Abstract
Unspecific bacterial reduction of azo dyes is a process widely studied in correlation with the biological treatment of colored wastewaters, but the enzyme system associated with this bacterial capability has never been positively identified. Several ascomycete yeast strains display similar decolorizing behaviors. The yeast-mediated process requires an alternative carbon and energy source and is independent of previous exposure to the dyes. When substrate dyes are polar, their reduction is extracellular, strongly suggesting the involvement of an externally directed plasma membrane redox system. The present work demonstrates that, in Saccharomyces cerevisiae, the ferric reductase system participates in the extracellular reduction of azo dyes. The S. cerevisiae Δfre1 and Δfre1 Δfre2 mutant strains, but not the Δfre2 strain, showed much-reduced decolorizing capabilities. The FRE1 gene complemented the phenotype of S. cerevisiae Δfre1 cells, restoring the ability to grow in medium without externally added iron and to decolorize the dye, following a pattern similar to the one observed in the wild-type strain. These results suggest that under the conditions tested, Fre1p is a major component of the azo reductase activity.
Research work on biodegradative processes for azo dyes usually exploits bacterial species, either isolated or in consortia (4, 36). Bacteria, under appropriate conditions (e.g., oxygen limitation and the presence of substrates utilized as carbon and energy sources), frequently reduce azo dyes, producing colorless amines. Nevertheless, many dyes are recalcitrant to conventional wastewater treatment processes with activated sludge (4). The overall impression in this research area is that many azo dyes can be reduced (and decolorized) by a considerable number of bacterial species, but as far as we know, the enzyme responsible for the unspecific primary reduction step has never been positively identified. What is currently postulated is that reductive decolorization of sulfonated azo dyes by living cells must occur extracellularly due to the impermeant nature of those compounds and that the primary reductant is a cytoplasmic electron donor, presumably NAD(P)H (36).
Previous studies (30, 31) have demonstrated that some nonconventional ascomycete yeasts are efficient azo dye decolorizers, acting, as many bacteria, by reducing the azo bond. Dye decolorization by yeasts is comparatively unspecific but is affected by the medium composition, by the yeast strain used, and by parameters such as pH and dissolved oxygen level. It also requires actively growing cells, being faster during the exponential growth phase, and displays an enzyme-like temperature profile, strongly suggesting its biotic nature. However, further information is required for successful application of yeasts in a wastewater treatment process. The present work was developed to demonstrate the participation of an externally directed plasma membrane redox system (PMRS) in azo dye reduction, linking an intracellular reductant to an extracellular electron acceptor. As a required first step, it was necessary to find a model yeast strain capable of decolorizing polar azo dyes. Among the screened strains, Saccharomyces cerevisiae CEN.PK113-7D proved to fulfill that condition.
In S. cerevisiae, the most extensively explored PMRS is the ferric/cupric reductase system which participates in the high-affinity uptake of iron. This activity can be assayed through the reduction of impermeable substrates like ferricyanide, iron(III)-citrate, iron(III)-EDTA, and a variety of other ferric chelates. In this complex system, the best-studied components are the metalloreductases encoded by the genes FRE1 (7) and FRE2 (15); the oxidase-permease complex encoded by FET3/FTR1 (reviewed in reference 9); the iron-dependent transcriptional regulators Aft1p (39, 41) and Aft2p (3, 40); and the copper-dependent transcriptional regulator Mac1p (16, 40). A potential Fe3+/Cu2+ reductase subunit is the cytoplasmic cofactor Utr1p (1).
FRE1 and FRE2 encode plasma membrane proteins (7, 15) and are both transcriptionally activated by Aft1p, whose intracellular location is dependent on the iron(III) level (42). FRE1 activation is also controlled by Aft2p (33) and Mac1p (40). Transcription of FRE2 through Aft1p (33) depends only on iron levels (14). The protein encoded by FRE1 contains several transmembrane domains (7) and shares 62% sequence similarity with the gp91phox subunit of cytochrome b558 (32). The protein motifs in gp91phox responsible for binding flavin adenine dinucleotide and NADPH are conserved in Fre1p (12, 23, 35). Fre1p and Fre2p together account for virtually all of the Fe3+/Cu2+ reductase activity of yeast cells but in varying proportions, depending both on iron and/or copper availability and on the growth phase of the cells (14, 15, 16). Typically, FRE2 is induced at a later stage. Fre1p and/or Fre2p reduces external Fe3+ (or Cu2+) prior to uptake, and this reduction is mediated by Fet3p/Ftr1p, where Fet3p is a multicopper oxidase and Ftr1p the permease component (10). The cytoplasmic cofactor Utr1p in S. cerevisiae has recently been shown to be a NAD kinase (21) which is regarded as the only enzyme catalyzing the synthesis of NADP.
The genome sequence of S. cerevisiae revealed the presence of five additional metalloregulated genes, FRE3 through FRE6 and FRE7, with sequence similarities to FRE1 and FRE2. The first four are transcriptionally regulated by the iron-responsive Aftp1p element and the fifth by the copper-dependent Mac1p (27). Fre3p and Fre4p are potential siderophore-iron reductases (43), but the function of the remaining gene products is unknown. Given their regulation pattern, they may participate in iron homeostasis (FRE5 and FRE6 products) and copper homeostasis (FRE7 product), possibly as internal metalloreductases (27).
