Abstract
In a previous study, tomato race 3 (T3) strains of Xanthomonas perforans became predominant in fields containing both X. euvesicatoria and X. perforans races T1 and T3, respectively. This apparent ability to take over fields led to the discovery that there are three bacteriocin-like compounds associated with T3 strains. T3 strain 91-118 produces at least three different bacteriocin-like compounds (BCN-A, BCN-B, and BCN-C) antagonistic toward T1 strains. We determined the relative importance of the bacteriocin-like compounds by constructing the following mutant forms of a wild-type (WT) T3 strain to evaluate the antagonism to WT T1 strains: Mut-A (BCN-A−), Mut-B (BCN-B−), Mut-C (BCN-C−), Mut-AB, Mut-BC, and Mut-ABC. Although all mutant and WT T3 strains reduced the T1 populations in in planta growth room experiments, Mut-B and WT T3 were significantly more effective. Mutants expressing BCN-B and either BCN-A or BCN-C reduced T1 populations less than mutants expressing only BCN-A or BCN-C. The triple-knockout mutant Mut-ABC also had a significant competitive advantage over the T1 strain. In pairwise-inoculation field experiments where plants were coinoculated with an individual mutant or WT T3 strain and the T1 strain, the mutant strains and the WT T3 strain were reisolated from more than 70% of the lesions. WT T3 and Mut-B were the most frequently reisolated strains. In field experiments where plants were group inoculated with Mut-A, Mut-B, Mut-C, Mut-ABC, and WT T1 and T3 strains, Mut-B populations dominated all three seasons. In greenhouse and field experiments, the WT and mutant T3 strains had a selective advantage over T1 strains. Bacterial strains expressing both BCN-A and BCN-C appeared to have a competitive advantage over all other mutant and WT strains. Furthermore, BCN-B appeared to be a negative factor, with mutant T3 strains lacking BCN-B having a selective advantage in the field.
Bacterial spot of tomatoes and peppers is caused by Xanthomonas campestris pv. vesicatoria. Pohronezny and Volin (34) estimated that as high as 50% loss of marketable fruit was due to bacterial spot on tomatoes. Control of bacterial spot of tomato is difficult when high temperatures and high moisture exist. Bactericides, such as fixed coppers and streptomycin, have provided the primary means of control (30, 45, 46); however, streptomycin-resistant mutants and copper-tolerant strains became prevalent on treated plants (46). Marco and Stall (30) showed that many Xanthomonas euvesicatoria strains were tolerant of copper and addition of mancozeb to copper sprays improved control of the copper-tolerant strains (6, 30). However, they also showed that this treatment is insufficient when conditions favorable for disease development exist.
On tomato, three races, designated tomato race 1 (T1), T2, and T3, were identified based on their reaction on three tomato genotypes (25, 26, 44). Recent reclassification of the xanthomonads associated with bacterial spot of tomato has changed the species classification of these races. Currently, T1 strains are classified as X. euvesicatoria, T2 strains are classified as X. vesicatoria, and T3 strains are now classified as Xanthomonas perforans (24). T1 and T2 were first identified in 1990 (55) and are distributed worldwide (3), although only T1 strains were present in Florida until the early 1990s. In 1991, T3 appeared in Florida fields (26) and eventually predominated (22). This new race had antagonistic activity toward T1 strains (8) and was determined to be a new group designated group C (23). Genetically, T3 strains are most closely related to T1 strains based on DNA similarity values (23).
In a previous study, wild type (WT) T3 strains were shown to be antagonistic toward WT T1 strains (8). Tudor-Nelson et al. (52) identified three cosmid clones, designated BCN-A, BCN-B, and BCN-C, which were found to confer the ability to inhibit a sensitive T1 strain in plate assays. These compounds were determined to have narrow inhibition spectra and fit the definition of a bacteriocin described by Reeves (36) based on the following criteria: (i) the presence of a biologically active protein moiety, (ii) inducibility with mitomycin C, and (iii) non-self inhibition (52). The three clones isolated by Tudor-Nelson et al. (52) were unique in activity and specificity based on differential inhibition of selected sensitive T1 and T2 strains (52). It has been speculated that the bacteriocins may confer in part or completely this competitive advantage.
Of the bacteriocins which have been well characterized in the field of bacteriology, the most studied of the gram-negative bacteria bacteriocins is the colicin family found in Escherichia coli. The genes encoding the bacteriocins of E. coli are found on plasmids. The modes of action of these colicins range from pore formation in the cell membrane to nuclease activity against DNA, rRNA, and tRNA (54). Most of the bacteriocins produced by gram-negative bacteria are large proteins and, in the case of the colicins, range in size from 449 to 629 amino acids for the pore-forming bacteriocins and 178 to 777 amino acids for the nuclease bacteriocins (54).
The extensive research on the colicin family of bacteriocins has offered many helpful insights into the evolution of bacteriocins and the relationships between the producing and sensitive strains. Theories have developed from studying this family related to the evolution of bacteriocins. It was hypothesized that new bacteriocin-producing strains arise through recombination (4, 47), as well as mutations (37, 38). It is likely that many closely related bacteria acquire bacteriocins by horizontal transfer. For instance, homology was detected between a bcnA-specific probe and genomic DNA of X. campestris pv. glycines strain 1717 (51). In addition, a subclone from X. perforans T3 strain 91-118 that conferred immunity to BCN-A also conferred immunity to X. campestris pv. glycines (16, 52).
There has been conjecture by Riley and Wertz (39) that bacteriocins may serve as anticompetitors enabling the strain to invade an established microbial community. This appears to suggest that bacteriocins may play a defensive role in limiting competition among bacteria, essentially giving the producing cells a competitive advantage over the nonproducing sensitive strains. The ability of bacteriocin-producing strains to reduce sensitive strains when grown together has been shown previously (13, 18, 33, 42, 49, 53).
In this study, we show the relative importance of bacteriocins of T3 strains of X. perforans for their antagonistic activity toward T1 strains in greenhouse and field studies and present information that one bacteriocin-like compound may be a negative factor in competition between strains.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The origin and relevant characteristics of the bacterial strains and plasmids used in this study are listed in Table 1. Strains of X. euvesicatoria and X. perforans were grown on nutrient agar (NA) medium (Difco Laboratories, Detroit, MI) at 28°C. Strains of E. coli were grown on Luria-Bertani (LB) medium at 37°C (32). All strains were stored in 20% glycerol in sterile tap water at −80°C. Antibiotics were used to maintain selection for resistance markers at the following concentrations: tetracycline, 12.5 μg/ml; rifampin, 100 μg/ml; spectinomycin, 50 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 34 μg/ml; streptomycin, 200 μg/ml; nalidixic acid, 50 μg/ml.
