABLE 1.
Characteristics of different membrane types used for the manufacturing of the in situ mesocosm systema
| Membrane type | Mesh width (μm) | % Open surface | Diffusion rate (t95%) of 3CBA (h)b | Mass transport coefficient (K) (liters h−1) of 3CBA | Diffusion rate (t95%) of Zn (h)b | Mass transport coefficient (K) (liters h−1) of Zn | Time (h) for equilibrium migration of one bacterial strain | Time (h) for equilibrium migration of a bacterial population |
|---|---|---|---|---|---|---|---|---|
| 72 RVS 25/25 | 25 | 25 | 12 | 0.07 | 7.1 | 0.13 | ND | ND |
| 72 RVS 140/32 | 140 | 32 | 4 | 0.22 | ND | ND | ND | ND |
| 49 PA 1/1 | 1 | 0.8 | 140 | 0.01 | ND | ND | ND | ND |
| 49 PA 6/5 | 6 | 5 | 55 | 0.08 | 35.5 | 0.03 | 25 | 25 |
| 49 PA 15/10 | 15 | 10 | 40 | 0.02 | 25.8 | 0.05 | 5 | ND |
| 49 PA 25/14 | 25 | 14 | 22 | 0.04 | ND | ND | ND | ND |
None of the membranes showed sorption of BTEX. Other membranes tested for sorption included polypropylene, ethylene-tetrafluorethylene, and polyester membranes with different mesh widths and amounts of open surface. None of those materials showed significant sorption. ND, not determined.
Number of hours after which 95% of the equilibrium (when the contaminant concentrations in both chambers were the same) was reached.
Sorption of BTEX to membrane materials.
Duplicate vials were filled with 5 g of sterile H2O containing 7.5 μg ml−1 of each BTEX compound, and the membrane material to be tested (1 cm2) was inserted. The vials were closed with Teflon-lined butyl septa and crimp caps and left at room temperature while they were regularly turned by hand. After 1 week, 1 ml of the aqueous phase was injected into 4 ml of H2O. After spiking the sample with 7.4 μl of an internal standard (d10-ethylbenzene [20 ng μl−1]), we measured the BTEX concentrations by headspace analysis on a MEGA 8000 TOP Series gas chromatograph (Thermoquest CE Instruments, Rodano-Milan, Italy), as described previously (9).
Diffusion of contaminants and migration of bacteria through membranes.
The diffusion of 3-chlorobenzoate (3CBA) and Zn through membranes was examined with a diffusion cell consisting of two rectangular 600-ml chambers separated by a test membrane as described previously (36). Both chambers were filled simultaneously, one with 600 ml of sterile water and the other with 600 ml of a 3 mM 3CBA or 3 mM ZnCl2 solution in water. The 3CBA and Zn concentrations in both chambers were measured over time by UV spectrometry (Ultraspec 3000, Novaspec II; Pharmacia Biotech, Cambridge, England) at 240 nm and plasma-emission spectrometry (Jarrell-Ash Atomcomp model 750; Thermo Optek, Brussels, Belgium) at 213.8 nm, respectively. The mass transport coefficient K and the diffusion rates of 3CBA and Zn, described as t95%, were calculated with the following equations: S1(t) = S1(0)/2 × [1 + exp(−2Kt95%/V)] and t95% = −ln 0.05V/2K, in which S1(t) is the 3CBA or Zn concentration in chamber 1 (mM liter−1) at time t (h), K is the mass transport coefficient (liters h−1), t95% is the time at which 95% of the equilibrium in the exponential diffusion profile is reached (h), and V is the volume of one chamber (liters). K was calculated by fitting the equation through the measured concentration values by means of nonlinear regression (36).
The migration of Pseudomonas putida P11, a ds-red-labeled derivative of P. putida mt-2 (PaW1) (45), through the membranes was studied with the same diffusion cell. Both chambers were filled with 600 ml of 10 mM MgSO4, and one chamber was inoculated with strain P11 at an A600 of 1. The concentration of strain P11 in both chambers was monitored over time by plating diluted samples on Tris minimal medium (30) containing 3 mM benzoate, 35 μg liter−1 nalidixic acid, and 25 μg liter−1 gentamicin. To study the migration of a bacterial aquifer community through the membrane, we suspended 60 g of aquifer material in 600 ml of 10 mM MgSO4 by stirring for 2 h at room temperature. After 2 h of settling of the soil particles, the suspension was transferred to the right chamber of the diffusion cell while the same volume of MgSO4 was poured into the left chamber. Eleven-milliliter samples were regularly removed from both chambers. Ten milliliters of each sample was centrifuged for 15 min at 3,000 rpm, and the pellet was frozen at −80°C for DNA extraction. From the residual 1 ml, a dilution series was plated on 0.1× 869 rich medium (30) for total CFU counting and on Tris minimal medium exposed to BTEX vapors to count the BTEX-degrading CFU. Both media were supplemented with 200 μg ml−1 cycloheximide. For growth in the presence of BTEX, the Tris minimal medium agar plates were incubated in closed 10-liter buckets in which the BTEX compounds were distributed via the gas phase from a vial containing 31.2 mg liter−1 benzene, 9.2 mg liter−1 toluene, and 1.1 mg liter−1 each of m-, p-, and o-xylene (Janssen Chemica, Beerse, Belgium) in 49.3 ml of vacuum oil (T.C.P.S. NV, Werchter, Belgium). The CFU were counted after 1 week of aerobic incubation in the dark at 30°C.
Mesocosm design and field implementation.
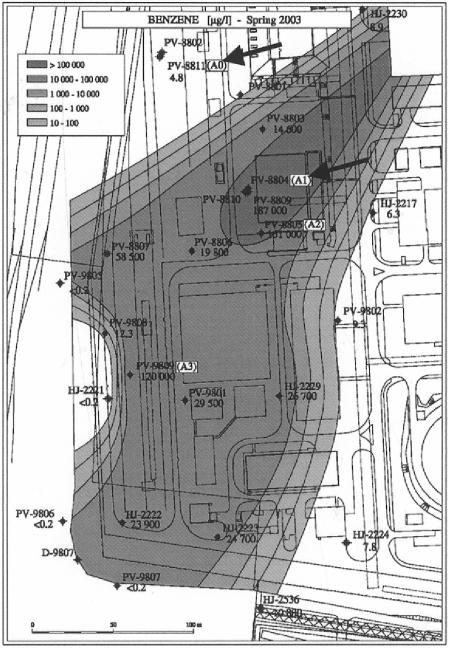
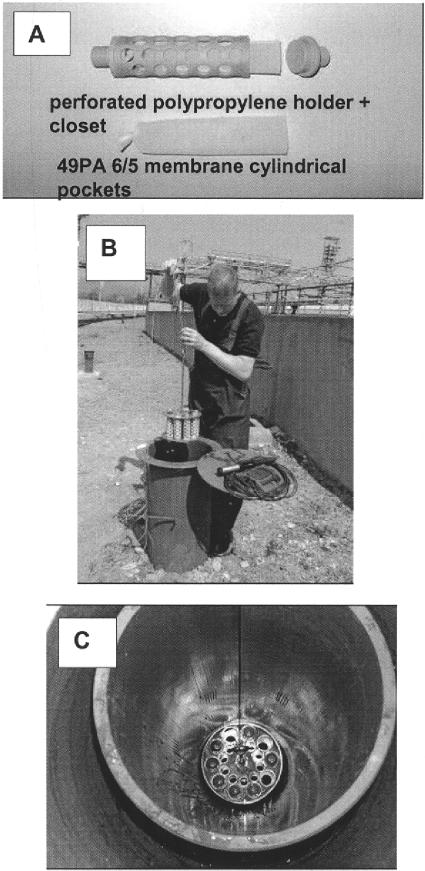
Figure 1 shows a map of the BTEX-contaminated site used for aquifer sampling and implementation of the in situ mesocosms. The site is an oil refinery site, situated in Northern Bohemia (Czech Republic), containing a BTEX (mainly benzene)-contaminated groundwater plume. The map shows the plume and the locations of sampling and monitoring wells of relevance. An aquifer sample (A0Z) was taken by core drilling with minimal heating of the core from the uncontaminated location A0 at a depth of 2.8 m on 11 December 2002. The soil was directly transferred from the boring equipment to plastic bags, and stones and large soil particles were removed under anaerobic conditions. The bags were sealed within a second plastic bag and stored at 4°C prior to use. Cylindrical membrane pockets were made from rectangular pieces (11 cm by 18 cm) of polyamide membrane 49 PA 6/5 and were closed at one side with a polyamide tie (Fig. 2A). Several identical pockets were filled in an anaerobic chamber with either a nonsterile or sterile A0Z aquifer sample. The sterile A0Z aquifer sample was obtained by autoclaving the nonsterile aquifer sample twice at 120°C for 50 min in a Falcone autoclave (LTE Scientific LTD, Greenfield, Oldham, Great Britain). After each pocket was filled, the other side of the pocket was closed. The pockets were inserted into polypropylene holders with a length of 12.5 cm, an outer diameter of 4 cm, and an inner diameter of 3.2 cm that were perforated with 44 holes of 1.3 cm in diameter (Fig. 2A). On both sides, the holders were closed with a cylindrical closet of 0.5-cm thickness by means of a stainless steel pin. On 9 January 2003, mesocosms with nonsterile and sterile material were positioned under the groundwater table in monitoring wells PV-8809 and PV-8810, respectively, both of which are located in the contaminated area A1. In addition, mesocosms containing