The present work shows that the azo reductase and ferric reductase activities of yeast cells assayed in different growth phases are closely parallel, being at the highest levels during the late exponential growth phase. This property of ferric reductase has been described in early studies (6, 15). Also, deletion of the FRE1 gene eliminates a major fraction of the azo reductase activity in intact cells of S. cerevisiae harvested in the late exponential growth phase, whereas the deletion of the FRE2 gene has a minor effect on that activity. We believe that our results will be relevant for biotechnological applications of this activity and also for a broader understanding of the unspecific redox activities associated with the yeast plasma membrane.
MATERIALS AND METHODS
Chemicals.
The azo dye used in the experiments was m-[(4-dimethylamino)phenylazo] benzenesulfonic acid, sodium salt, and was synthesized as described for methyl orange (13).
Yeast strains and plasmids.
The yeast strains and the plasmids used in this work are listed respectively in Tables 1 and 2. The cultures were maintained on slants of YPD—yeast extract (1%, wt/vol), peptone (1%, wt/vol), glucose (2%, wt/vol), and agar (2%, wt/vol). Growth on solid media was carried out at 30°C.
TABLE 1.
S. cerevisiae strains used in this work
| Strain | Description | Reference or Source |
|---|---|---|
| CEN.PK113-7D | Wild type (MATaMAL2-8c SUC2) | 11 |
| Y04163 | BY4741; MATahis3 leu2 met15 ura3 YLR214W::KanMX4 | Euroscarf |
| Y07039 | BY4741; MATahis3 leu2 met15 ura3 YKL220C::KanMX4 | Euroscarf |
| SP1 | Δfre1 (CEN.PK YLR214W::KanMX4) | This work |
| SP2 | Δfre2 (CEN.PK YKL220C::KanMX4) | This work |
| SP3 | BY4741; YKL220C::HphMX4 | This work |
| SP4 | Δfre1 Δfre2 (CEN.PK YLR214W::KanMX4 YKL220C::HphMX4) | This work |
| SPcmp-FRE1 | Δfre1 (pSP3) (CEN.PK YLR214W::KanMX4 plus plasmid pSP3) | This work |
TABLE 2.
Plasmids used in this work
Cell growth in liquid medium.
The attenuance of appropriately diluted cell suspensions (as described in reference 30) was measured at 640 nm with a Spectronic 21 Bausch & Lomb spectrophotometer using a 1-cm-path-length cell.
Decolorization in liquid media.
Decolorization experiments with growing cultures of S. cerevisiae CEN.PK113-7D (hereafter referred to as the wild-type strain) were typically performed in 250-ml cotton-plugged Erlenmeyer flasks with 100 ml of sterile medium (normal decolorization medium [NDM]) containing yeast extract (0.25%, wt/vol), glucose (2%, wt/vol), and 0.2 mmol liter−1 of the tested dye in a mineral salts base of the composition previously described (31) incubated at 26°C and 120 rpm in a Stuart Scientific S150 orbital incubator. Whenever required, iron(III) was added to the medium as the EDTA chelate, from a 100 mM stock solution in FeCl3 and EDTA. For the mutant strains, which show impaired growth in our standard medium, cells were grown for 137 h in NDM supplemented with 2 mM iron(III) as the EDTA chelate. For controls, wild-type cells were grown under similar conditions. The cells were then harvested by centrifugation at 16.1 × g, washed several times with sterile distilled water, and resuspended in NDM to produce cell suspensions with 3.8 ± 0.2 attenuance units (4.2 ± 0.2 g of cells [dry weight] liter−1). Throughout this work, decolorizing activity refers to the decolorization capability of growing yeast cultures.
Cell counting.
Cell suspensions (diluted to an attenuance of ca. 0.5 units) were diluted 1:25,000 and 1:250,000. From each dilution, 100 μl was spread onto YPD agar plates. The plates were incubated at 37°C for 2 days, and after that time, the number of isolated colonies was counted. All plates with more than 300 colonies or fewer than 30 were not considered. All the dilutions were prepared in triplicate.
Ferric reductase assay.
Cells were grown for ca. 6 h in NDM, harvested by centrifugation, washed twice with sterile distilled water, and resuspended in assay buffer consisting of 0.05 M sodium citrate, pH 6.5, with 5% glucose at a cell density of ca. 1.3 ± 0.1 attenuance units (1.4 ± 0.1 g of cells [dry weight] liter−1). The assays were performed in triplicate at two different cell densities obtained with either 780 μl of suspension or 390 μl of suspension plus 390 μl of assay buffer. The cell suspensions were preincubated for 10 min at room temperature. The final assay mixtures contained, in a total volume of 1 ml, 2 mM ferrozine {[3-(2-pyridyl)-5,6-bis-(4-phenylsulfonic acid)-1,2,4-triazine]} and 0.2 mM iron(III) as ferric chloride. The mixtures were allowed to react at room temperature (20 ± 2°C) for 5 or 10 min. Cells were then harvested by centrifugation, and the absorbance at 562 nm was measured against a blank prepared similarly but without cells. The ferrous iron concentration was estimated by using a molar absorbance of 27,900 M−1 cm−1 for the iron(II)ferrozine complex (17).
Azo reductase assays.
Azo reductase assays were performed as the ferric reductase assays but using acetate buffer at 0.05 M, pH 4.0, and 5% glucose. The assay mixture contained a cell suspension of 1 or 2 attenuance units (1.1 ± 0.1 or 2.2 ± 0.1 g of cells [dry weight] liter−1) and 0.05 mM dye and was allowed to react for 15 to 20 min. Within this period, the decrease in absorbance was linear with time. The absorbances of the final supernatants were read at dye λmax (461 nm). The amount of dye reduced was determined from a molar absorbance of 21,440 M−1 cm−1, obtained from a calibration curve. Throughout this work, azo reductase specific activity refers to the results of activity assays within a short period of time, being expressed as μmol (g of cells [dry weight])−1 min−1.
