Abstract
Phylogenetic analyses of 16S rRNA sequences of sponge-associated cyanobacteria showed them to be polyphyletic, implying that they derived from multiple independent symbiotic events. Most of the symbiont sequences were affiliated to a group of Synechococcus and Prochlorococcus species. However, other symbionts were related to different groups, such as the Oscillatoriales.
Although both cyanobacteria and sponges have a very long evolutionary history (1, 3), little is known about the identity and phylogeny of cyanobacterial sponge symbionts. For marine sponges, symbiosis with cyanobacteria can be obligate or nonobligate according to the sponge species (2). Thus far, all attempts at culturing sponge-associated cyanobacteria have failed (13; unpublished data). It is thus not known whether sponge-associated cyanobacteria are able to survive outside their host. The most common sponge-associated cyanobacterium was not found in water samples (13, 15). In Chondrilla australiensis, this cyanobacterium is transmitted vertically (through sponge eggs) (14). Additionally, a study showing cospeciation between Dysidea species and their associated cyanobacteria (12) supports the hypothesis that sponges and associated cyanobacteria are coevolving. Few studies sequenced the 16S rRNA gene of sponge-associated cyanobacteria: Diaz (5) identified the first two cyanobacteria from marine sponges, Webb and Maas (16) found that the cyanobacteria inhabiting Mycale hentscheli were phylogenetically related to Cyanobacterium stanieri and species of Synechocystis and Prochloron, and Hentschel et al. (7) found that seven sequences from cyanobacteria inhabiting the sponges Aplysina aerophoba and Theonella swinhoei could be divided into two clades (Synechococcus/Prochlorococcus and Pleurocapsa). The aim of the present study was to increase our understanding of the diversity of sponge-associated cyanobacteria and to determine their phylogenetic position.
Sponge samples from 16 species were collected from four locations: the Caribbean (Bahamas, 26°33′N, 77°52′W), Mediterranean (Rapallo, Italy, 44°18′N, 9°12′E), Red Sea (Elat, 31°35′N, 34°54′E), and Western Indian Ocean (Zanzibar, 06°09′S, 39°11′E). Aposymbiotic specimens (specimens that do not contain cyanobacterial symbionts), growing in dark caves or overhangs, were also collected for two sponge species (Petrosia ficiformis and Xestospongia muta). Those samples, collected at short distances from symbiotic specimens, were used as negative controls, to ensure that 16S rRNA sequences were derived from true symbionts and not from surface-associated cyanobacteria or digested cyanobacteria. Photosynthetic activity inside the living sponge tissue was tested by pulse amplitude modulated fluorometry (Diving PAM, Walz, Germany). Tissue samples (1 cm3) were rinsed twice in 100% ethanol, and kept in 100% ethanol at 4°C. DNA was extracted following the procedure of Bernatzky and Tanksley (4). 16S rRNA was amplified with the primers 361F (5′-GAATTTTCCGCAATGGGC-3′) and 1459R (5′-GGTAAYGACTTCGGGCRT-3′) (5). Fragments 1,060 bp long were cloned in the PTZ57R/T vector (Fermentas). Twenty clones per individual were amplified using M13 universal primers. The PCR products were digested with restriction enzymes ApaI and HaeIII. One clone was sequenced for each pattern present in more than 10% of the clones.
Neither photosynthetic activity nor amplification of the 16S rRNA gene was obtained for aposymbiotic specimens. On the contrary, photosynthetic activity was recorded for all the other samples. Up to three different cyanobacterial clones were sequenced per individual sponge investigated (with a sequence homology of 90 to 99.7%). Similarly, Webb and Maas (16) showed four closely related (99.1 to 99.8%) cyanobacterial clones in the sponge Mycale hentscheli. Different cyanobacterial types from the same individual were usually phylogenetically close to each other (<1 to 2% sequence divergence), but in one case (Lendenfeldia dendyi) the symbionts were very divergent: two types were in group 5, while the third type was in group 2, with 10% sequence divergence from the former two (Fig. 1). Closely related cyanobacterial types should not be the result of Taq polymerase errors or cloning bias, since only patterns present in more than one clone were sequenced. However, the true cyanobacterial diversity in sponges might be underestimated. Nevertheless our results are in agreement with other studies (13, 16). For example, Usher et al. (13) also identified only one cyanobacterial type in the sponge Petrosia ficiformis.