nonsterile material were installed in monitoring well PV-8811, located in the uncontaminated A0 area. The wells had an inner diameter of 25 cm, and several mesocosms were simultaneously implemented in the wells by use of a device consisting of a stainless steel screw with two perforated stainless steel plates of 23 cm in diameter. Each plate contained 10 holes (4.1-cm diameter in the upper plate and 3.0-cm diameter in the bottom plate) to hold the mesocosms (Fig. 2B). The top of the screw was attached to a stainless steel cable to insert the system into and recover it from the well. At specified times, the mesocosms were lifted together from their in situ positions, and one mesocosm was transported to the laboratory on dry ice while the rest were directly reinserted into the well. Samples were taken for plating, while the rest of the sample was immediately stored at −80°C. The frozen aquifer in the membrane was crushed, and after mixing with a sterile spatula, divided into Falcon tubes (2 g of frozen aquifer) and stored at −80°C prior to DNA extraction. On 20 May 2003, fresh aquifer material was sampled at locations A1 (A1Z material) and A0 by drilling just adjacent to the wells containing the microcosms.
FIG. 1.
Map of BTEX (mainly benzene)-contaminated site in the Czech Republic showing the benzene contamination plume and indicating relevant sampling locations and monitoring wells. The arrows indicate the locations of A0 and A1. Numbers indicated with PV-, D-, and HJ- show the positions of groundwater monitoring wells, with the number below reporting the groundwater benzene concentration measured in that well in March 2003.
FIG. 2.
(A) In situ mesocosm system consisting of a perforated polypropylene holder and a permeable polyamide membrane (49 PA 6/5) pocket. (B and C) Implementation of mesocosms filled with aquifer into monitoring well PV8809.
Monitoring of groundwater characteristics.
Groundwater samples were regularly taken from monitoring wells. BTEX concentrations in the groundwater were determined by the modified US EPA 8260 method. The NO32− and SO42− concentrations were measured by method CSN EN ISO 10304, and the concentrations of NH4+ and PO42− were measured by the modified CSN ISO 7150-1 and CSN EN ISO 1189 methods, respectively. The groundwater O2 concentration, temperature, redox, and pH were measured in situ by use of a multimeter (pH/Oxi 340i; WTW, Weilheim, Germany). In situ measurements of the conductivity were performed by use of a conductivity meter (LF 320; WTW, Weilheim, Germany). The chemical oxygen demand was determined by means of the semimicro method HACH, TNV 75 7520, and the dissolved organic carbon was measured with a special analyzer (total carbon monitor TCM 480; Carlo Erba Instruments). The groundwater level was measured continuously by use of an interface meter (IM-1; Geotechnical Instruments, Warwickshire, United Kingdom).
DNA extraction from soil.
DNAs were extracted from 2 g of soil by use of the following protocol, which was modified from the work of El-Fantroussi et al. (10). Soil was suspended in 4 ml of Tris-glycerol buffer (10 mM Tris, 15% glycerol, pH 7), and cells were lysed by mechanical disruption by glass beads (0.10 to 0.11 mm) for 30 s (0°C) in a bead beater apparatus (B. Braun Biotech International GmbH, Melsungen, Germany) followed by enzymatic lysis with lysozyme (2 mg ml−1; 30 min at 37°C). Sodium dodecyl sulfate and proteinase K were added to final concentrations of 0.6% (wt/vol) and 0.1 mg ml−1, respectively, and the vials were incubated for 30 min at 50°C. Two milliliters of a NaKPO4 buffer consisting of 5.34 g Na2HPO42H2O and 4.08 g KH2PO4 in 500 ml H2O adjusted to pH 8.0 was added. The vials were placed on ice and subjected to a second bead-beating step (30 s). Glass beads, soil, and cell debris were removed by centrifugation (3 min at 18,000 × g), and the crude DNA extract was further purified by a single extraction with phenol-chloroform-isoamyl alcohol (25:24:1). One gram of acid-washed polyvinylpolypyrrolidone (Sigma-Aldrich NV/SA, Bornem, Belgium) was added to the DNA solution, and after incubation for 30 min on ice, the polyvinylpolypyrrolidone was removed by centrifugation (3 min at 18,000 × g). The DNA was precipitated with 2 volumes of 100% ethanol (Merck KGaA, Darmstadt, Germany) and 0.1 volume of 3 M sodium acetate, pH 5.2 (Merck KGaA, Darmstadt, Germany), left overnight at −20°C, and subsequently washed with 70% ethanol. The DNA pellet was resuspended in 400 μl of Tris-EDTA buffer and cleaned by use of a Wizard column (Wizard DNA clean-up system; Promega Corporation, Madison, Wis.). The DNA concentration in the 50-μl soil extract was measured spectrophotometrically (Ultrospec 3000 UV/visible spectrophotometer; Pharmacia Biotech, Cambridge, England), and ca. 6 to 31 μg DNA g−1 soil was obtained.
PCR.
PCRs with the extracted DNAs were performed in 50-μl reaction mixtures. A 496-bp eubacterial 16S rRNA gene fragment was amplified by use of the primer set GC-63F/518R as described previously (11). The primer sets used for the detection of the BTEX-catabolic genotypes are reported in Table 2 and were applied in PCRs as follows. The xylM gene fragment was amplified with the primer set TOL-F/TOL-R as described previously (3). The other primer sets were newly designed primer sets that are described elsewhere (17). For the TBMD-F/TBMD-R, TMOA-F/TMOA-R, TOL-F/TOL-R, XYLA-F/XYLA-R, TODC1-F/TODC1-R, TBUE-F/TBUE-R, and TODE-F/TODE-R primer sets, each PCR mixture contained 1.25 U exTaq polymerase (TaKaRa Ex Taq; TaKaRa Shuzo Co. Biomedical Group, Japan), 10 pmol of the forward primer, 10 pmol of the reverse primer, a 200 μM concentration of each deoxynucleoside triphosphate, and 5 μl of 10× exTaq reaction buffer (20 mM MgCl2). For the XYLE-F/XYLE-R, XYLE2-F/XYLE2-R, and CDO-F/CDO-R primer sets, each PCR mixture contained 67 mM Tris-HCl (pH 8.8), 16.6 mM (NH4)2SO4, 0.45% Triton X-100, 0.2 mg gelatin ml−1, a 120 μM concentration of each deoxynucleoside triphosphate, 2 mM MgCl2, 10 pmol primer F, 10 pmol primer R, and 1.25 U Taq DNA polymerase. The primers were synthesized by Westburg (Leusden, The Netherlands). The PCR temperature/time profile used for all primer sets, except for the XYLE2-F/XYLE2-R pair, was an initial denaturation of 5 min at 95°C followed by 35 cycles of one denaturation step for 1 min at 94°C, one annealing step for 1 min at the annealing temperature used for the particular primer set (Table 2), and one elongation step for 2 min at 72°C. The last step was an extension for 10 min at 72°C. The profile used for the XYLE2-F/XYLE2-R primer set consisted of an initial denaturation step of 5 min at 95°C followed by 35 cycles of one denaturation step for 15 s at 94°C, one annealing step for 30 s at 64°C, and one elongation step for 45 s at 72°C. The last step was an extension for 3 min at 72°C. PCRs were performed on a Biometra (Göttingen, Germany) or Perkin-Elmer (Conn.) PCR machine. Checking of the PCR product sizes by agarose gel electrophoresis was performed as described previously (38).