Transformation of S. cerevisiae cells.
Transformation of S. cerevisiae cells was done by the lithium acetate/single-stranded carrier DNA/polyethylene glycol method (18). When required, transformants were recovered at 30°C in YPD medium for 4 h before being plated onto YPD solid medium containing either 200 mg/liter Geneticin (G418 from Life Technologies) or 30 μg/ml phleomycin (CAYLA, Toulouse, France). Transformants were obtained after 2 to 3 days of incubation at 30°C. To purify transformants from background, each large colony was restreaked onto fresh YPD-Geneticin or YPD-phleomycin plates. Only those clones that grew after the double selection were further analyzed, by analytical PCR as described by Kruckeberg et al. (22), as potentially correct transformants.
Cloning of the FRE1 and FRE2 genes.
The FRE1 gene was amplified by PCR with the Pfu Turbo DNA polymerase (Stratagene) using the primers Fre1forw and Fre1rev and genomic DNA isolated from S. cerevisiae CEN.PK 113-7D. The PCR fragment was cloned into the plasmid pGEM-T Easy vector (Promega), producing the plasmid pSP1 (Table 2). The primers Fre2forw and Fre2rev were used to amplify the FRE2 gene by following the same procedure described for the FRE1 gene. The PCR product was cloned into the pGEM-T Easy vector, producing the plasmid pSP2 (Table 2). DNA cloning and manipulation were performed according to standard protocols (34).
FRE1 knockout.
The S. cerevisiae Y04163 strain with the gene FRE1 (YLR214W) deleted was obtained from the Euroscarf collection. Two primers, A-YLR214W and D-YLR214W (Table 3), were used to amplify by PCR the YLR214W::KanMX4 allele of the S. cerevisiae strain Y04163. The PCR product was used to transform wild-type cells. Cells were plated onto YPD solid medium containing 200 mg/liter Geneticin. Successful integration of the YLR214W::KanMX4 cassette was scored by the presence of the YLR214W::KanMX4 band (2,352 bp) and the absence of the YLR214W wild-type band (2,796 bp) following analytical PCR on whole cells with the same primers. Internal primers for the kanamycin cassette (K2 and K3 [see Table 3]) were also used to reconfirm the disruption. This strain was named SP1.
TABLE 3.
Oligonucleotides used for cloning, gene deletion, and verification by PCR
| Primer | Sequence |
|---|---|
| A-YLR214W | AAAAATGTATTTAGGTTGCTTGACG |
| D-YLR214W | TATGAATTAAGGTTAGTGACGAGGC |
| A-YKL220C | ACAGGAAAACAAGTAAATTTTGACG |
| D-YKL220C | CAATTAACGTTTCATAAAATTTGCC |
| Fre1forw | ATGGTTAGAACCCGTGTATTATTC |
| Fre1rev | TTACCATGTAAAACTTTCTTC |
| Fre2forw | ATGCATTGGACGTCCATCTTG |
| Fre2rev | TCACCAGCATTGATACTCTTC |
| K2 | CGATAGATTGTCGCACCTG |
| K3 | CCATCCTATGGAACTGCCTC |
| CMPfre1forw | CATGGATCCAAAATGGTTAGAACCCGTG |
| CMPfre1rev | CATGTCGACTTACCATGTAAAACTTTCTTC |
| GAL1p_c | ATTGTTAATATACCTCTATACTTTAAC |
FRE2 knockout.
The procedure followed to disrupt the gene FRE2 (YKL220C) was similar to the one described above. Primers A-YKL220C and D-YKL220C (Table 3) were used to amplify by PCR the YKL220C::KanMX4 allele in the S. cerevisiae strain Y07039. The PCR product was used to transform the S. cerevisiae CEN.PK 113-7D strain, and correct integration of the cassette was scored by the presence of the YKL220C::KanMX4 band (2,323 bp) and the absence of the YKL220C wild-type band (2,842 bp) following analytical PCR on whole cells with the same primers. This strain was named SP2.
FRE1/FRE2 double knockout.
The vector pAG32, containing the hygromycin resistance gene HphMX4, was digested with the restriction enzymes BglII and EcoRV. The digested DNA was used to switch the selective marker of the gene replacement cassette in S. cerevisiae Y07039 from KanMX4 to HphMX4, resulting in strain SP3. The replacement of KanMX with HphMX4 was confirmed by PCR. SP3 chromosomal DNA was used to amplify the YKL220C::HphMX4 cassette, which was used to transform the SP1 strain (already carrying the YLR214W::KanMX4 cassette), resulting in the double mutant SP4.
RNA analysis.
Total cellular mRNA was prepared from yeast cells grown for 6 h in NDM, electrophoresed on 1.5% (wt/vol) agarose MOPS (morpholinepropanesulfonic acid)-formaldehyde gels (29), and blotted onto nylon membranes by vacuum transfer. Hybridization was carried out using a PstI fragment of 718 bp from pSP1 as a probe for FRE1 or a HindIII fragment of 682 bp from pSP2 as a probe for FRE2. The probes were labeled according to standard procedures (34). Densiometer scanning was performed using the Integrated Density Analysis program from the EagleSight software, version 3.2 (Stratagene, CA).