TABLE 1.
Xanthomonas euvesicatoria, X. perforans strains and mutants, E. coli, and plasmids
| Strain or plasmid | Relevant characteristics | Source or referencea |
|---|---|---|
| X. euvesicatoria | ||
| E3-1 | Nalr Smr | This study |
| 91-106 | 50 | |
| X. perforans | ||
| 91-118R | Rifr | 52 |
| Mut-A | BCN− A− B+ C+ Rifr Spr | This study |
| Mut-B | BCN− A+ B− C+ Rifr Kmr | 52 |
| Mut-C | BCN− A+ B+ C− Rifr Cmr | This study |
| Mut-AB | BCN− A− B− C+ Rifr Spr Kmr | This study |
| Mut-BC | BCN− A+ B− C− Rifr Kmr Cmr | This study |
| Mut-ABC | BCN− A− B− C− Rifr Spr Kmr Cmr | This study |
| E. coli | ||
| DH5α | F−recA | BRL |
| C2110 | Nalr | BRL |
| λPIR | Sprori R6K sacB RK2 replicon | UB |
| Plasmids | ||
| pBluescript-KS+ | Phagemid, pUC derivative; Ampr | Stratagene |
| pLAFR3 | Tcrrlx+ RK2 replicon | BJS |
| pRK2073 | Sprtra+mob+ | 11 |
| pXV120 | pLAFR3 cosmid clone from X. campestris pr. vesicatoria strain 91-118 (BCN-C+) | 50 |
| pXV442 | pLAFR3 cosmid clone from X. campestris pr. vesicatoria strain 91-118 (BCN-B+) | 50 |
| pXV519 | pLAFR3 cosmid clone from X. campestris pr. vesicatoria strain 91-118 (BCN-A+ IMM-A+) | 51 |
| pXV12.1 | pLAFR119 EcoRI subclone derived from pXV519 (BCN-A+ IMM-A+) | 51 |
| pXV8.0 | pLAFR119 KpnI/EcoRI subclone (8.0 kb) derived from pXV519 (BCN-A+ IMM-A+) | 51 |
| pXV8.9 | pLAFR119 KpnI subclone (8.9 kb) derived from pXV442 (BCN-B+) | 51 |
| pXV5.1 | pLARF119 HindIII/EcoRI subclone (5.1 kb) derived from pXV120 (BCN-C+) | 51 |
| pXV6.0 | pLAFR119 KpnI/EcoRI subclone (6.0 kb) derived from pXV8.9 (BCN-B+) | 51 |
| pXV1.7 | pLAFR119 SalI/EcoRI subclone (1.7 kb) derived from pXV5.1 (BCN-C+) | 51 |
| pXV12.1-92 | pXV12.1 with Spr cassette insertion lacking BCN-A activity | 51 |
| pXV442-255 | pXV6.0 with Kmr cassette insertion lacking BCN-B activity | 51 |
| pXV1.7-515 | pXV1.7 with Cmr cassette insertion lacking BCN-C activity | This study |
BRL, Bethesda Research Laboratories, Gaithersburg, MD; Stratagene, Stratagene Inc., La Jolla, CA; BJS, B. J. Staskawicz, University of California, Berkeley; UB, U. Bonas, Martin- Luther-Universität, Halle, Germany.
DNA manipulations.
Standard techniques for molecular cloning were used as described by Sambrook et al. (40). Restriction endonuclease digestions were performed using the manufacturer's specifications. All enzymes were obtained from Promega (Madison, WI). All DNA extractions were as described by Sambrook et al. (40). T4 DNA ligase was used according to the manufacturer's specifications (Promega, Madison, WI). Constructs were transformed into competent E. coli DH5α cells that were prepared as described by Sambrook et al. (40) and stored at −80°C until used for transformation.
Transposon mutagenesis.
Insertion mutagenesis of BCN-A+ was performed using a transposon Tn3HoHo1 derivative of pHOKmGus (2). E. coli strain DH5α carrying pShe (Cmr) and pDooey (Ω Spr-encoding gene cassette) was transformed with the BCN-A+-containing plasmid pXV519 (Table 1). Transformants (Tcr Spr) were mated with E. coli C2110 (Nalr) using pRK2073 as the helper plasmid. Matings were incubated on LB agar plates for 8 h at 37°C and then plated on LB medium amended with tetracycline, spectinomycin, and nalidixic acid to select for mutated plasmids. Insertion derivatives were mobilized into X. euvesicatoria strain ME-90 (Rifr Spr) as previously described and screened for loss of BCN+ activity against strain 91-106. The insertion of the Spr-encoding gene in pBCN-A−-92 (Fig. 1) was located within the region amplified by two PCR primers (132 and 135) specific to open reading frame 4 identified previously (51).
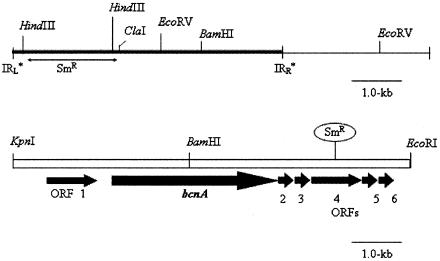
FIG. 1.
Illustration of transposable element in plasmid pDooey and the 8.0-kb BCN-A+ fragment (pXV8.0 BCN-A+ 8.0-kb KpnI/EcoRI). (Top) Location of pBCN-A−-92, used for insertion of spectinomycin resistance in the transposable element region to knock out BCN-A+. (Bottom) Partial physical map of pXV8.0 subclone and approximate location of the Tn3-Spr insert. The insertion resulted in a loss of BCN-A+ function. The approximate location of the insertion was determined using primers p132 and p135 from sequencing (51). IRL and IRR, left and right inverted repeats, respectively; ORF, open reading frame.
Generation of BCN-A mutants.