TABLE 2.
Primer sets used for this study
| Primer pair | Protein or gene targeted | Sequences (5′-3′) | PCR annealing temp (°C) | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| TBMD-F/TBMD-R | Subfamily 1 of α-subunit of hydroxylase component of multicomponent mono-oxygenases | GCCTGACCATGGATGC(C/G)TACTGG, CGCCAGAACCACTTGTC(A/G)(A/G)TCCA | 65.5 | 640 | 17 |
| (GC-)TMOA-F/TMOA-Ra | Subfamily 2 of α-subunit of hydroxylase component of multicomponent mono-oxygenases | CGAAACCGGCTT(C/T)ACCAA(C/T)ATG, ACCGGGATATTT(C/T)TCTTC(C/G)AGCCA | 61.2 | 505 | 17 |
| TOL-F/TOL-R | Subfamily 5 of hydroxylase component of two-component side chain mono-oxygenases | TGAGGCTGAAACTTTACGTAGA, CTCACCTGGAGTTGCGTAC | 55 | 475 | 3 |
| XYLA-F/XYLA-R | Electron transfer component of two-component side chain mono-oxygenases | CCAGGTGGAATTTTCAGTGGTTGG, AATTAACTCGAAGCGCCCACCCCA | 64 | 291 | 17 |
| TODC1-F/TODC1-R | Subfamilies D.1.B, D.1.C, D.2.A, D.2.B, and D.2.C of α-subunits of type D iron-sulfur multicomponent aromatic dioxygenases | CAGTGCCGCCA(C/T)CGTGG(C/T)ATG, GCCACTTCCATG(C/T)CC(A/G)CCCCA | 66 | 510 | 17 |
| XYLE-F/XYLE-R | Subfamily 1.2.A of catechol extradiol dioxygenases | CCGCCGACCTGATC(A/T)(C/G)CATG, TCAGGTCA(G/T)CACGGTCA(G/T)GA | 61.5 | 242 | 17 |
| XYLE2-F/XYLE2-R | Subfamily 1.2.B of catechol extradiol dioxygenases | GTAATTCGCCCTGGCTA(C/T)GTICA, GGTGTTCACCGTCATGAAGCG(C/G/T)TC | 64 | 906 | 17 |
| CDO-F/CDO-R | Cdo (U01826) of subfamily 1.2.C of catechol extradiol dioxygenases | CATGTCAACATGCGCGTAATG, CATGTCTGTGTTGAAGCCGTA | 58 | 255 | 17 |
| TBUE-F/TBUE-R | TbuE (U20258) of subfamily 1.2.C of catechol extradiol dioxygenases | CTGGATCATGCCCTGTTGATG, CCACAGCTTGTCTTCACTCCA | 60 | 444 | 17 |
| TODE-F/TODE-R | TodE (Y18245), TodE (Y18245), and TobE (AF180147) of subfamily 1.3.B of catechol extradiol dioxygenases | GGATTTCAAACTGGAGACCAG, GCCATTAGCTTGCAGCATGAA | 58 | 246 | 17 |
| (GC-)63F/518Ra | Eubacterial 16S rRNA gene | CAGGCCTAACACATGCAAGTC, TTACCGCGGCTGCTGG | 55 | 455 | 11 |
A 40-bp GC clamp (5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG-3′) was attached to the 5′ end of forward primers TMOA-F and 63F (47).
Denaturing gradient gel electrophoresis.
Eubacterial 16S rRNA gene PCR products obtained with the primer set GC-63F/518R were analyzed by denaturing gradient gel electrophoresis (DGGE) in 8% polyacrylamide gels with a denaturing gradient of 35% to 65% (100% denaturant gels contain 7 M urea and 40% formamide) as described by Muyzer et al. (31). PCR products obtained with the GC-TMOA-F/TMOA-R primer set were analyzed in 6% polyacrylamide gels with a denaturing gradient of 40% to 70% (17). In both cases, DGGE was performed at a constant voltage of 120 V for 15 h in 1× Tris-acetate-EDTA running buffer at 60°C on an INGENYphorU-2DGGE apparatus (INGENY International BV, The Netherlands). The gels were stained with 1× SYBR gold nucleic acid gel stain (Molecular Probes Europe BV, Leiden, The Netherlands) and photographed as described previously (27). Photofiles were processed and analyzed with Bionumerics software (version 2.50; Applied Maths, Kortrijk, Belgium).
Sequence analysis of PCR-amplified 16S rRNA gene and xylE1 gene fragments.
PCR products obtained with the 16S rRNA gene primer set GC-63F/518R and with primer set XYLE-F/XYLE-R were cloned into the plasmid vector pCR2.1-TOPO by use of a TOPO TA cloning kit and TOP10 One Shot chemically competent cells (N.V. Invitrogen SA, Merelbeke, Belgium) as described previously (27). A selection of 16S rRNA gene clones and all xylE1 clones were sequenced by Westburg (Leusden, The Netherlands) on both strands by use of the primers M13Forward and M13Reverse. The DNA sequences obtained from the cloned PCR products with the eubacterial 16S rRNA gene primers were submitted to the “Chimera Check” program of the Ribosomal Database Project II (Michigan State University, Mich.) to detect possible chimeras that could have been formed during PCR (7). A similarity analysis of the DNA sequences was obtained by applying the Advanced BLAST search program available from GenBank (National Center for Biotechnology Information, Bethesda, Md.).
Nucleotide sequence accession numbers.
The partial sequences of the sequenced 16S rRNA gene clones reported in this study are available in GenBank under accession numbers AY465120 to AY465169, and the partial sequences of the sequenced xylE1 clones are available under accession numbers AY513289 to AY513296.
RESULTS
Selection of membrane type for mesocosm manufacturing.
The sorption of BTEX to the material used for mesocosm manufacturing has to be avoided, as it will influence the supply of BTEX to the bacteria inside the mesocosm. None of the materials showed significant sorption of BTEX (Table 1 and data not shown). A polyamide (PA) membrane was selected for further research because of the durability of the material and because this membrane type was available in various mesh widths (from 1 μm to 4,000 μm) and degrees of open surface (from 0.8% to 64.0%).
The diffusion of pollutants and the migration of bacteria through different polyamide membranes was examined by using 3CBA and Zn as model contaminants. 3CBA, like BTEX, consists of an aromatic ring but is less volatile and easier to work with. Table 1 shows the diffusion rates and mass transport coefficients for 3CBA and Zn obtained with the different membrane types. As expected, the higher the percentage of open surface, the faster the diffusion of 3CBA and Zn. Polyamide membranes with a mesh width of 6 μm and a percentage of open surface of 5% (49 PA 6/5) and with a mesh width of 15 μm and a percentage of open surface of 10% (49 PA 15/10) were chosen to test the migration of bacteria through the membrane, since these membrane characteristics allowed acceptable containment of the aquifer material and acceptable diffusion rates of 3CBA and Zn. For membranes 49 PA 15/10 and 49 PA 6/5, the P11 populations in the left and right chambers reached equilibrium after 5 and 25 h, respectively (data not shown). In the case of the microbial community in the aquifer, the numbers of CFU on diluted 869 medium and on Tris minimal medium exposed to BTEX vapors were identical at both sites with membrane 49 PA 6/5 after ca. 25 h (data not shown). In addition, after 23 h the 16S rRNA gene DGGE fingerprints of the bacterial community were identical for both chambers. However, some bacterial populations seemed to move more rapidly than others through the membranes, as not all bands appeared at the same moment in the DGGE profile for the uninoculated chamber (data not shown).