Construction of the pSH65-FRE1 vector.
The open reading frame (ORF) of FRE1 was amplified by PCR with the primers CMPfre1forw and CMPfre1rev. CMPfre1forw contains one BamHI site, and CMPfre1rev contains one SalI site; the FRE1 ORF was cloned into the vector pSH65 (20) by using the same restriction sites. The FRE1 ORF was directionally cloned between the GAL1-10 promoter and the CYC1 terminator in the vector pSH65, which is a CEN6/ARSH4 low-copy-number vector carrying the Bler phleomycin resistance gene for selection in yeast. Correct clones were verified by sequencing. A clone named pSP3 (Table 2) was selected for further studies.
Transformation of the Δfre1 strain with the plasmid pSP3 (pSH65-FRE1).
Cells of the strain SP1 were transformed with the plasmid pSP3 and plated onto YPD solid medium containing 30 μg/ml phleomycin. Ten colonies were checked by analytical PCR using the primers GAL1p_c and CMPfre1rev. The method described in the “The SixPack Guidelines” of the EUROFAN project (http://mips.gsf.de/proj/eurofan/eurofan_1/b0/home_requisites/guideline/sixpack.html) was used. The GAL1p_c and CMPfre1rev primers form a 2.1-kb PCR product only if the FRE1 ORF is present in the correct orientation with respect to the GAL1-10 promoter in pSH65. One of the positive strains was named SPcmp-FRE1 (Table 1) and was used in further studies.
RESULTS
Decolorization by growing yeast cultures.
Growing cultures of S. cerevisiae completely decolorized the tested azo dye in ca. 8.5 h. Figure 1A illustrates the yeast cells' growth curve and the pH variation and dye absorbance in the supernatant medium. A diauxic growth was observed, with a specific growth rate of 0.175 h−1 when growing in glucose and of 0.013 h−1 after the switch to ethanol utilization. The decolorization progress was unaffected by previous exposure of the cells to the dye (results not shown). Similar observations have been described earlier for Candida zeylanoides (31) and Issatchenkia occidentalis (30). The confirmation that color loss was due to the reductive cleavage of the azo bond in the dye molecules was provided by the detection of the related aromatic amines by high-performance liquid chromatography analysis, as shown in previous work (31).
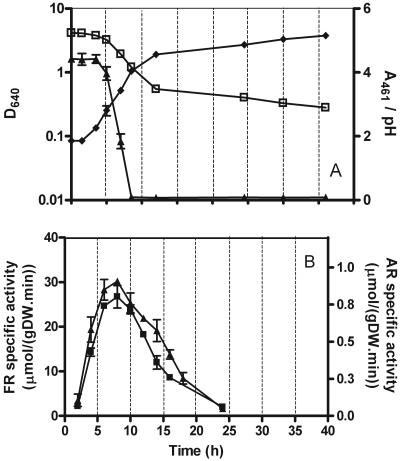
FIG. 1.
Decolorization progress and effect of growth stage on ferric reductase and azo reductase specific activities. (A) Time course of cell growth, measured as attenuance at 640 nm (D640 [⧫]); pH variation (pH [□]); and progress of decolorization, measured as dye absorbance at 461 nm (A461 [▴]). S. cerevisiae was grown at 26°C and 120 rpm in NDM containing 0.2 mM dye. (B) Variation of ferric reductase (FR [▪]) and azo reductase (AR [▴]) specific activities in cells of S. cerevisiae harvested at the specified times, expressed as μmol (g of cells [dry weight])−1 min−1. The cells were grown in NDM at 26°C and 120 rpm.
The effect of the growth phase on specific ferric and azo reductase activities was determined by assaying cells harvested from growing cultures at different incubation times. The results are shown in Fig. 1B, and despite the difference in the absolute values, the two curves are closely parallel at all times. Both have an activity peak in the late exponential growth phase, which is also when the fastest decrease of dye concentration in the incubation medium is observed.
Effect of iron concentration on specific ferric and azo reductase activities.
The progress of decolorization by growing cultures was measured in incubation media with different concentrations of iron(III), supplied as the EDTA chelate. Increasing iron concentrations resulted in much-delayed decolorization. As seen in Fig. 2A, total decolorization required over 50 h in the presence of 1.0 mM iron(III), in contrast with the 8.5 h required in NDM without iron addition. In media containing 2.5 mM iron(III), dye concentration decreased only ca. 20% in 50 h. For concentrations above 2.5 mM iron(III), we observed precipitation of the iron in the medium. The reduced decolorizing activity of the cells grown at higher iron concentrations was not due to impaired growth or loss of cell viability since cell counting in aliquots of the different cultures, collected after 28 h of growth, produced identical numbers of viable cells.
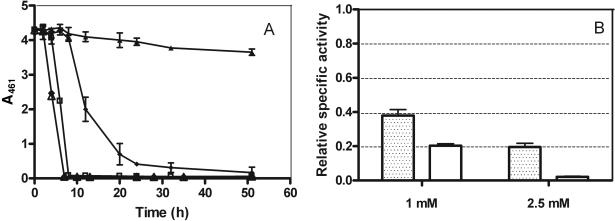
FIG. 2.