Triparental matings were performed using pRK2073Sp or pRK2013Km as the helper (Table 1), E. coli DH5α containing pXV12. 1-92 (pXV519Tc with insertion of an Spr cassette for inactivation of BCN-A) as the donor, and 91-118Rif, Mut-BRif Sp, and Mut-BCRif Km Cm as the recipients. Marker exchange was achieved using standard methods (40). The candidate colonies were screened for loss of BCN+ activity and confirmed for insertion by Southern hybridization (43) (using subclones of BCN-A, BCN-B, and BCN-C as the probes) and/or PCR using primers BCN-1 (5′-CCCAAGCTTCACGGTCGTCGCCCGCACC-3′, where the HindIII restriction sequence is in boldface type) and BCN-2 (5′-CCCAAGCTTACTGCAGCGCCGACGCACC-3′).
Generation of BCN-B mutants.
Triparental matings were performed using pRK2013Km as the helper (Table 1), E. coli DH5α containing pXV442-255 (pXV442Tc with insertion of a Kmr cassette for inactivation of BCN-B) as the donor, and 91-118Rif as the recipient. Marker exchange was achieved using standard methods (40). The candidate colonies were screened for loss of BCN+ activity and confirmed for insertion by Southern hybridization (using subclones of BCN-A, BCN-B, and BCN-C as the probes) and/or PCR (with primers BCN-1 and BCN-2).
Generation of BCN-C mutants.
Activity in the BCN-C+ in pXV1.7 (Table 1) was knocked out using PCR deletion rearrangement mutagenesis as described by Kanifa et al. (27). A 515-bp region of the 1.7-kb fragment within pXV1.7 was deleted using divergent primers BCN-3 (5′-CACCAGCGGCAACTACACCATCC-3′) and BCN-4 (5′-CATAGACGCGGTCCTGGGTGCC-3′) to create pXV1. 7-515 (Table 1).
A gene cassette with chloramphenicol resistance (35) was ligated into the BCN-C− fragment using HindIII sites added by divergent primers BCN-3 and BCN-4. After addition of the chloramphenicol cassette, the final product was 2.0 kb long. The fragment was excised from pBluescript using flanking restriction sites (ApaI and BamHI) and ligated into suicide vector pOK1 (19). The WT T3 strain was transformed using a suicide vector technique described by Kanifa et al. (27). To confirm replacement of the BCN-C region, PCR primers were designed to amplify flanking regions of the crossover event within primers BCN-1 and BCN-2. Successful replacement mutants yielded a PCR product of ∼800 bp, whereas the WT strain yielded an ∼515-bp fragment. The BCN-C knockout candidates were screened for BCN activity after conjugation into X. euvesicatoria strain ME90 (Rifr Kmr) (T1 strain) using the previously described triparental mating technique.
Southern hybridization analysis.
DNA digested with restriction enzymes for Southern analysis was electrophoresed in 0.75% agarose gel according to standard protocols (40). DNA was denatured in 0.4 N NaOH-0.6 M NaCl for 30 min and neutralized for 30 min in 0.5 M Tris-1.5 M NaCl. The DNA was transferred from gels to Nytran membranes (Schleicher & Schuell, Keene, NH) by Southern blotting (40). Hybridized DNA was detected with the Genius nonradioactive DNA labeling and detection kit (Boehringer Mannheim Biochemicals, Indianapolis, IN) as described by the manufacturer. BCN+ clones were used as the probes in cross-hybridization experiments with each other. Probes were labeled by the random primer method (9) for incorporation of digoxigenin-dUTP.
Population dynamics of WT T1 and T3 strains and mutant T3 strains in leaf tissue.
Strains were grown in nutrient broth (NB; Difco Laboratories, Detroit, MI) for 18 h, harvested by centrifugation, and resuspended in sterile tap water or 0.01 M MgSO4. Mutant and WT T3 strains were inoculated at 5 ×106 CFU/ml into leaflets of 6-week-old seedlings of the tomato cultivar Bonny Best. Leaflets were infiltrated (15 leaflets each) using a hypodermic syringe and needle as described previously (22). Following inoculation, plants were incubated at 24°C to 28°C. Three samples were taken for each treatment at each time interval. Populations were quantified by macerating 1-cm2 leaf disks in 1 ml sterile tap water and dilution plating onto NA medium amended with the appropriate antibiotic. Plates were incubated at 28°C, and colonies were counted after 48 to 72 h. Samples were assayed at 24-h intervals for 96 h. Population data were transformed to log10 values, and standard errors were determined. The overall growth rate was determined by calculating the area under the population progress curve (AUPPC). The AUPPC is a modification of the area under the disease progress curve (AUDPC) which has been used to analyze disease progress (41): standardized AUPPC = Σ [(xi + xi − 1)/2](ti − ti − 1), where x is population density in log CFU per milliliter and t is time in hours.
Significant differences were determined using the standardized AUPPC, and statistical analyses were done using SAS (SAS Inc., Cary, NC). Each experiment was conducted three times.
Effects of T3 and mutant strains on internal leaf populations of a T1 strain.
Strains were grown in NB for 18 h, harvested by centrifugation, and resuspended in sterile tap water or 0.01 M MgSO4. The WT T3 or a mutant strain and a T1 strain at 5 × 107 CFU/ml and 5 × 106 CFU/ml, respectively, were infiltrated into leaflets (15 leaflets each) of 6-week-old seedlings of the tomato cultivar Bonny Best. Prior experiments were conducted to maximize the timing and concentration of populations to optimize the bacteriocin activity (52). The WT T3 or mutant strain was inoculated 12 h prior to inoculation with the T1 strain. Each treatment consisted of three replications. Following inoculation, plants were incubated at 24°C to 28°C. Populations were determined as described above. Samples were assayed at 12-h intervals for 96 h. Population data were analyzed following log10 transformation, and standard errors were determined. Significant differences were determined using the standardized AUPPC, and statistical analyses were done using SAS (SAS Inc., Cary, NC). Each experiment was conducted three times.
Pairwise inoculation of WT T3 or a mutant T3 strain with a T1 strain in the field.
In the first experiment, individual knockout mutants (Mut-A, Mut-B, and Mut-C), a triple-knockout mutant (Mut-ABC), and the WT T3 strain (91-118) were evaluated individually for antagonism toward a T1 (E3-1) strain. Strains were grown in NB for 24 h, harvested by centrifugation, and resuspended in a solution of 0.01 M MgSO4. The bacterial suspensions were adjusted to 5 ×107 CFU/ml, and the surfactant Silwet (Loveland Industries Inc., Greeley, CO) was added to a final concentration of 0.025%. Six-week-old seedlings of the tomato cultivar FL47 were used in the field experiments. Plants were dipped into suspensions of the WT T1 strain 24 h prior to inoculation with the T3 mutant or WT strains to give an advantage to the T1 strains.