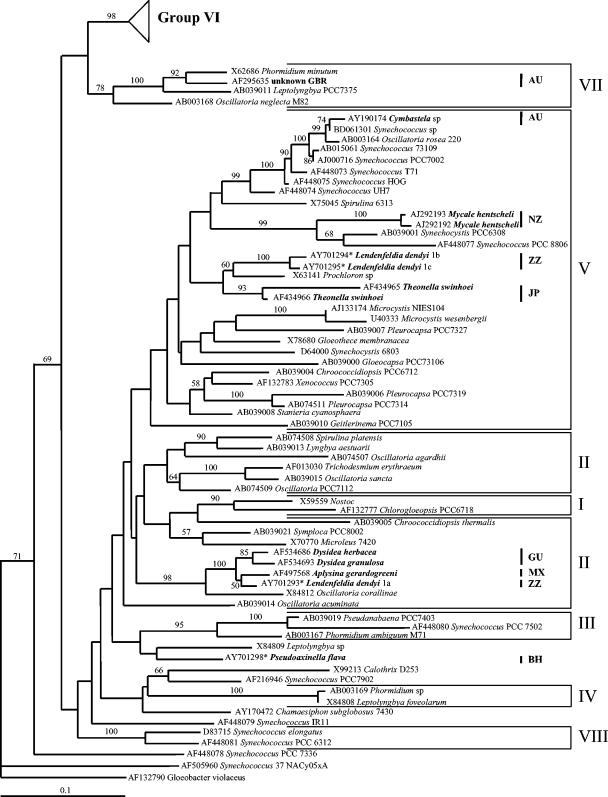
FIG. 1.
Maximum-likelihood tree of sponge-associated cyanobacteria based on 16S rRNA. Bootstrap values above 50% are indicated. Sponge-associated cyanobacteria (which were named following their host species) are indicated in bold. The group numbers follow Honda et al. (8) and Robertson et al. (10). The triangle at the top represents group 6, detailed in Fig. 2. The individual identification number is given at the end of the sequence; different letters represent different clones obtained for the same sponge individual. Sequences from this work are indicated by a star after the accession number. The sampling localities of sponge-associated cyanobacteria are indicated by bars: AU, Australia; BH, Bahamas; GU, Guam; JP, Japan; MX, Mexican Pacific; NZ, New Zealand; ZZ, Zanzibar.
A maximum-likelihood (ML) analysis (6) was done based on 147 taxa and 1,396 nucleotides. The sequences were chosen to include (i) representatives of cyanobacterium diversity, (ii) all sponge-associated cyanobacterium sequences overlapping with the new 16S rRNA sequences, and (iii) representatives of the Synechococcus and Prochlorococcus diversity because sponge symbionts have been suggested to belong to these genera. BLAST searches were also conducted for each sponge sequence in order to include in the analysis the most similar sequences available in the databanks. The GTR model with a gamma distribution (four categories), and a proportion of the invariant site was found to be the most appropriate using Modeltest version 3.06 (9). The ML tree was reconstructed in an iterative way using PAUP* version 4.0b10 (11). First a heuristic search was conducted using the ML parameters identified by Modeltest; Modeltest topology as a starting tree as well as NNI branch swapping. The topology found at the end of the search was used to identify new parameters. These new parameters and topology were used to conduct a new search until convergence. The best ML parameters found were then used to compute 500 bootstrap replicates starting with a neighbor-joining tree and setting the maximum number of tree to 1 to reduce computation time.
The phylogenetic tree obtained in this study was in agreement with the results of other studies that divided cyanobacteria into seven or eight major lineages (8, 10), except that group 2 (8) was here paraphyletic (Fig. 1). Sponge-associated cyanobacteria were found to be polyphyletic. They were divided into 13 lineages spread among various groups of cyanobacteria (Fig. 1 and 2). The major clade of sponge-associated cyanobacteria (37 strains from 18 different sponge species collected from a wide range of geographic regions: Australia, Caribbean, French and Italian Mediterranean coast, Red Sea, and Zanzibar) was strongly affiliated with group 6 (Prochlorococcus, Synechococcus, and Microcystis) (8). Three additional sponge-associated cyanobacteria (from Chondrilla spp. and Haliclona sp.) were also part of group 6, but were closer to free-living Synechococcus than to the other sponge symbionts (Fig. 2). A second group of sponge-associated cyanobacteria (consisting of four sequences from Dysidea herbacea, Dysidea granulosa, Lendenfeldia dendyi, and Aplysina gerardogreeni, originating from Guam, the Mexican Pacific, and Zanzibar) were affiliated with group 2 (morphologically classified in the order Oscillatoriales). A few other sponge-associated cyanobacteria were affiliated with group 5 (Chroococcales, Oscillatoriales, Pleurocapsales, and Prochlorales) (8), but they did not form a monophyletic group (Fig. 1). One additional cyanobacterium (AF295635) from an Australian sponge was found affiliated with marine group 7 (8). Finally, a sequence of a cyanobacterium associated with the sponge Pseudoaxinella flava appeared unrelated to any major group of cyanobacteria (Fig. 1). The polyphyletic origin of sponge-associated cyanobacteria indicates that they derived from multiple independent symbiotic events, involving several different cyanobacterial types and/or host sponges.