Use of in situ mesocosm system to monitor the effect of BTEX contamination on the in situ dynamics of an oligotrophic bacterial community and its aerobic BTEX-catabolic gene composition.
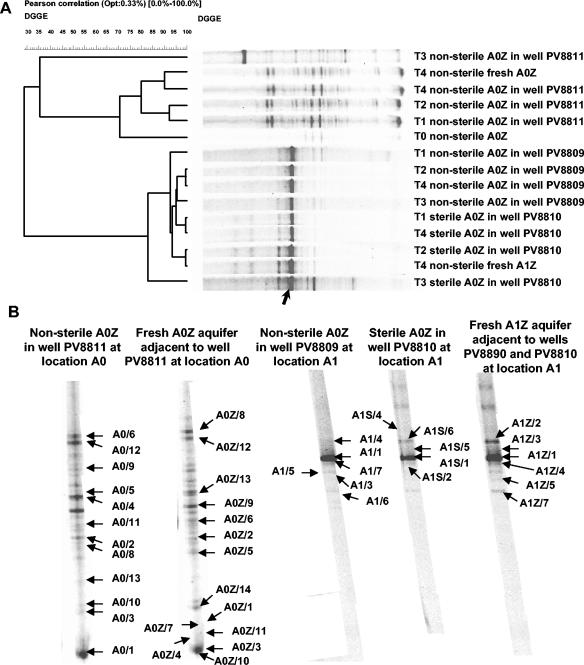
On 9 January 2003, mesocosm pockets manufactured from membrane type 49 PA 6/5 were filled with either nonsterile or sterile A0Z aquifer material which was sampled from the uncontaminated area A0. The mesocosms were positioned in the groundwater in monitoring wells PV-8809 (nonsterile A0Z material) and PV-8810 (sterile A0Z material), which were located in the contaminated area A1. Sterile and nonsterile material was inserted to distinguish between adaptation of the original community and colonization by bacteria from outside the mesocosm. Nonsterile A0Z aquifer material was also inserted into monitoring well PV-8811 at the noncontaminated location A0 (Fig. 2). Eight to 10 identically treated mesocosms were installed in each monitoring well. After 7 days (16 January 2003), 21 days (30 January 2003), 56 days (10 March 2003), and 122 days (20 May 2003), one mesocosm was harvested from each well, the DNAs were extracted, and the eubacterial community structure and BTEX-catabolic gene composition were examined by PCR amplification of either the 16S rRNA gene or BTEX-catabolic genes, followed by DGGE analysis. The data were compared with those obtained from fresh aquifer samples drilled on 20 May 2003 just adjacent to the wells containing the mesocosms. The unweighted-pair group method using average linkages (UPGMA) clustering of the bacterial 16S rRNA gene DGGE patterns showed three main clusters (Fig. 3). One cluster contained the DGGE profiles derived from the materials (initial A0Z material, mesocosm material, and adjacent fresh aquifer material) recovered from the uncontaminated location A0. The second cluster consisted of the profiles for the materials (mesocosm material and fresh aquifer material) recovered from the contaminated aquifer at location A1, and the third cluster contained the profile for the mesocosm material inserted at the uncontaminated location and sampled on 10 March 2003. This result showed that at location A1, both the microbial community of the inserted sterile material and the nonsterile A0Z material evolved into a rather stable community which was highly similar to that of the adjacent aquifer but significantly different from the communities at location A0. This indicates that the bacterial community present in the contaminated A1 aquifer rapidly colonized and dominated the A0Z material within the mesocosms when installed in the contaminated A1 location. At location A0, the bacterial community in the mesocosms remained similar to the original community throughout most of the incubation period and showed a profile identical to that for material drilled freshly from the adjacent aquifer on March 20. Over time, the community patterns were quite similar, except for that for the samples harvested in March. This was especially the case for the sample obtained from the uncontaminated area, but some additional bands were also observed for the mesocosm material sampled from the contaminated area. A community analysis of samples taken from three different parts of the mesocosms recovered on 10 March 2003 showed that the community was the same all over each mesocosm (data not shown).
FIG. 3.
(A) UPGMA clustering of eubacterial 16S rRNA gene community fingerprints obtained from mesocosm aquifer material and fresh aquifer material at different times. Clustering was based on the Pearson moment-based similarity coefficient. T0, 9 January 2003; T1, 16 January 2003; T2, 30 January 2003; T3, 10 March 2003; T4, 20 May 2003. PV8809 is the contaminated well at location A1 into which microcosms containing nonsterile uncontaminated aquifer material were placed. PV8810 is the contaminated well at location A1 into which microcosms containing sterile uncontaminated aquifer material were placed. PV8811 is the well located in the uncontaminated area A0 into which microcosms containing nonsterile uncontaminated aquifer material were placed. The arrow indicates the dominant 16S rRNA gene band whose DNA showed 99% identity with the 16S rRNA gene sequence of Pseudomonas sp. strain SE22#1a. (B) Eubacterial 16S rRNA gene community fingerprints for samples obtained on 20 May 2003, with the cloned bands indicated (arrows). Based on BLASTN analysis, the 16S rRNA gene sequences corresponding to the indicated bands matched best with other 16S rRNA gene sequences as follows (accession numbers with the best match are given in parentheses): A0/1, Rhodococcus fascians ATCC 12974T (X81930); A0/2, Acidovorax sp. strain BSB421 (Y18617); A03, Rhodococcus sp. strain 5/14 (AF181690); A0/4, Rhodococcus sp. strain KU18 (AJ517832); A0/5, uncultured β-bacterium clone KD5-43 (AY218738); A0/6, Pseudomonas umsongensis Ps 3-10 (AF468450); A0/8, Pseudomonas sp. strain MSB2071 (AY275482); A0/9, Pseudomonas lanceolata ATCC 14669T (AB021390); A0/10, Arthrobacter sp. strain CS16 (AY371258); A0/11, Zoogloea sp. strain BAL15 (U63941); A0/12, Pseudomonas sp. strain MWH1 (AJ556801); A0/13, Rhodococcus sp. strain P_wp0233 (AY188941); A0Z/1, Rhodococcus fascians ATCC 12974T (X81930); A0Z/2, Bradyrhizobium sp. strain Hambi 2145 (AJ132567); A0Z/3, Rhodococcus erythropolis DCL14 (AJ131637); A0Z/4, Rhodococcus fascians ATCC 12974T (X81930); A0Z/5, Arthrobacter oxydans CF-46 (AJ243423); A0Z/6, uncultured beta-proteobacterium clone NMS8.125WL (AY043788); A0Z/7, Rhodococcus sp.strain P_wp0233 (AY188941); A0Z/9, uncultured beta-proteobacterium clone C47.33PG (AF431314); A0Z/10, Rhodococcus sp. strain P_wp0233 (AY188941); A0Z/11, Acidovorax sp. strain G8B1 (AJ012071); A0Z/12, Pseudomonas sp. strain MWH1 (AJ556801); A0Z/13, Rhodococcus sp. strain KU18 (AJ517832); A0Z/14, Arthrobacter sp. strain CS16 (AY371258); A1/1, Pseudomonas sp. strain SE22#1a (AY263477); A1/3, Rhodococcus sp. strain Lgg15.6 (AJ489362); A1/4, Pseudomonas fluorescens DSM50108 (D86002); A1/5, Pseudomonas sp. strain GOBB3-103 (AF321030); A1/6, Zoogloea sp. strain BAL15 (U63941); A1/7, Pseudomonas sp. strain NAF88 (AJ271413); A1S/1, Pseudomonas sp. strain SE22#1a (AY263477); A1S/2, Pseudomonas sp. strain NAF88 (AJ271413); A1S/4, Pseudomonas stutzeri API-2-142 (AJ410871); A1S/5, uncultured bacterium clone ZZ14C11 (AY214196); A1S/6, Pseudomonas fluorescens DSM50108 (D86002); A1Z/1, Pseudomonas sp. strain SE22#1a (AY263477); A1Z/2, Pseudomonas fluorescens DSM50108 (D86002); A1Z/3, uncultured bacterium clone ZZ14C11 (AY214196); A1Z/4, Pseudomonas sp. strain NAF88 (AJ271413); A1Z/5, Pseudomonas sp. strain MSB2074 (AY275484); A1Z/7, Pseudomonas sp. strain QSSC1-9 (AF170731).