The experimental design was a completely randomized block design consisting of four replications. Raised beds were 0.91 m wide and were covered with black plastic mulch. Plots were arranged in paired rows that were 1.83 m apart from center to center, and the paired rows were spaced 7.32 m apart. Plots within the paired rows were spaced at 6.1 m. Plantings were spaced 45.7 cm apart and contained 20 plants.
The experiment consisted of five treatments: (i) WT T3 and WT T1, (ii) Mut-A and WT T1, (iii) Mut-B and WT T1, (iv) Mut-C and WT T1, and (v) Mut-ABC and WT T1. Plants were transplanted 2 days after the second dip inoculation. Lesions were sampled by removing symptomatic leaflets from the uppermost portion of the tomato plants and isolating bacteria from 30 lesions from 10 to 15 leaflets to determine the incidence of WT or mutant T3 strains and WT T1 strains in each lesion. Individual lesions were macerated in 50 μl of sterile 0.01 M MgSO4 solution and quadrant streaked for isolation of individual colonies for identification. Significant differences for both seasons (spring and fall 2001) were determined using the area under the incidence progress curve (AUIPC). The AUIPC is also a modified AUDPC to analyze bacterial populations recovered from lesions rather than disease progress: standardized AUIPC = Σ [(xi + xi − 1)/2](ti − ti − 1), where x is the arcsin of the percent recovery and t is hours after inoculation.
Asymptomatic leaf tissue was also sampled every 2 weeks beginning 50 days after transplanting to determine epiphytic populations in each plot. Three samples were taken from each plot, each containing three or more leaflets. Each sample was weighed, placed in a sterile Falcon 50-ml polystyrene tube (Becton Dickinson, Rutherford, NJ) containing 10 ml of peptone buffer (Difco Laboratories, Detroit, MI) per g of tissue (31), and shaken at 200 rpm for 30 to 45 min. Serial 10-fold dilutions were made in sterile 0.01 M MgSO4 solution. A 50-μl aliquot of each dilution was plated onto Tween B medium (31). After incubation at 28°C for 4 to 5 days, three to five colonies typical of X. euvesicatoria were selected and plated on medium amended with appropriate antibiotics to select for mutant, WT T1, and WT T3 strains. Significance was determined by using the area under the epiphytic progress curve (AUEPC). The AUEPC is another modified AUDPC to analyze the bacterial epiphytic populations rather than disease progress: standardized AUEPC = Σ [(xi + xi − 1)/2](ti − ti − 1), where x is the arcsin of the percent recovery and t is days after inoculation.
In a group inoculation experiment, all the T3 mutant strains (Mut-A, Mut-B, Mut-C, and Mut-ABC), the WT T3 (91-118) strain, and the WT T1 (E3-1) strain were inoculated into the same plants. This experiment consisted of four replications. For the first two seasons, the plants were dip inoculated with each strain 2 days prior to being transplanted. In the third season, each strain was inoculated on individual leaflets by leaf infiltration to eliminate any problems due to multiple dip inoculations. Symptomatic leaf tissue was collected as indicated in the first experiment. Significant differences for the three seasons (spring and fall in 2001 and fall in 2002) were determined using SAS (SAS Inc., Cary, NC) for the AUIPC.
Symptomatic leaflets from each plot were sampled every 2 weeks to determine the incidence of the WT T1 strain, the WT T3 strain, or the mutant strain in lesions. Thirty lesions were sampled from 10 to 20 leaflets randomly collected in each plot. Bacteria were reisolated by macerating a lesion in 75 μl of sterile deionized water and streaking onto NA medium amended with pentachloronitrobenzene at 134 μg/ml and/or cycloheximide at 50 μg/ml. Individual colonies were plated onto two media, NA medium amended with the appropriate antibiotics for the mutant T3 strains (91-118Rif, Mut-ARif Sp, Mut-BRif Km, Mut-CRif Cm, and Mut-ABCRif Sp Km Cm) and NA medium amended with antibiotics for the T1 strain (E3-1Nal Sm). The percent recovery was determined based on the ratio of WT or mutant T3 strains compared to the total lesions as follows: percent recovery of mutant strains = no. of mutant strains isolated/total no. of strains isolated.
RESULTS
Relative importance of BCN+ in planta.
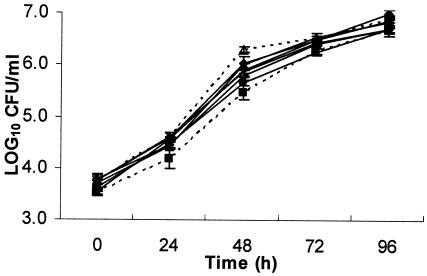
Greenhouse experiments were conducted to determine the effects of mutations on growth rate and their ability to reduce T1 populations in planta. In the growth curve assay, WT T3 had the highest overall growth rate; however, it was not significantly different from Mut-A, Mut-BC, or Mut-ABC (Table 2; Fig. 2). According to the AUPPC, WT T3 was only significantly different from Mut-B, Mut-C, and Mut-AB.
TABLE 2.
In planta population analysis of T3 and T1 strains within leaf tissue
| Strain | AUPPC
|
|
|---|---|---|
| Growth curve assay | Antagonism assay | |
| H2O | 598b (a)c | |
| wt T3 | 567a (a) | 266 (f) |
| Mut-A | 546 (ab) | 545 (b) |
| Mut-B | 534 (b) | 313 (f) |
| Mut-C | 533 (b) | 413 (d) |
| Mut-AB | 535 (b) | 474 (cd) |
| Mut-BC | 552 (ab) | 368 (e) |
| Mut-ABC | 553 (ab) | 504 (bc) |
AUPPC for growth rate assay for each strain over a 96-h period.
AUPPC for the assay based on WT T1 strain populations recovered over a 96-h period from leaflets containing both a test strain and a T1 strain.
Values followed by the same letter are not significantly different according to Duncan's multiple-range test. (P = 0.05) using SAS (Cary, NC) program 8.1.
FIG. 2.