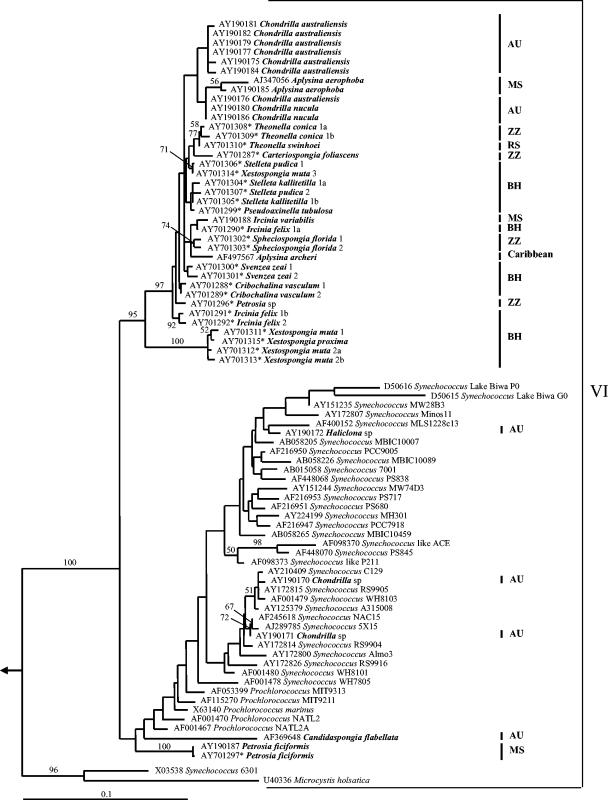
FIG. 2.
“Group 6” detailed 16S rRNA-based maximum-likelihood tree of sponge-associated cyanobacteria. An arrow indicates the outgroup (cf. Fig. 1). Bootstrap values above 50% are indicated. Sponge-associated cyanobacteria (which were named following their host species) are indicated in bold. The individual identification number is given at the end of the sequence; different letters represent different clones obtained for the same sponge. Sequences from this work are indicated by a star after the accession number. The sampling localities of sponge-associated cyanobacteria are indicated by bars: AU, Australia; BH, Bahamas; MS, Mediterranean Sea; RS, Red Sea; ZZ, Zanzibar.
The phylogenetic relationships between the cyanobacterial sequences of group 6 suggest only a partial specificity of cyanobacteria to the host sponges. Two specimens of Petrosia ficiformis, one collected in Italy (this study) and another in France (13), were found to be 99% identical, and closely related (bootstrap percentage [BP] = 100%). Three cyanobacterial types from two Bahamian Xestospongia muta specimens were strongly affiliated with each other and with that from the congeneric species Xestospongia proxima (BP = 100%). Cyanobacteria from two congeneric Theonella species (T. swinhoei and T. conica) were affiliated to each other (BP = 77%). Finally, six cyanobacteria associated with Chondrilla australiensis (13, 15) were closely affiliated (BP = 49%). These results may also support the idea of coevolution between sponges and their symbionts. However, cyanobacteria associated with Theonella swinhoei from Japan and the Red Sea clustered in different groups (Fig. 1 and 2). In this large-scale phylogenetic analysis, sponge-associated cyanobacterium clades deriving from distant geographic location (as the clades in group 2 and 6) are most probably true symbionts, while the symbiotic nature of sponge cyanobacteria that are closely related to free-living species (e.g. those in Pseudoaxinella flava) remains uncertain.
In the future, it will be interesting to examine whether cyanobacterial sponge symbionts from different lineages perform diverse functions in this symbiosis. In addition, reconstruction of sponge molecular phylogeny will enable testing for coevolution of cyanobacteria and their host sponges.
Nucleotide sequence accession numbers.
The sequences of the clones used in this study have been deposited in the GenBank database under accession no. AY701287 to AY701315.
Acknowledgments
This work was supported by grant 2000-321 from the United States-Israel Binational Science Foundation (BSF) to M. Ilan, S. Beer, and J. R. Pawlik. L. Steindler received a scholarship from the Rieger Foundation for Environmental Studies that assisted in the travel costs for sampling sponge specimens.