Iron(III)-dependent decolorization and activities of ferric reductase and azo reductase. (A) Time course of dye decolorization in the presence of 1.0 mM (⧫) and 2.5 mM (▴) iron(III). Cells were grown at 26°C and 120 rpm in NDM with 0.2 mM dye, and iron was supplied as the EDTA chelate at the specified concentrations. Control experiments were performed without the addition of iron to the medium (□) and in media supplemented with EDTA, either at 1 mM (⋄) or 2.5 mM (▵). The effect was monitored by measuring dye absorbance at 461 nm (A461). (B) Specific activity assays of ferric reductase (dotted bars) and azo reductase (plain bars) were performed with cells harvested after 6 h of growth in NDM at 26°C and 120 rpm. Growth media contained either 1.0 mM or 2.5 mM iron(III). Specific activities were calculated relative to those of cells grown without additional iron(III). Error bars show the standard deviations of results from three independent determinations.
Azo and ferric reductase activities were also measured in cells harvested after 6 h of growth from media with different iron concentrations. Cells were collected at this point because of the peak in activities of both enzymes around this time. The results in Fig. 2B show that the production of both activities was repressed by iron, in a concentration-dependent manner: azo reductase activities were reduced to ca. 20% at 1 mM iron and to 2% at 2.5 mM iron, despite the growth stimulation at higher Fe concentrations (data not shown). These observations point to an additional link between these two activities.
Effect of deletion of FRE1 and FRE2 genes on the activities of ferric and azo reductases.
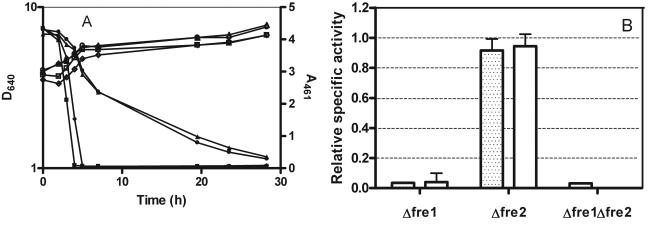
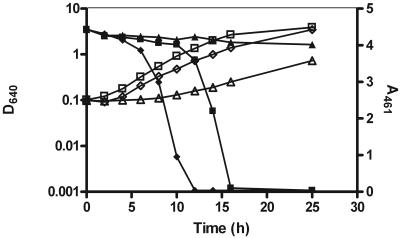
The S. cerevisiae Δfre1, Δfre2, and Δfre1 Δfre2 mutant strains have impaired growth in iron-deficient media. In order to overcome this problem, decolorization assays with the mutant strains were performed with high-density suspensions of pregrown cells, as described in Materials and Methods. Under these conditions, both the wild-type strain and the Δfre2 mutant achieved complete decolorization in ca. 5 h. Therefore, deletion of the FRE2 gene had a negligible effect on the decolorization process under our experimental conditions. In contrast, the Δfre1 and Δfre1 Δfre2 strains showed much-reduced decolorizing activities, requiring more than 45 h to completely remove the color from the medium (Fig. 3A). The azo reductase activity assays with the different strains allowed similar conclusions. As seen in Fig. 3B, the specific activity of the Δfre2 mutant reached the same order of magnitude as that of the wild type whereas those of the Δfre1 and Δfre1 Δfre2 strains were negligible. The ferric reductase assays produced very similar results, as seen in Fig. 3B. These results demonstrate the importance of the FRE1 gene product in the decolorizing activity of the yeast cells.
FIG. 3.
Deletion of FRE1 and FRE2 genes affects decolorization progress and ferric reductase and azo reductase activities. (A) Cells were grown at 26°C and 120 rpm in NDM with 0.2 mM dye. Cell growth was measured as attenuance at 640 nm (D640; open symbols), and decolorization progress was assessed by measuring dye absorbance at 461 nm (A461; closed symbols): wild type, diamonds; Δfre1 strain, triangles; Δfre2 strain, squares; and Δfre1 Δfre2 strain, circles. (B) Activities of the ferric reductase (dotted bars) and azo reductase (plain bars) of FRE mutant strains were calculated relative to those of cells of the reference strain, all grown in NDM at 26°C and 120 rpm and harvested after 6 h of growth.
FRE1 expression in S. cerevisiae.
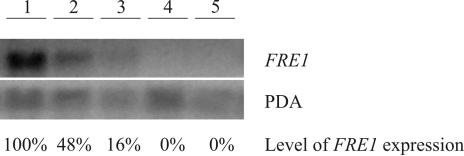
The expression of FRE1 was monitored by Northern blot analysis (Fig. 4). In cells of wild-type strain S. cerevisiae CEN.PK 113-7D grown in the absence of added iron, a strong mRNA signal against an FRE1 probe was revealed, proving the expression of this gene. Wild-type cells grown in the presence of added iron showed decreased FRE1 mRNA levels with increasing iron concentrations in the range between 1.0 and 2.5 mM. Therefore, iron seems to regulate the expression of the FRE1 gene. As expected, in cells of S. cerevisiae Δfre1 and Δfre1 Δfre2 deletion strains, no FRE1 mRNA was detected.
FIG. 4.
Northern blot analysis of FRE1 transcriptional level. Cells used for RNA extraction were harvested after 6 h of growth in NDM at 26°C and 120 rpm, with or without iron addition. Each lane contained 20 μg of total RNA, and PDA1 (38) served as an internal standard. Lane 1, wild type; lane 2, wild type with 1 mM iron(III) added to the growth medium; lane 3, wild type with 2.5 mM iron(III) added to the growth medium; lane 4, Δfre1 strain; lane 5, Δfre1 Δfre2 strain. The percentage of FRE1 expression (average of results from two independent experiments) is relative to that in the wild-type strain grown in NDM without externally added iron.
Recovery of FRE1 activity.