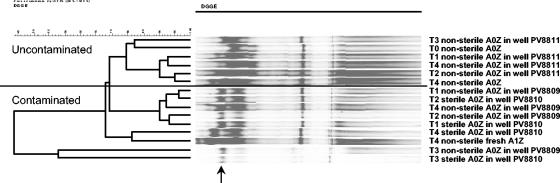
In order to compare the evolution of genotypes involved in aerobic BTEX catabolism at the different locations, we performed PCRs with previously designed primer sets (Table 2). The results are shown in Table 3. For both the sterile and nonsterile A0Z aquifer samples inserted at location A1, mainly xylM/xylE-like genotypes along with the tmoA-like genotype were detected. These results corresponded well with the results obtained for fresh aquifer samples taken in May. In contrast, for location A0, the A0Z mesocosm samples and aquifer showed tmoA-like genotypes but not xylM/xylE1-like genotypes. Differences in tmoA diversity between samples were analyzed by DGGE analysis of the tmoA gene amplicons (Fig. 4). The original A0Z tmoA gene DGGE profile did not change throughout the experiment for microcosms implemented in the noncontaminated well PV8811 and was similar to that for the adjacent aquifer. In contrast, one strong additional DGGE band appeared at the top of the profile (Fig. 4, arrow) for the A0Z material inserted in the contaminated wells, generating a profile similar to that for the surrounding aquifer. This extra band remained over time. As observed for the community fingerprints for both contaminated and uncontaminated samples, a difference in tmoA-like gene diversity profiles of the contaminated samples was found between the samples harvested in March and those sampled in January and May.
TABLE 3.
Results of PCRs assessing the BTEX-catabolic gene compositions in the in situ mesocosm material and aquifer samples obtained at different time pointsa
| Primer pair (detected gene family) | PCR result for uncontaminated area A0
|
PCR result for contaminated area A1
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sterile A0Z material | Nonsterile A0Z material
|
Nonsterile A0Z material in mesocosms in well PV8811
|
Nonsterile A0Z material in mesocosms in well PV8809
|
Sterile A0Z material in mesocosms in well PV8810
|
A1Z material
|
|||||||||||
| Jan 9 | Jan 9 | May 20 | Jan 16 | Jan 30 | March 10 | May 20 | Jan 16 | Jan 30 | March 10 | May 20 | Jan 16 | Jan 30 | March 10 | May 20 | May 20 | |
| TBMD-F/TBMD-R (tbmD) | − | − | − | − | − | − | − | (+) | (+) | − | − | − | − | − | − | − |
| TMOA-F/TMOA-R (tmoA) | − | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| TOL-F/TOL-R (xylM) | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + |
| XYLE-F/XYLE-R (xylEl) | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + |
All sample data were from 2003. ++, strong signal; +, signal; (+), weak signal; −, no signal. No signals were obtained with the primer sets XYLA-F/XYLA-R (xylA), TODC1-F/TODC1-R (todC1), XYLE2-F/XYLE2-R (xylE2), TBUE-F/TBUE-R (tbuE), TODE-F/TODE-R (todE), and CDO-F/CDO-R (cdo).
FIG. 4.
UPGMA clustering of DGGE fingerprints of the tmoA-like PCR amplicons obtained with primer set GC-TMOA-F/TMOA-R from DNAs extracted from the aquifer materials in the mesocosms and from fresh aquifer materials at different times. Clustering was performed by using the Pearson moment-based similarity coefficient. T0, 9 January 2003; T1, 16 January 2003; T2, 30 January 2003; T3, 10 March 2003; T4, 20 May 2003. The wells of origin are indicated as described in the legend to Fig. 3. The arrow indicates the DGGE band which appeared when uncontaminated aquifer material from A0Z was placed into the wells located in the contaminated area A1.
Cloning and sequencing of amplified 16S rRNA gene fragments.
Clone libraries of the 16S rRNA gene PCR amplicons obtained with the primer set GC-63F/518R were constructed from all samples harvested on 20 May 2003. Sixteen and 22 clones were recovered from the microcosm and the adjacent aquifer of the uncontaminated area, respectively. For the contaminated area, 21 clones were recovered from the microcosms in monitoring well PV8809, 24 clones were recovered from the microcosm in well PV8810, and 20 clones were recovered from the aquifer adjacent to the wells. The DGGE profiles of the clones were matched with the DGGE community profiles from the materials of origin to identify the corresponding bands (Fig. 3B). For all samples, almost all dominant bands were recovered in the clone libraries. Most bands of the DGGE profile obtained with material from the A0 area were represented by only one clone. In contrast, many clones from the A1 area matched the same bands in the A1 community patterns. In particular, the dominant band indicated with an arrow in Fig. 3A was represented many times in the clone libraries derived from all three A1 samples (see Table S1 in the supplemental material). A representative clone for each band was sequenced for each soil sample. In cases of dominant bands, more than one clone was sequenced.
The differences in the DGGE fingerprints observed between samples recovered from the uncontaminated and contaminated areas were translated into clear differences in 16S rRNA gene clone sequences between those samples (see Table S1 in the supplemental material). The richness of 16S rRNA gene sequences was higher in the uncontaminated location than in the contaminated location. Actinobacteria (14 of 27 sequenced clones) and Proteobacteria (13 of 27 sequenced clones) seemed to constitute the two main groups of resident bacteria in the absence of pollution. The Actinobacteria included bacteria related to Rhodococcus (11 sequenced clones) and Arthrobacter (3 sequenced clones). The recovered 16S rRNA gene sequences of Proteobacteria were related to those of β-Proteobacteria (Acidovorax, Zoogloea, Pseudomonas, and Aquaspirillum; seven sequenced clones) and γ-Proteobacteria (six sequenced clones). The microcosm material and fresh aquifer material showed similar ratios of Actinobacteria to Proteobacteria. Several identical sequences were found in the mesocosms and in the aquifers adjacent to the monitoring wells. The 16S rRNA gene sequences represented by bands A0/3, A0/13, A0Z/7, and A0Z/10 in the DGGE profiles were related to sequences of known hydrocarbon degraders (Rhodococcus sp. strain 5/14 [4] and Rhodococcus sp. strain P_wp0233 [accession no. AY188941; Ma et al., unpublished]), while the sequence of band A0Z/9 was related to the sequence of an uncultured bacterial 16S rRNA gene clone, GOUTB17, derived from an in situ reactor treating monochlorobenzene-contaminated groundwater (1).
The sequences recovered from location A1 were clearly less diverse than those from location A0. Most 16S rRNA gene sequences were related to those of Proteobacteria. Only one clone sequence, i.e., the clone corresponding to band A1/3, was closely related to a Rhodococcus 16S rRNA gene sequence. Most Proteobacteria sequences were similar to those of γ-Proteobacteria (Pseudomonas) (18 of 22 sequenced clones), and a few were similar to those of β-Proteobacteria (Acidovorax and Zoogloea) (3 sequenced clones). All sequenced clones that matched the dominant bands of the three soil samples (bands A1/1, A1S/1, and A1Z/1) showed identical sequences, which were 99% related to the 16S rRNA gene sequence of Pseudomonas sp. strain SE22#1a, a cold-tolerant, alpine soil Pseudomonas sensu stricto strain belonging to the fluorescens group (accession no. AY263477; unpublished data). In addition to those clones, the sequences of several other clones recovered from inside the mesocosms and from the adjacent aquifer were identical. The 16S rRNA gene sequences represented by bands A1S/5 and A1Z/3 were related to the sequence of clone ZZ14C11, which was obtained from benzene-contaminated groundwater (accession no. AY214196; A. Alfreider, unpublished data), while the sequences of the clones represented by bands A1/7, A1S/2, A1S/3, and A1Z/4 were related to that of Pseudomonas sp. strain NAF88, a polyaromatic hydrocarbon-degrading bacterial strain (accession no. AJ271413; Duran, unpublished data).