Growth rates of X. perforans mutant T3 (Mut-A, ▴; Mut-B, ×; Mut-C, ⋄; Mut-AB, •; Mut-BC, +; Mut-ABC, Δ), WT T3 (⧫), and WT T1 (▪) strains in tomato cultigen Bonny Best over time.
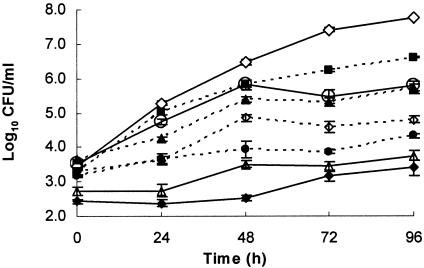
A second in planta assay was conducted to determine antagonistic activity of WT and mutant T3 strains toward the T1 strain (Fig. 3). The AUPPCs were based on the WT T1 strain population recovered. The T1 population that was coinoculated with water exhibited a normal growth curve over the 96-h sampling period and, based on the AUPPC (Table 2), had a significantly higher population level than all other treatments. The WT T3 strain and Mut-B provided the greatest reduction in T1 populations. Mut-A and Mut-ABC reduced T1 populations only slightly compared to the water control and were not considered significantly different from one another. Interestingly, Mut-AB and Mut-BC were both able to reduce T1 populations significantly more than Mut-A and Mut-C, respectively. In both cases, expression of BCN-B appeared to result in a less antagonistic bacterium.
FIG. 3.
Growth of X. euvesicatoria T1 strain 91-106 in tomato cultigen Bonny Best over time when inoculated 12 h after treatment with an X. perforans T3 mutant (Mut-A, ▪; Mut-B, Δ; Mut-C, +; Mut-AB, ▴; Mut-BC, •; Mut-ABC, ○), WT T3 (⧫), or water (⋄). Results are averages of three independent experiments.
In a separate experiment, we tested the effect of prior inoculation of a T1 strain or Mut-ABC against our WT T1 indicator strain to determine if the Mut-ABC strain behaved like a non-bacteriocin-expressing pathogenic tomato strain. We found that there were no significant differences between the Mut-ABC and WT T1 strains in reducing the indicator T1 strain (data not shown). The experiment was repeated once.
Pairwise inoculation of WT T3 or a mutant T3 strain with a T1 strain in the field.
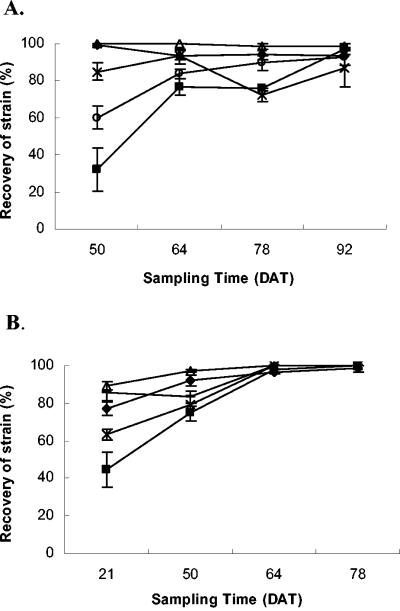
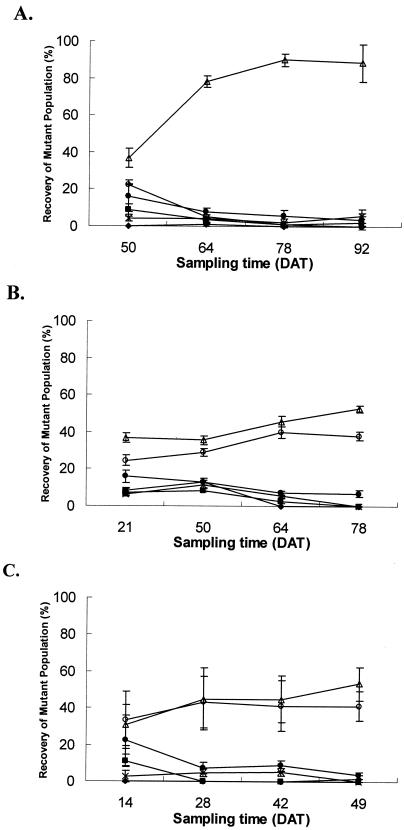
In the pairwise inoculation experiment, WT T1 populations were significantly reduced by all strains by the second sampling date (Fig. 4). For the first season, an average of 95 to 99% of WT T3 or Mut-B was recovered over WT T1 strains (Table 3). The lowest recovery of mutant strains occurred in plots receiving Mut-A and T1, although it still averaged over 70%. Mut-B and WT T3 91-118 strains gave the highest recovery overall.
FIG. 4.
Percent recovery (over four sampling periods) of mutant strains (Mut-A, ▪; Mut-B, Δ; Mut-C, +; Mut-ABC, ○) or WT T3 (⧫) from lesions of plants coinoculated with individual mutant strains or WT T3 and a T1 strain in the field. A, spring 2001; B, fall 2001. DAT stands for days after transplanting.
TABLE 3.
AUIPC and percent recovery in pairwise inoculation field experiments
| Strain | Pairwise inoculation
|
|||
|---|---|---|---|---|
| Spring 2001
|
Fall 2001
|
|||
| AUIPC | % Recovery | AUIPC | % Recovery | |
| WT T3 | 5,349 (a)a | 95 | 4,807 (bc) | 91 |
| Mut-A | 3,372 (c) | 70 | 4,095 (d) | 79 |
| Mut-B | 5,879 (a) | 99 | 5,385 (a) | 97 |
| Mut-C | 4,505 (b) | 85 | 4,557 (c) | 86 |
| Mut-ABC | 4,066 (b) | 81 | 5,001 (ab) | 92 |
Letters in parentheses indicate significance using proc analysis of variance (P = 0.05 using SAS).
In the second season, WT T3 and mutant strains were predominantly recovered (Fig. 4B). The AUIPC of WT T3 was not significantly different from Mut-ABC or Mut-C. Mut-B was recovered from approximately 97% of the lesions and was not significantly different from Mut-ABC. Mut-C was recovered from 86% of the lesions and was not significantly different from the WT T3 strain. Mut-A plots exhibited the lowest recovery of the mutant strain, with an average recovery of 70%, and had a significantly reduced AUIPC compared to all other strains.