To confirm that under our experimental conditions the recovery of the azo reductase activity is mainly associated with FRE1, the progress of decolorization was monitored in cultures of the wild-type strain, the Δfre1 strain, and the Δfre1 strain transformed with the plasmid pSP3 containing FRE1 under the promoter GAL1-10. The cells were grown in media with 20 g/liter galactose as a carbon source for activation of the GAL1-10 promoter. As seen in Fig. 5, the FRE1 gene complemented the phenotype of S. cerevisiae Δfre1 cells, restoring the ability to grow in medium without externally added iron, following a pattern similar to the one observed in the wild-type strain. In this assay, the wild-type and Δfre1 strains behaved as expected regarding decolorization abilities, with total removal by the wild type and negligible removal by the mutant strain. The transformed Δfre1(pSP3) strain, although with a small delay in the start of the decolorization process, was able to fully decolorize the dye. This small difference could be due to distinct regulatory properties of the two promoters. These experiments provide the evidence that FRE1 is responsible for the azo reductase activity of the intact yeast cells under our operational conditions.
FIG. 5.
Restoration of FRE1 activity. Cells were grown at 26°C and 120 rpm in NDM with 0.2 mM dye and with 20 g/liter galactose as a carbon source for activation of the GAL1-10 promoter. Cell growth was measured as attenuance at 640 nm (D640; open symbols), and decolorization progress was assessed by measuring dye absorbance at 461 nm (A461; closed symbols): wild type, diamonds; Δfre1 strain, triangles; and SPcmp-FRE1, squares.
DISCUSSION
PMRSs are ubiquitous, being expressed in all living cells, including bacteria and cyanobacteria, yeasts, algae, and plant and animal cells (8, 26). These systems are linked to several vital cellular functions, including growth control, iron uptake, apoptosis, bioenergetics, transformation, and hormone responses (2, 5, 28). Some of these roles may be linked to the maintenance of appropriate NAD(P)+/NAD(P)H cytoplasmic ratios. In fact, an increase in the glycolytic flux, leading to an accumulation of NADH in the cytoplasm, induces an increase of PMRS activity (28). A number of such systems have been described, such as NADH:ascorbate free-radical oxidoreductase, NADH:ubiquinone oxidoreductase, and ferric reductase, among others (26, 28). However, it is not clear whether different phenomenological enzyme activities correspond to different PMRSs. On the contrary, it is generally accepted that several PMRSs are multifunctional (5, 8, 28).
The FRE1-dependent ferric reductase activity of intact yeast cells is inversely regulated by iron(III) concentration through the transcriptional activators Aft1p and Aft2p (33, 42). Our decolorization experiments with media containing additional iron revealed a considerable increase in the time required for complete dye removal and a negative effect of iron(III) on the azo reductase activity of yeast cells. Ferric reductase activities also decreased, as expected, but the effect of increased iron concentration on the azo reductase activities was more pronounced.
Both ferric reductase (23) and yeast azo reductase display an activity peak in the exponential growth phase. This is not an unexpected observation since many enzymes involved in cell growth have peak activities in this phase, when concentrations of intracellular reductants are also high.
The use of the strains defective in the genes encoding structural components of the transmembrane ferric reductase, FRE1 and FRE2, unequivocally demonstrated that Fre1p is a major component of the azo reductase system. In contrast, Fre2p was less important in azo reduction, at least under our assay conditions. Our observation is in agreement with previous reports that the FRE1 gene accounts for 80 to 98% of the ferric reductase activity (6, 7). Nevertheless, growing cultures of the Δfre1 strain and of the double-deletion mutant still showed low decolorizing capabilities. A residual ferric reductase activity has been explained by postulating the existence of an excreted reductase activity (15) which, however, has never been described. An alternative explanation has been provided by Lesuisse and colleagues (25), who have shown that the excretion of anthranilic and 3-hydroxyanthranilic acids was correlated with the extracellular ferric reductase activity. Whether those or other extracellular reductants participate in azo dye reduction requires further investigation. The insignificant participation of Fre2p in the ferric and azo reductase activities measured in this work (cells harvested after 6 h of growth) is probably due to the fact that the FRE2 gene is expressed primarily after 8 to 10 h of growth whereas the expression of FRE1 is highest in cells grown for up to 6 h (14). Therefore, the effect of FRE2 was not investigated at the present stage of our work.
It must be taken into account that the ferric reductase activity of intact yeast cells does not depend exclusively on one or more transmembrane proteins encoded by FRE genes. The in vivo association of the Fre1p component with the NAD-phosphorylating kinase Utr1p (21) is now generally accepted, since increased ferric reductase activity is observed only when FRE1 and UTR1 are overexpressed together (23). It has therefore been suggested that Utr1p is the supplier of NADP to the ferric reductase system (26). This is also consistent with the existence of the NADPH binding motif in Fre1p (12, 23, 35), suggesting that NADPH is the electron donor for iron reduction.
In conclusion, this work strongly indicates that the Fre1p-dependent reductase system of the yeast plasma membrane is an important component of the azo reductase activity in intact S. cerevisiae cells harvested between mid- and late exponential growth phases. Further information on the azo reductase system will be provided by examining the effect of known inhibitors of the ferric reductase, by establishing the nature of the electron donor, and by searching other components affecting the in vivo fully functional system. For example, it has been demonstrated that the ferric reductase activity in isolated plasma membranes is due to NADPH dehydrogenase (diaphorase) activity and that Fre1p, per se, has no reductase activity (23). Additionally, it has been shown that activation of the in vivo ferric reductase system requires the integrity of the Ras/cyclic AMP pathway (24). Interestingly, among several laboratory strains of S. cerevisiae, the only strain with decolorizing activity was the CEN.PK113-7D strain, which has a mutation on the CYR1 gene encoding the enzyme adenylate cyclase (37).