S. Beer is thanked for his editorial comments on the manuscript, and T. Pupko is acknowledged for providing access to his computers. We thank J. R. Pawlik (University of North Carolina, Wilmington), who invited L. Steindler, M. Ilan, and S. Beer to participate in research cruises aboard the R/V Seward Johnson, which enabled us to collect many samples for this study. The staff of the Institute of Marine Sciences (Zanzibar, Tanzania) is thanked for helpful hospitality. We are grateful to C. Cerrano (Universita'degli Studi di Genova, Genova, Italy), who helped to collect Petrosia ficiformis specimens. S. Zea (Universidad Nacional de Colombia) is acknowledged for assisting in identification of some of the Bahamian sponges. L. Steindler expresses her appreciation to S. Schuster, who first introduced her to the “molecular world.”
REFERENCES
- 1.Adell, T., V. A. Grebenjuk, M. Wiens, and W. E. G. Muller. 2003. Isolation and characterization of two T-box genes from sponges, the phylogenetically oldest metazoan taxon. Dev. Genes Evol. 213:421-434. [DOI] [PubMed] [Google Scholar]
- 2.Arillo, A., G. Bavestrello, B. Burlando, and M. Sara. 1993. Metabolic integration between symbiotic cyanobacteria and sponges—a possible mechanism. Mar. Biol. 117:159-162. [Google Scholar]
- 3.Awramik, S. M. 1992. The oldest records of photosynthesis. Photosynth. Res. 33:75-89. [PubMed] [Google Scholar]
- 4.Bernatzky, R., and S. D. Tanksley. 1986. Methods for detection of single or low copy sequences in tomato on Southern blots. Plant Mol. Biol. Rep. 4:37-41. [Google Scholar]
- 5.Diaz, M. C. 1997. Molecular detection and characterization of specific bacterial groups associated with tropical sponges, p. 1399-1402. In H. A. Lessios and I. G. Macintyre (ed.), Proceedings of the 8th International Coral Reef Symposium, Panama, 24 to 29 June 1996, vol. 2. Smithsonian Tropical Research Institute, Balboa, Ancon, Panama. [Google Scholar]
- 6.Felsenstein, J. 1985. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 7.Hentschel, U., J. Hopke, M. Horn, A. B. Friedrich, M. Wagner, J. Hacker, and B. S. Moore. 2002. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68:4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honda, D., A. Yokota, and J. Sugiyama. 1999. Detection of seven major evolutionary lineages in cyanobacteria based on the 16S rRNA gene sequence analysis with new sequences of five marine Synechococcus strains. J. Mol. Evol. 48:723-739. [DOI] [PubMed] [Google Scholar]
- 9.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 10.Robertson, B. R., N. Tezuka, and M. M. Watanabe. 2001. Phylogenetic analyses of Synechococcus strains (cyanobacteria) using sequences of 16S rDNA and part of the phycocyanin operon reveal multiple evolutionary lines and reflect phycobilin content. Int. J. Syst. Evol. Microbiol. 51:861-871. [DOI] [PubMed] [Google Scholar]
- 11.Swofford, D. L. 2000. PAUP*. Phylogenetic analysis using parsimony (* and other methods). Version 4b4a. Sinauer Associates, Sunderland, Mass.
- 12.Thacker, R. W., and S. Starnes. 2003. Host specificity of the symbiotic cyanobacterium Oscillatoria spongeliae in marine sponges, Dysidea spp. Mar. Biol. 142:643-648. [Google Scholar]
- 13.Usher, K. M., J. Fromont, D. C. Sutton, and S. Toze. 2004. The biogeography and phylogeny of unicellular cyanobacterial symbionts in sponges from Australia and the Mediterranean. Microb. Ecol. 48:167-177. [DOI] [PubMed] [Google Scholar]
- 14.Usher, K. M., J. Kuo, J. Fromont, and D. C. Sutton. 2001. Vertical trans-mission of cyanobacterial symbionts in the marine sponge Chondrilla australiensis (Demospongiae). Hydrobiologia 461:15-23, 35. [Google Scholar]
- 15.Usher, K. M., S. Toze, J. Fromont, J. Kuo, and D. C. Sutton. 2004. A new species of cyanobacterial symbiont from the marine sponge Chondrilla nucula. Symbiosis 36:183-192. [Google Scholar]
- 16.Webb, V. L., and E. W. Maas. 2002. Sequence analysis of 16S rRNA gene of cyanobacteria associated with the marine sponge Mycale (Carmia) hentscheli. FEMS Microbiol. Lett. 207:43-47. [DOI] [PubMed] [Google Scholar]