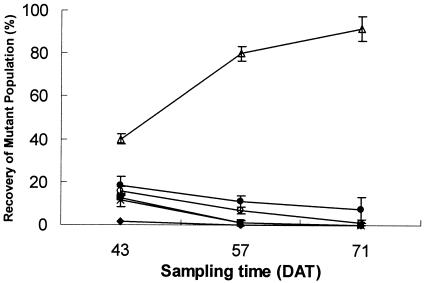
In the group inoculation experiment, both symptomatic (Fig. 5A; Table 4) and epiphytic (Fig. 6; Table 4) populations were sampled for the first season (spring 2001). The WT T1 strain had the lowest AUIPC for both sampling techniques (Table 4). Mut-B was recovered most frequently. The trends observed in the epiphytic populations in spring 2001 (Fig. 6) were similar to the symptomatic data in the spring of 2001 (Fig. 5A). The strain recovered at the highest frequency was Mut-B. WT T3 was not significantly different than Mut-ABC, Mut-A, or Mut-C. The AUEPC of the WT T1 strain was the lowest of all strains tested; however, it was not significantly different than Mut-A or Mut-C.
FIG. 5.
Percent recovery of symptomatic leaf populations of strains in plots with a mixture of mutant strains (Mut-A, ▪; Mut-B, Δ; Mut-C, ×; Mut-ABC, ○) and with a WT T1 (⧫) and T3 (•) strain in the field. A, spring 2001; B, fall 2001; C, fall 2002. DAT stands for days after transplanting.
TABLE 4.
AUIPC for group inoculation field experiments
| Strain | Symptomatic
|
Epiphytic, spring 2001 | ||
|---|---|---|---|---|
| Spring 2001 | Fall 2001 | Fall 2002 | ||
| WT T3 | 1,259.0 (b)a | 1,364.9 (c) | 1,026.8 (b) | 736.3 (b) |
| WT T1 | 75.4 (c) | 87.9 (e) | 26.4 (b) | 103.9 (c) |
| Mut-A | 835.7 (bc) | 885.8 (de) | 408.9 (b) | 374.6 (c) |
| Mut-B | 3,556.1 (a) | 2,641.8 (a) | 1,901.8 (a) | 1,949.2 (a) |
| Mut-C | 768.2 (bc) | 973.8 (d) | 508.7 (b) | 364.3 (c) |
| Mut-ABC | 1,173.8 (b) | 1,909.0 (b) | 1,875.8 (a) | 582.7 (bc) |
Letters in parentheses indicate significance using proc analysis of variance P = 0.05 using SAS.
FIG. 6.
Percent recovery of epiphytic populations of mutant strains in plots with a mixture of mutant strains (Mut-A, ▪; Mut-B, Δ; Mut-C, ×; Mut-ABC, ○) and with WT T1 (⧫) and T3 (•) strains. DAT stands for days after transplanting.
For the fall 2001 and 2002 seasons, only Mut-B and Mut-ABC were recovered at a frequency of over 20% of the samples. For the fall 2001 season, Mut-B was recovered at a significantly higher percentage than Mut-ABC; however, this significance was not found in fall 2002. Similarly WT T1, WT T3, Mut-A, and Mut-C had significance between strains only in fall 2001. For 2001, WT T3 and WT T1 were significantly higher and lower, respectively, than Mut-A and Mut-C; however, in the fall 2002 experiment there were no significant differences detected between these same strains.
DISCUSSION
In this study, we determined that three bacteriocin-like compounds, BCN-A, BCN-B, and BCN-C, provided the T3 strains with a competitive advantage over T1 strains. Furthermore, certain combinations of the bacteriocins were more effective in reducing T1 populations. The importance of the bacteriocins in antagonism toward T1 strains was observed when looking at the greenhouse experiments. The growth rate of the WT T3 strain was the highest of all the strains tested; however, there could be no correlation made between growth rate and bacteriocin expression.
It appears that BCN-B has a negative effect on WT T3 strains in the greenhouse antagonism experiment and in the field. This does not appear to result from a negative effect on growth rate. This was observed when comparing Mut-A and Mut-AB or Mut-C and Mut-BC in the growth room experiment. Mut-AB and Mut-BC both reduced the WT T1 strain more significantly than Mut-A and Mut-C. This effect was also observed in both field experiments, where Mut-B exhibited a higher percent recovery than all other strains, including WT T3, and when all mutants were competing against each other, Mut-B and Mut-ABC (the only BCN-B− mutants) predominated. This positive effect on antagonism by loss of BCN-B did not occur when strains lost either BCN-A or BCN-C. This leads to several questions as to the effect that BCN-B production has on the producing T3 strain. There are a few possible explanations: (i) BCN-B may affect virulence factors in the T3 strain; (ii) it may be toxic to the producing strains; or (iii) it may also affect the production or delivery of BCN-A and/or BCN-C in the producing T3 strain, lowering its antagonistic properties. Further research is necessary to determine if any of these explanations is correct.
It is unlikely that the first two theories are correct based on the in planta growth rate assay experiment. There were no consistent significant differences related to the presence or absence of BCN-B. There is, however, evidence in support of the third explanation in the in planta antagonism assay experiment, in which Mut-BC and Mut-AB (only expressing BCN-A and BCN-C, respectively) gave significantly lower AUPPCs compared to Mut-C and Mut-A (expressing BCN-AB and BCN-BC, respectively). Tudor-Nelson et al. (51, 52) previously noted the WT T3 strains produced a smaller zone of inhibition than a T1 strain expressing the BCN-A clone or Mut-B in plate assays. This may be an indication that BCN-B has an adverse effect on expression of BCN-A in a WT T3 background. Negative effects on producing strains have been observed previously in E. coli. Colicin V has been shown to cause a sickly appearance in E. coli strains not properly delivering the bacteriocin across the outer membrane (4).
The increase in antagonism toward expression of BCN-A and BCN-C together observed in the in planta assays was also observed in the field experiments, although less dramatically. In the field experiments with the pairwise inoculation of WT T3 or a mutant and a T1 strain, Mut-A and Mut-C strains reduced T1 populations less than the WT. This was consistently observed for Mut-A, although Mut-C only showed this reduced effect in the second season.