No identical sequences were recovered from the uncontaminated A0 area and the contaminated A1 area, showing that the dominant populations at both sites are clearly different. The similarity of the DGGE fingerprints and the identical sequences found both inside and outside the mesocosms at the contaminated site strongly indicate that the bacterial community outside the mesocosm colonized the sterile and nonsterile uncontaminated mesocosm material after its insertion into the contaminated groundwater.
Cloning and sequencing of subfamily I.2.A catechol 2,3-dioxygenase (C23O) (xylE-like) gene fragments.
To compare the xylE-like sequences from the inserted aquifer material obtained on 20 May 2003 from wells PV8809 and PV8810 with other xylE-like gene sequences recovered from xylE clones and from BTEX-degrading organisms from the same site in other studies (17, 18, 19), we cloned the xylE fragments obtained in this study. All nine recovered clones were sequenced, and the sequences were analyzed by BLASTX analysis. Clustering with previously described C23O protein sequences showed that the deduced protein sequences of the clones clustered around the DmpB C23O sequence of P. putida CF600 (data not shown). This cluster also contained the putative XylE protein sequences of three BTEX-degrading strains, i.e., Pseudomonas veronii A1X/4B, P. putida A1Y11, and isolate 3YdBTEX2, which were previously isolated from the contaminated A1 area of the site (17, 18, 19). However, the deduced C23O sequences obtained from the majority of BTEX-degrading isolates reported by Junca and Pieper (19) from the same site clustered in a second major C23O cluster (cluster 2) containing typical C23O proteins, such as NahH of P. putida PpG7, XylE of P. putida mt-2 (PaW1), and PhlH and XylE of P. putida H.
Physicochemical parameters of the groundwater.
Most of the physicochemical parameters of the groundwater, except the temperature, were quite stable over time at both the contaminated and uncontaminated locations (see Table S2 in the supplemental material). The temperature of the groundwater behaved in a similar way in all wells during the period January to June 2003. In January, the groundwater temperature was about 9°C and then decreased to 7°C during February and March. From April on, the temperature increased, reaching 11°C in June (data not shown). In the contaminated well, anaerobic conditions prevailed (oxygen concentration, 0.04 to 0.23 mg liter−1), while at the uncontaminated location, oxygen was present at levels of 7.3 mg liter−1 in January and 1.67 mg liter−1 in May. The groundwater levels were different but stable for the uncontaminated (about 2.25 m) and contaminated (about 2.75 m) wells throughout the experimental period. At the uncontaminated area, nitrate concentrations reached 40 mg liter−1, while at the contaminated area they were never higher than 1 mg liter−1. All wells demonstrated significant SO42− concentrations.
DISCUSSION
In this study, we describe the design of an in situ mesocosm system and its use in combination with the application of molecular ecology techniques to study bacterial community dynamics in aquifers. More specifically, we examined the effect of a BTEX-contaminated groundwater plume on the in situ dynamics of an oligotrophic aquifer community and on the dynamics of specific BTEX-catabolic genotypes within this community.
The present study demonstrates that the mesocosm system allowed us to monitor the on-site dynamics of an aquifer microbial community over time since the same microbial community and BTEX-catabolic genotypes were found inside the mesocosms and in the adjacent aquifer at both the contaminated and uncontaminated areas. We believe that the relevance of the mesocosm system is specified by the membrane used for the construction of the membrane pockets. The polyamide membrane used for this study allowed the migration of bacteria, nutrients, and contaminants between the material inside the mesocosm and the bulk aquifer. This membrane is available with many different mesh widths, which can be used to control the migration of specific biotic micro- and mesofaunal species (21). For example, mesocosms can be used to examine the role of subsurface protozoa on the microbial community structure or to determine the effects of in situ physicochemical conditions on genetically engineered microorganisms used for aquifer bioaugmentation by containing them within the mesocosm.
Our results indicated that (i) downstream aquifer material will be rapidly colonized by bacteria from the contaminated upstream area, (ii) the new community remained stable over an extended time period, (iii) Pseudomonas spp. were the dominant species involved in the colonization, (iv) obligate anaerobic bacteria such as sulfate-reducing bacteria (SRB) did not dominate the community, and (v) changes in the BTEX-catabolic genotype structure reflected the community change.
The rapid colonization of the mesocosm material by the community present in the aquifer environment suggests that bacteria that are present in an upstream pollutant plume will colonize downstream nonpolluted material when it becomes contaminated. However, in our case, the bacterial community from the uncontaminated aquifer inserted in mesocosms installed in the contaminated area was directly exposed to extremely high BTEX concentrations. This might have caused a direct elimination of the resident bacteria and an overgrowth by bacteria from the A1 area which were already adapted to the high BTEX concentrations, for example, by displaying high solvent tolerance mechanisms (22, 23, 39, 40) . In reality, a more gradual increase in the contaminant concentration occurs, allowing a more gradual adaptation of the original population to the contamination. The fact that the colonization occurred so rapidly might be an indication that resident bacteria were indeed eliminated. On the other hand, the fact that the same bacteria colonized both the sterile and nonsterile aquifers in the mesocosms cannot exclude that a community similar to the community of A1 evolved from the inserted A0 community.
The observation that the new bacterial community remained stable over a 122-day period was possibly due to the fact that most environmental parameters remained quite stable during the incubation period. Interestingly, in the samples from March, the bacterial community seemed slightly different from that at other time points, which may have been due to the lower groundwater temperature recorded in March 2003.
A characterization of the bacterial communities revealed a much richer community in the uncontaminated area than in the contaminated location. For the uncontaminated area, sequences related to both Proteobacteria and Actinobacteria (Rhodococcus and Arthrobacter) were obtained, while at the contaminated location almost all sequences were related to Proteobacteria (mostly fluorescent Pseudomonas). This is in contrast to the results of Shi et al. (41), who found that fuel-contaminated and noncontaminated aquifer materials demonstrated similar relative abundances of Proteobacteria and gram-positive bacteria. Both Pseudomonas (13, 41, 42) and Rhodococcus (8, 14, 16, 22) have been reported to include members that are able to degrade BTEX and other organic pollutants. Fluorescent Pseudomonas strains, especially P. putida strains, are often isolated as BTEX degraders from BTEX- and gasoline-contaminated sites. This study shows that they are also relevant in subsurface environments under conditions of high BTEX contamination and, hence, high growth substrate concentrations and a low availability of macronutrients. In particular, members of Pseudomonas fluorescens and the closely related P. veronii were recovered. Strongly related strains (99% 16S rRNA gene identity) have been previously isolated from the same aquifer from drilled samples (17, 18, 19). Interestingly, although P. fluorescens is mainly found in plant ecosystems, it has also been recovered from another aquifer that is highly contaminated with BTEX (D. Springael, unpublished results). Pseudomonas spp. are known for their rapid growth, while gram-positive strains such as Rhodococcus are slow growers (13, 43). The high availability of BTEX growth substrates might have been favorable for Pseudomonas, which may also generally be more resistant to solvents (39, 40). A large number of the reported organic solvent-tolerant bacteria are indeed Pseudomonas strains, especially P. putida strains, although benzene-tolerant gram-positive bacteria have also been reported (34, 44).