Acknowledgments
P.A.R. gratefully acknowledges a scholarship from the European BIOEFTEX Project.
We thank Sónia Barbosa for technical help with the Northern blot experiments and Björn Johansson for expert help in all stages of this work. P.A.R. thanks André Gouffeau for a fruitful discussion and critical reading of the manuscript. We acknowledge the help of Paulo Silva with the artwork.
REFERENCES
- 1.Anderson, G. J., A. Dancis, D. G. Roman, and R. D. Klausner. 1994. Ferric iron reduction and iron uptake in eucaryotes: studies with the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Adv. Exp. Med. Biol. 356:81-89. [DOI] [PubMed] [Google Scholar]
- 2.Baker, M. A., and A. Lawen. 2000. Plasma membrane NADH-oxidoreductase system: a critical review of the structural and functional data. Antioxid. Redox Signal. 2:197-212. [DOI] [PubMed] [Google Scholar]
- 3.Blaiseau, P. L., E. Lesuisse, and J. M. Camadro. 2001. Aft2p, a novel iron-regulated transcription activator that modulates, with Aft1p, intracellular iron use and resistance to oxidative stress in yeast. J. Biol. Chem. 276:34221-34226. [DOI] [PubMed] [Google Scholar]
- 4.Chung, K. T., and S. E. Stevens, Jr. 1993. Degradation of azo dyes by environmental microorganisms and helminths. Environ. Toxicol. Chem. 12:2121-2132. [Google Scholar]
- 5.Crane, F. L., I. L. Sun, M. G. Clark, C. Grebing, and H. Low. 1985. Transplasma-membrane redox systems in growth and development. Biochim. Biophys. Acta 811:233-264. [DOI] [PubMed] [Google Scholar]
- 6.Dancis, A., R. D. Klausner, A. G. Hinnebusch, and J. G. Barriocanal. 1990. Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:2294-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dancis, A., D. G. Roman, G. J. Anderson, A. G. Hinnebusch, and R. D. Klausner. 1992. Ferric reductase of Saccharomyces cerevisiae: molecular characterization, role in iron uptake, and transcriptional control by iron. Proc. Natl. Acad. Sci. USA 89:3869-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.del Castillo-Olivares, A., I. Nunez de Castro, and M. A. Medina. 2000. Dual role of plasma membrane electron transport systems in defense. Crit. Rev. Biochem. Mol. Biol. 35:197-220. [DOI] [PubMed] [Google Scholar]
- 9.Eide, D. J. 2000. Metal ion transport in eukaryotic microorganisms: insights from Saccharomyces cerevisiae. Adv. Microb. Physiol. 43:1-38. [DOI] [PubMed] [Google Scholar]
- 10.Eide, D. J. 1998. The molecular biology of metal ion transport in Saccharomyces cerevisiae. Annu. Rev. Nutr. 18:441-469. [DOI] [PubMed] [Google Scholar]
- 11.Entian, K., and P. Koetter. 1998. Yeast mutant and plasmid collections, p. 431-449. In A. Brown and M. Tuite (ed.), Methods in microbiology, vol. 26. Academic Press, San Diego, Calif. [Google Scholar]
- 12.Finegold, A. A., K. P. Shatwell, A. W. Segal, R. D. Klausner, and A. Dancis. 1996. Intramembrane bis-heme motif for transmembrane electron transport conserved in a yeast iron reductase and the human NADPH oxidase. J. Biol. Chem. 271:31021-31024. [DOI] [PubMed] [Google Scholar]
- 13.Furniss, B. S., A. J. Hannaford, W. G. Smith, and A. R. Tatchell. 1989. Vogel's textbook of practical organic chemistry, 5th ed. Longman Group, London, United Kingdom.