This study may suggest a possibility for developing a biological control strategy utilizing these compounds. The first step in developing a biological control strategy is to show a competitive advantage associated with the bacteriocins. In this study, X. perforans T3 strain bacteriocins have been shown to directly inhibit T1 strains in the greenhouse and the field. Bacteriocins have been observed in lab settings for many phytopathogenic bacteria, including Agrobacterium (28, 48), Burkholderia solanacearum (12), Corynebacterium (7, 15) Erwinia (5, 7, 20, 21), Pseudomonas (12, 14, 42), Xanthomonas (1, 10, 17, 56), and Vibrio (18); however, few bacteria have been shown to have a competitive advantage over the nonproducing strains in the field. Pseudomonas syringae strain PSW-1 was shown to express a bacteriocin that can suppress sensitive strains in bean hypocotyls at certain concentrations, and the bacteriocin activity can be isolated from infected hypocotyls of bean (42). Agrocin 84 is commercially available and is produced by Agrobacterium radiobacter strain K84 and has also been found to suppress sensitive strains by a bacteriocin that is actually a fraudulent adenine nucleoside analog with no protein component (49).
In this study, we have shown that T3 strains produce three bacteriocins, each of which is antagonistic toward T1 strains in the greenhouse and in the field; however, more work is necessary to characterize each bacteriocin in order to determine the exact genes responsible for the production and delivery of each bacteriocin, as well as the individual mode of action. As for the development of a biological control strategy utilizing these bacteriocins, there are several possibilities: (i) direct incorporation of these genes into the tomato genome, (ii) purification of BCN-A and BCN-C from supernatant may provide a source of prophylactic control of T1 strains as a foliar spray in the field, and (iii) expression of these bacteriocin-like compounds in a bacterium that colonizes plants epiphytically or in an attenuated strain. A study has demonstrated that a nonpathogenic T3 strain of X. perforans was able to reduce the severity of bacterial spot disease in the field (29). This promising result, along with the findings of this study, warrants further investigation into the development of a biological control strategy for bacterial spot disease.
Acknowledgments
This research was supported by the Florida Agricultural Experiment Station and USDA T-STAR (J. B. Jones, M. T. Momol, P. D. Roberts, and A. P. Hert, USDA 2003-34135-14077-S).
Footnotes
Journal series no. R-10634 from the Florida Agricultural Experiment Station.
REFERENCES
- 1.Ahn, E. J., and Y. S. Cho. 1996. Cloning of the bacteriocin gene from Xanthomonas campestris pv. glycines 8ra. Korean J. Plant Pathol. 47:480-487. [Google Scholar]
- 2.Bonas, U., R. Stall, and B. Staskawicz. 1989. Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol. Gen. Genet. 218:127-136. [DOI] [PubMed] [Google Scholar]
- 3.Bouzar, H., J. B. Jones, R. E. Stall, N. C. Hodge, and N. Daouzli. 1994. Physiological, chemical, serological, and pathologenic analyses of worldwide collection of Xanthomonas campestris pv. vesicatoria strains. Phytopathology 31:753-755. [Google Scholar]
- 4.Braun, V., H. Pilsl, and P. Groβ. 1994. Colicins: structures, modes of actions, transfer through membranes, and evolution. Arch. Microbiol. 161:199-206. [DOI] [PubMed] [Google Scholar]
- 5.Chaung, D. Y., A. G. Kyeremeh, Y. Gunji, Y. Takahara, Y. Ehara, and T. Kikumoto. 1999. Identification and cloning of an Erwinia carotovora subsp. carotovora bacteriocin regulator gene by insertion mutagenesis. J. Bacteriol. 181:1953-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conover, R. A., and N. A. Gerhold. 1981. Mixtures of copper and maneb or mancozeb for control of bacterial spot of tomato and their compatibility for control of fungus diseases. Proc. Fla. State Hortic. Soc. 94:154-156. [Google Scholar]
- 7.Echandi, E. 1976. Bacteriocin production by Corynebacterium michiganense. Phytopathology 66:430-432. [Google Scholar]
- 8.El-Morsy, G. A., G. C. Somodi, J. W. Scott, R. E. Stall, and J. B. Jones. 1994. Aggressiveness of Xanthomonas campestris pv. vesicatoria tomato race strains over tomato race 1 strains: evidence for antagonism. Phytopathology 84:551-556. [Google Scholar]
- 9.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 10.Fett, W. F., M. F. Dunn, G. T. Maher, and B. E. Maleef. 1987. Bacteriocins and temperate phage of Xanthomonas campestris pv. glycines. Curr. Microbiol. 16:137-144. [Google Scholar]
- 11.Figurski, D., and D. R. Helsinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frey, P., J. J. Smith, L. Albar, P. Prior, C. Marie, G. S. Saddler, D. Trigalet-Demery, and A. Triglet. 1996. Bacteriocin typing of Burkholdaria (Pseudomonas) solanacearum race 1 of the French West Indies and the correlation with genomic variation of the pathogen. Appl. Environ. Microbiol. 62:473-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goel, A. K., S. S. Sindhu, and K. R. Dadarwal. 1999. Bacteriocin-producing native rhizobia of green gram (Vigna radiata) having competitive advantage in nodule occupancy. Microbiol. Res. 154:43-48. [Google Scholar]
- 14.Govan, J. R. W. 1974. Studies on the pyocins of Pseudomonas aeruginosa: production of contractile and flexous pyocins in Pseudomonas aeruginosa. J. Gen. Microbiol. 80:17-30. [DOI] [PubMed] [Google Scholar]
- 15.Gross, D. C., and A. K. Vidaver. 1979. Bacteriocins of phytopathogenic Corynebacterium species. Can. J. Microbiol. 25:367-374. [DOI] [PubMed] [Google Scholar]
- 16.Heu, S., and Y. S. Cho. 1997. The characterization of genes which are responsible for the bacteriocin production in Xanthomonas campestris pv. glycines 8ra. Phytopathology 87:S41. [Google Scholar]