Although the dissolved oxygen concentrations were low, we did not detect 16S rRNA gene sequences related to those of obligate anaerobic bacteria such as sulfate-reducing bacteria (SRB) and Fe(III) reducers belonging to the genus Geobacter. In addition, no denitrifying bacteria belonging to the genera Thauera and Azoarcus (both β-Proteobacteria) were detected. All of these different bacterial groups contain isolates with the ability to degrade BTEX under anaerobic or anoxic conditions (2, 37, 46). In the uncontaminated zone, oxygen was detected in the groundwater. Oxygen was also present upstream of the contamination plume (M. Cernik, unpublished results). Oxygen inhibits the activity of anaerobes but is typically only available at the fringe of the contaminant plume (28). Rapid consumption of the incoming oxygen possibly occurs, sustaining aerobic BTEX degradation (32, 33). Furthermore, the concentration of nitrate was low at the contaminated area but high at the uncontaminated well, indicating that the rapid consumption of incoming nitrate occurred in the contaminated area, which might have sustained the oxygen-requiring but nitrate-enhanced degradation of BTEX under hypoxic conditions, i.e., conditions with low concentrations of dissolved oxygen, as recently described for Ralstonia pickettii PKO1 and other aerobic BTEX degraders (25). Although sulfate was present at concentrations sufficient for the degradation of BTEX by SRB, the absence of SRB might be explained by the toxic concentrations of BTEX. BTEX degradation by SRB is mostly reported for relatively low BTEX concentrations of ca. 10 ppm and lower (46). Moreover, SRB, methanogenic bacteria, and Fe(III)-reducing bacteria are only active at redox potentials of <−200 mV (28, 37, 46), and the redox potential of the groundwater in our study was highly variable but always positive (data not shown). Our data show that aerobic bacteria were dominant in the aquifer. The presence of anaerobic organisms, however, cannot be excluded. This can be examined by using PCR detection methods targeting specific anaerobic organisms such as SRB or targeting genes involved in anaerobic degradation (5).
Regarding BTEX-catabolic genes, the community in the contaminated area was especially characterized by the appearance of xylM/xylE (C23O)-like genotypes, as these genotypes could only be recovered from the contaminated area. Both locations contained tmoA-like genotypes. No other BTEX-catabolic genotypes were detected. The occurrence of the xylM/xylE-like genotypes can be related to the dominant bacteria present at the contaminated site, i.e., fluorescent pseudomonads. Indeed, xylM/xylE-like genotypes have been found for fluorescent pseudomonads such as P. putida (18). Recently, gene fragments corresponding to toluene 4-mono-oxygenase (TmoA) and catechol 2,3-dioxygenase (XylE) were detected by PCR in BTEX-polluted groundwater samples (6) and were associated with indigenous BTEX-degrading Pseudomonas strains. The site used for the present study contained mainly benzene, which cannot be degraded by XylM/XylA-like mono-oxygenases, but rather is degraded by TmoA-like mono-oxygenases and TodC1-like dioxygenases. Therefore, rather than the appearance of the xylE/xylM genotype, the stronger tmoA PCR signal and the additional DGGE band appearing in the DGGE profiles of mesocosm and aquifer materials from the contaminated area may represent the adaptation of the community to benzene. The appearance of xylE/xylM genotypes in the community upon contamination might be due to the fact that the contamination plume at A1 also contains toluene. Interestingly, many BTEX-degrading isolates from that location with similar xylE genes were able to consume both toluene and benzene (17, 18, 19). Previously, Guo et al. (15) found that the concentrations of the tmoABCDE and xylE genes were significantly higher in contaminated subsurface samples collected along a BTEX concentration gradient at a fuel oil-contaminated site than in noncontaminated samples. The site was mainly contaminated with m- and p-xylene. The clear appearance of the tmoA- and xylM/xylE-like genotypes in these different studies suggests the contribution of these genes to community adaptation to BTEX in the field.
In contrast to the 16S rRNA gene-based DGGE patterns, the DGGE patterns of tmoA-like genes of samples from A0 and A1 showed some common bands. This incongruence between 16S rRNA and tmoA-like gene-based DGGE fingerprints suggests the presence of very similar or identical tmoA-like genes in phylogenetically distinct bacteria. The occurrence of highly similar functional genes in different bacteria has been shown before and can be explained by the horizontal transfer, either as a recent or an ancient event, of these catabolic genes among bacteria in the aquifer ecosystem (26).
In conclusion, the designed in situ mesocosm system, in combination with PCR-DGGE analysis of the eubacterial 16S rRNA genes and the BTEX-catabolic genotypes, was shown to be an appropriate tool for monitoring bacterial community dynamics in a BTEX-contaminated aquifer. This new in situ monitoring technique can be applied to more easily study the dynamics of actively degrading microbial populations in aquifers in situ and to monitor their metabolic markers during in situ bioremediation. As an example, using the in situ mesocosm approach, we are currently looking at the response of the resident BTEX-degrading community at the Czech site to biostimulation by adding N and P nutrients.
Supplementary Material
Acknowledgments
This work was supported by project QLK3-CT2000-00731 of the European Commission. B.H. was supported by a PhD fellowship from Vito.
We thank J. Vos, L. Raets, and L. Timmers for their assistance with the design of the in situ mesocosm system, L. Houtmeyers for her contribution to the experimental work, S. Van Roy for assistance with the diffusion cell experiments, K. Vanbroekhoven for assistance with the clustering analysis, P. De Vos for help with 16S rRNA gene analysis, and M. Hausner for providing strain P11.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alfreider, A., C. Vogt, and W. Babel. 2002. Microbial diversity in an in situ reactor system treating monochlorobenzene contaminated groundwater as revealed by 16S ribosomal DNA analysis. Syst. Appl. Microbiol. 25:232-240. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R. T., and D. R. Lovley. 2000. Anaerobic bioremediation of benzene under sulfate-reducing conditions in a petroleum-contaminated aquifer. Environ. Sci. Technol. 34:2261-2266. [Google Scholar]
- 3.Baldwin, B. R., C. H. Nakatsu, and L. Nies. 2003. Detection and enumeration of aromatic oxygenase genes by multiplex and real-time PCR. Appl. Environ. Microbiol. 69:3350-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bej, A. K., D. Saul, and J. Aislabie. 2000. Cold tolerant alkane-degrading Rhodococcus species from Antarctica. Polar Biol. 23:100-105. [Google Scholar]
- 5.Beller, H. R., S. Kane, T. C. Legler, and P. J. J. Alvarez. 2002. A real-time polymerase chain reaction method for monitoring anaerobic, hydrocarbon-degrading bacteria based on a catabolic gene. Environ. Sci. Technol. 36:3977-3984. [DOI] [PubMed] [Google Scholar]
- 6.Cavalca, L., E. Dell'Amico, and V. Andreoni. 2004. Intrinsic bioremediability of an aromatic hydrocarbon-polluted groundwater: diversity of bacterial population and toluene monooxygenase genes. Appl. Microbiol. Biotechnol. 64:576-587. [DOI] [PubMed] [Google Scholar]
- 7.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and T. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deeb, R. A., and L. Alvarez-Cohen. 1999. Temperature effects and substrate interactions during the aerobic biotransformation of BTEX mixtures by toluene-enriched consortia and Rhodococcus rhodochrous. Biotechnol. Bioeng. 62:526-536. [PubMed] [Google Scholar]
- 9.Dries, J., L. Bastiaens, D. Springael, S. N. Agathos, and L. Diels. 2004. Competition for sorption and degradation reactions in batch zero-valent iron systems. Environ. Sci. Technol. 38:2879-2884. [DOI] [PubMed] [Google Scholar]
- 10.El-Fantroussi, S., J. Mahillon, H. Naveau, and S. N. Agathos. 1997. Introduction and PCR detection of Desulfomonile tiedjei in soil slurry microcosms. Biodegradation 8:125-133. [DOI] [PubMed] [Google Scholar]
- 11.El-Fantroussi, S., L. Verschuere, W. Verstraete, and E. M. Top. 1999. Effect of phenylurea herbicides on soil microbial communities estimated by analysis of 16S rRNA gene fingerprints and community-level physiological profiles. Appl. Environ. Microbiol. 65:982-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallego, J. L. R., J. Loredo, J. F. Llamas, F. Vazquez, and J. Sanchez. 2001. Bioremediation of diesel-contaminated soils: evaluation of potential in situ techniques by study of bacterial degradation. Biodegradation 12:325-335. [DOI] [PubMed] [Google Scholar]
- 13.Galli, E., S. Silver, and B. Witholt. 1992. Pseudomonas: molecular biology and biotechnology. American Society for Microbiology, Washington, D.C.