- 14.Georgatsou, E., and D. Alexandraki. 1999. Regulated expression of the Saccharomyces cerevisiae Fre1p/Fre2p Fe/Cu reductase related genes. Yeast 15:573-584. [DOI] [PubMed] [Google Scholar]
- 15.Georgatsou, E., and D. Alexandraki. 1994. Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:3065-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgatsou, E., L. A. Mavrogiannis, G. S. Fragiadakis, and D. Alexandraki. 1997. The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J. Biol. Chem. 272:13786-13792. [DOI] [PubMed] [Google Scholar]
- 17.Gibbs, C. R. 1976. Characterisation and application of ferrozine iron reagent as a ferrous iron indicator. Anal. Chem. 48:1197-1201. [Google Scholar]
- 18.Gietz, R. D., and R. A. Woods. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87-96. [DOI] [PubMed] [Google Scholar]
- 19.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 20.Gueldener, U., J. Heinisch, G. J. Koehler, D. Voss, and J. H. Hegemann. 2004. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed]
- 21.Kawai, S., S. Suzuki, S. Mori, and K. Murata. 2001. Molecular cloning and identification of UTR1 of a yeast Saccharomyces cerevisiae as a gene encoding an NAD kinase. FEMS Microbiol. Lett. 200:181-184. [DOI] [PubMed] [Google Scholar]
- 22.Kruckeberg, A. L., L. Ye, J. A. Berden, and K. van Dam. 1999. Functional expression, quantification and cellular localization of the Hxt2 hexose transporter of Saccharomyces cerevisiae tagged with the green fluorescent protein. Biochem. J. 339(Pt. 2):299-307. [PMC free article] [PubMed] [Google Scholar]
- 23.Lesuisse, E., M. Casteras-Simon, and P. Labbe. 1996. Evidence for the Saccharomyces cerevisiae ferrireductase system being a multicomponent electron transport chain. J. Biol. Chem. 271:13578-13583. [DOI] [PubMed] [Google Scholar]
- 24.Lesuisse, E., B. Horion, P. Labbe, and F. Hilger. 1991. The plasma membrane ferrireductase activity of Saccharomyces cerevisiae is partially controlled by cyclic AMP. Biochem. J. 280(Pt. 2):545-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesuisse, E., M. Simon, R. Klein, and P. Labbe. 1992. Excretion of anthranilate and 3-hydroxyanthranilate by Saccharomyces cerevisiae: relationship to iron metabolism. J. Gen. Microbiol. 138(Pt. 1):85-89. [DOI] [PubMed] [Google Scholar]
- 26.Ly, J. D., and A. Lawen. 2003. Transplasma membrane electron transport: enzymes involved and biological function. Redox Rep. 8:3-21. [DOI] [PubMed] [Google Scholar]
- 27.Martins, L. J., L. T. Jensen, J. R. Simon, G. L. Keller, and D. R. Winge. 1998. Metalloregulation of FRE1 and FRE2 homologs in Saccharomyces cerevisiae. J. Biol. Chem. 273:23716-23721. [DOI] [PubMed] [Google Scholar]
- 28.Medina, M. A., A. del Castillo-Olivares, and I. Nunez de Castro. 1997. Multifunctional plasma membrane redox systems. Bioessays 19:977-984. [DOI] [PubMed] [Google Scholar]
- 29.Newman, A. 1994. Analysis of pre-mRNA splicing in yeast. IRL Press, Oxford, United Kingdom.
- 30.Ramalho, P. A., M. H. Cardoso, A. Cavaco-Paulo, and M. T. Ramalho. 2004. Characterization of azo reduction activity in a novel ascomycete yeast strain. Appl. Environ. Microbiol. 70:2279-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramalho, P. A., H. Scholze, M. H. Cardoso, M. T. Ramalho, and A. M. Oliveira-Campos. 2002. Improved conditions for the aerobic reductive decolourisation of azo dyes by Candida zeylanoides. Enzyme Microb. Technol. 31:848-854. [Google Scholar]
- 32.Rotrosen, D., C. L. Yeung, T. L. Leto, H. L. Malech, and C. H. Kwong. 1992. Cytochrome b558: the flavin-binding component of the phagocyte NADPH oxidase. Science 256:1459-1462. [DOI] [PubMed] [Google Scholar]
- 33.Rutherford, J. C., S. Jaron, and D. R. Winge. 2003. Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J. Biol. Chem. 278:27636-27643. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Shatwell, K. P., A. Dancis, A. R. Cross, R. D. Klausner, and A. W. Segal. 1996. The FRE1 ferric reductase of Saccharomyces cerevisiae is a cytochrome b similar to that of NADPH oxidase. J. Biol. Chem. 271:14240-14244. [DOI] [PubMed] [Google Scholar]
- 36.Stolz, A. 2001. Basic and applied aspects in the microbial degradation of azo dyes. Appl. Microbiol. Biotechnol. 56:69-80. [DOI] [PubMed] [Google Scholar]
- 37.Vanhalewyn, M., F. Dumortier, G. Debast, S. Colombo, P. Ma, J. Winderickx, P. Van Dijck, and J. M. Thevelein. 1999. A mutation in Saccharomyces cerevisiae adenylate cyclase, Cyr1K1876M, specifically affects glucose- and acidification-induced cAMP signalling and not the basal cAMP level. Mol. Microbiol. 33:363-376. [DOI] [PubMed] [Google Scholar]
- 38.Wenzel, T. J., A. W. Teunissen, and H. Y. de Steensma. 1995. PDA1 mRNA: a standard for quantitation of mRNA in Saccharomyces cerevisiae superior to ACT1 mRNA. Nucleic Acids Res. 23:883-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamaguchi-Iwai, Y., A. Dancis, and R. D. Klausner. 1995. AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 14:1231-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamaguchi-Iwai, Y., M. Serpe, D. Haile, W. Yang, D. J. Kosman, R. D. Klausner, and A. Dancis. 1997. Homeostatic regulation of copper uptake in yeast via direct binding of MAC1 protein to upstream regulatory sequences of FRE1 and CTR1. J. Biol. Chem. 272:17711-17718. [DOI] [PubMed] [Google Scholar]
- 41.Yamaguchi-Iwai, Y., R. Stearman, A. Dancis, and R. D. Klausner. 1996. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 15:3377-3384. [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaguchi-Iwai, Y., R. Ueta, A. Fukunaka, and R. Sasaki. 2002. Subcellular localization of Aft1 transcription factor responds to iron status in Saccharomyces cerevisiae. J. Biol. Chem. 277:18914-18918. [DOI] [PubMed] [Google Scholar]
- 43.Yun, C. W., M. Bauler, R. E. Moore, P. E. Klebba, and C. C. Philpott. 2001. The role of the FRE family of plasma membrane reductases in the uptake of siderophore-iron in Saccharomyces cerevisiae. J. Biol. Chem. 276:10218-10223. [DOI] [PubMed] [Google Scholar]