- 14.Greene, E. A., J. G. Kay, K. Jaber, L. G. Stehmeier, and G. Voordouw. 2000. Composition of soil microbial communities enriched on a mixture of aromatic hydrocarbons. Appl. Environ. Microbiol. 66:5282-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo, C., W. Suo, J. B. Harsh, and A. Ogram. 1997. Hybridization analysis of microbial DNA from fuel oil-contaminated and noncontaminated soil. Microb. Ecol. 34:178-187. [DOI] [PubMed] [Google Scholar]
- 16.Haddad, S., D. M. Eby, and E. L. Neidle. 2001. Cloning and expression of the benzoate dioxygenase genes from Rhodococcus sp. strain 19070. Appl. Environ. Microbiol. 67:2507-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrickx, B. 2004. Dynamics of the microbial community structure and the catabolic gene composition in BTEX-contaminated aquifers using molecular techniques combined with in situ mesocosm stocks, p. 215. Ph.D. thesis. Faculty of Agricultural en Applied Biological Sciences, University of Ghent, Ghent, Belgium.
- 18.Junca, H., and D. H. Pieper. 2003. Amplified functional DNA restriction analysis to determine catechol 2,3-dioxygenase gene diversity in soil bacteria. J. Microbiol. Methods 55:697-708. [DOI] [PubMed] [Google Scholar]
- 19.Junca, H., and D. H. Pieper. 2004. Functional gene diversity analysis in BTEX contaminated soils by means of PCR-SSCP DNA fingerprinting: comparative diversity assessment against bacterial isolates and PCR-DNA clone libraries. Environ. Microbiol. 6:95-110. [DOI] [PubMed] [Google Scholar]
- 20.Kampichler, C., A. Bruckner, and E. Kandeler. 2001. Use of enclosed model ecosystems in soil ecology: a bias towards laboratory research. Soil Biol. Biochem. 33:269-275. [Google Scholar]
- 21.Kandeler, E., C. Kampichler, R. G. Joergensen, and K. Mölter. 1999. The impact of mesofauna on the soil microbial community and N cycling in field mesocosms. Soil Biol. Biochem. 31:1801-1810. [Google Scholar]
- 22.Kim, D., Y. S. Kim, S. K. Kim, S. W. Kim, G. J. Zylstra, Y. Min Kim, and E. Kim. 2002. Monocyclic aromatic hydrocarbon degradation by Rhodococcus sp. strain DK17. Appl. Environ. Microbiol. 68:3270-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, K., S. Lee, K. Lee, and D. Lim. 1998. Isolation and characterization of toluene-sensitive mutants from the toluene-resistant bacterium Pseudomonas putida GM73. J. Bacteriol. 180:3692-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirk, J. L., L. A. Beaudette, M. Hart, P. Moutoglis, J. N. Klironomos, H. Lee, and J. T. Trevors. 2004. Methods of studying soil microbial diversity. J. Microbiol. Methods 58:169-188. [DOI] [PubMed] [Google Scholar]
- 25.Kukor, J. J., and R. H. Olsen. 1996. Catechol 2,3-dioxygenase functional in oxygen-limited (hypoxic) environments. Appl. Environ. Microbiol. 62:1728-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence, J. G., and H. Ochman. 2002. Reconciling the many faces of lateral gene transfer. Trends Microbiol. 10:1-4. [DOI] [PubMed] [Google Scholar]
- 27.Leys, N. M. E. J., A. Ryngaert, L. Bastiaens, W. Verstraete, E. M. Top, and D. Springael. 2004. Occurrence and phylogenetic diversity of Sphingomonas strains in soils contaminated with polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 70:1944-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lovley, D. R. 2001. Bioremediation. Anaerobes to rescue. Science 293:1444-1446. [DOI] [PubMed] [Google Scholar]
- 29.Mandelbaum, R. T., M. R. Shati, and D. Ronen. 1997. In situ microcosms in aquifer bioremediation studies. FEMS Microbiol. Rev. 20:489-502. [DOI] [PubMed] [Google Scholar]
- 30.Mergeay, M., D. H. Nies, H. G. Schlegel, J. Gerits, P. Charles, and F. Van Gijsegem. 1985. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 162:328-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muyzer, G., E. C. de Waal, and A. Uitterlinden. 1993. Profiling of complex microbial populations using denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olsen, R. H., M. D. Mikesell, and J. J. Kukor. 1994. Enumeration and characterization of BTEX-degrading bacteria from hypoxic environments functional with mixed electron acceptors. Res. Microbiol. 145:47-49. [DOI] [PubMed] [Google Scholar]
- 33.Olsen, R. H., M. D. Mikesell, J. J. Kukor, and A. M. Byrne. 1995. Physiological attributes of microbial BTEX degradation in oxygen-limited environments. Environ. Health Perspect. 103:49-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paje, M., B. Neilan, and I. Couperwhite. 1997. A Rhodococcus species that thrives on medium saturated with liquid benzene. Microbiology 143:2975-2981. [DOI] [PubMed] [Google Scholar]
- 35.Parales, R. E., N. C. Bruce, A. Schmid, and L. P. Wackett. 2002. Biodegradation, biotransformation, and biocatalysis (B3). Appl. Environ. Microbiol. 68:4699-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peys, K. 1999. A new membrane biofilm reactor for the degradation of organic pollutants in waste water, p. 212. Ph.D. thesis. Faculty of Agricultural and Applied Biological Sciences, Leuven, Belgium.
- 37.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 1999. Microbial communities associated with anaerobic benzene mineralization in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Sardessai, Y., and S. Bhosle. 2002. Tolerance of bacteria to organic solvents. Res. Microbiol. 153:263-268. [DOI] [PubMed] [Google Scholar]
- 40.Segura, A., E. Duque, G. Mosqueda, J. L. Ramos, and F. Junker. 1999. Multiple responses of gram-negative bacteria to organic solvents. Environ. Microbiol. 1:191-198. [DOI] [PubMed] [Google Scholar]
- 41.Shi, Y., M. D. Zwolinski, M. E. Schreiber, J. M. Bahr, G. W. Swell, and W. J. Hickey. 1999. Molecular analysis of microbial community structures in pristine and contaminated aquifers: field and laboratory microcosm experiments. Appl. Environ. Microbiol. 65:2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shim, H., and S. T. Yang. 1999. Biodegradation of benzene, toluene, ethylbenzene, and o-xylene by a coculture of Pseudomonas putida and Pseudomonas fluorescens immobilized in a fibrous-bed bioreactor. J. Biotechnol. 67:99-112. [DOI] [PubMed] [Google Scholar]
- 43.Tongpim, S., and M. A. Pickard. 1996. Growth of Rhodococcus S1 on anthracene. Can. J. Microbiol. 42:289-294. [DOI] [PubMed] [Google Scholar]
- 44.Tsitko, I. V., G. M. Zaitsev, A. G. Lobanok, and M. S. Salkinoja-Salonen. 1999. Effect of aromatic compounds on cellular fatty acid composition of Rhodococcus opacus. Appl. Environ. Microbiol. 65:853-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venkata Nancharaiah, Y., P. Wattiau, S. Wuertz, S. Bathe, S. Venkata Mohan, P. A. Wilderer, and M. Hausner. 2003. Dual labeling of Pseudomonas putida fluorescent proteins for in situ monitoring of conjugal transfer of the TOL plasmid. Appl. Environ. Microbiol. 69:4846-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Widdel, F., and R. Rabus. 2001. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr. Opin. Biotechnol. 12:259-276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.