- 17.Heu, S., J. Oh, Y. Kang, S. Ryu, S. K. Cho, Y. Cho, and M. Cho. 2001. gly gene cloning and expression and purification of glycinecin A, a bacteriocin produced by Xanthomonas campestris pv. glycines 8ra. Appl. Environ. Microbiol. 67:4105-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoyt, P. R., and R. K. Sizemore. 1982. Demonstration of competitive dominance by a bacteriocin-producing strain of Vibrio harveyi. Appl. Environ. Microbiol. 44:653-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huguet, E., K. Hahn, K. Wengelnik, and U. Bonas. 1998. hpaA mutants of Xanthomonas campestris pv. vesicatoria are affected in pathogenicity but retain the ability to induce host-specific hypersensitive reaction. Mol. Microbiol. 29:1379-1390. [DOI] [PubMed] [Google Scholar]
- 20.Itoh, Y. J., Sugiura, K. Izaki, and H. Takahashi. 1982. Enzymological and immunological properties of pectin lyases from bacterocinogenic strains of Erwinia carotovora. Agric. Biol. Chem. 46:199-205. [Google Scholar]
- 21.Jabrane, A., L. Ledoux, P. Thonart, T. Deckers, and P. Lepoivre. 1996. The efficacy in vitro and in vivo of bacteriocin against Erwinia amylovora: comparison of biological and chemical control of fire blight. Acta Hortic. 411:355-359. [Google Scholar]
- 22.Jones, J. B., H. Bouzar, G. C. Somodi, R. E. Stall, K. Pernezny, G. El-Morsy, and J. W. Scott. 1998. Evidence for the preemptive nature of tomato race 3 of Xanthomonas campestris pv. vesicatoria in Florida. Phytopathology 88:33-38. [DOI] [PubMed] [Google Scholar]
- 23.Jones, J. B., H. Bouzar, R. E. Stall, E. C. Almira, P. D. Roberts, B. W. Bowen, J. Sudberry, P. M. Strickler, and J. Chun. 2000. Systematic analysis of xanthomonads (Xanthomonas spp.) associated with pepper and tomato lesions. Int. J. Syst. Evol. Microbiol. 50:1211-1219. [DOI] [PubMed] [Google Scholar]
- 24.Jones, J. B., G. H. Lacy, H. Bouzar, R. E. Stall, and N. W. Schaad. 2004. Reclassification of the Xanthomonas associated with bacterial spot disease of tomato and pepper. Syst. Appl. Microbiol. 27:755-762. [DOI] [PubMed] [Google Scholar]
- 25.Jones, J. B., R. E. Stall, and H. Bouzar. 1998. Diversity among xanthomonads pathogenic on pepper and tomato. Annu. Rev. Phytopathol. 36:41-58. [DOI] [PubMed] [Google Scholar]
- 26.Jones, J. B., R. E. Stall, J. W. Scott, G. C. Somondi, H. Bouzar, and N. C. Hodge. 1995. A third tomato race of Xanthomonas campestris pv. vesicatoria. Plant. Dis. 79:395-398. [Google Scholar]
- 27.Kanifa, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 28.Kerr, A., and K. Htay. 1974. Biological control of crown gall through bacteriocin production. Physiol. Plant Pathol. 4:37-44. [Google Scholar]
- 29.Liu, T. 1998. Biological control of tomato bacterial spot with a Hrp− mutant of Xanthomonas campestris pv. vesicatoria. M.S. thesis. University of Florida, Gainesville.
- 30.Marco, G. M., and R. E. Stall. 1983. Control of bacterial spot of pepper initiated by strains of Xanthomonas campestris pv. vesicatoria which differ in sensitivity to copper. Plant Dis. 67:779-781. [Google Scholar]
- 31.McGuire, R. G., and J. B. Jones. 1986. Tween media for semiselective isolation of Xanthomonas campestris pv. vesicatoria from soil and plant material. Plant Dis. 70:887-891. [Google Scholar]
- 32.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Mishra, C., and J. Lambert. 1996. Production of anti-microbial substances by probiotics. Asia Pacific J. Clin. Nutr. 5:20-24. [PubMed] [Google Scholar]
- 34.Pohronezny, K., and R. B. Volin. 1983. The effect of bacterial spot on yield and quality of fresh market tomatoes. Hortscience 18:69-70. [Google Scholar]
- 35.Reece, K. S., and G. J. Phillips. 1995. New plasmids carrying antibiotic-resistance cassettes. Gene 165:141-142. [DOI] [PubMed] [Google Scholar]
- 36.Reeves, P. 1972. The Bacteriocins. Molecular biology, biochemistry, and biophysics. Springer Verlag, New York, N.Y.
- 37.Riley, M. A. 1993. Molecular mechanisms of bacteriocin evolution. Annu. Rev. Genet. 32:255-278. [DOI] [PubMed] [Google Scholar]
- 38.Riley, M. A. 1993. Positive selection for colicin diversity in bacteria. Mol. Biol. Evol. 10:1380-1395. [DOI] [PubMed] [Google Scholar]
- 39.Riley, M. A., and J. E. Wertz. 2002. Bacteriocins: evolution, ecology, and application. Annu. Rev. Microbiol. 56:117-137. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Shaner, G., and R. Finney. 1997. The effect of nitrogen fertilization on the expression of slow mildewing resistance in Knox wheat. Phytopathology 67:1051-1056. [Google Scholar]
- 42.Smidt, M. L., and A. K. Vidaver. 1982. Bacteriocin production by Pseudomonas syringae PsW-1 in plant tissue. Can. J. Microbiol. 28:600-604. [DOI] [PubMed] [Google Scholar]
- 43.Southern, E. 1975. Detection of specific sequences among NA fragments separated for the random generations of galactosidase gene fusions: application to analysis of gene expression in Agrobacterium. EMBO J. 4:891-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stall, R. E. 1995. Xanthomonas campestris pv. vesicatoria, p. 167-181. In U. S. Singh, R. P. Singh, and K. Kohmoto (ed.), Pathogenesis and host specificity in plant diseases: histopathological, biochemical, genetic, and molecular bases. Elsevier Science, New York, N.Y.
- 45.Stall, R. E., D. C. Loschke, and J. B. Jones. 1986. Linkage of copper resistance and avirulence loci on a self-transmissible plasmid in Xanthomonas campestris pv. vesicatoria. Phytopathology 76:240-243. [Google Scholar]
- 46.Stall, R. E., and P. L. Thayer. 1962. Streptomycin resistance of the bacterial spot pathogen and control with streptomycin. Plant Dis. Rep. 46:389-392. [Google Scholar]
- 47.Tan, Y., and M. A. Riley. 1997. Positive selection and recombination: major molecular mechanisms in colicin diversification. Trends Ecol. Evol. 12:348-351. [DOI] [PubMed] [Google Scholar]
- 48.Tate, K., and I. W. Sutherland. 2002. Antagonistic interactions amongst bacteriocin-producing enteric bacteria in dual species biofilms. J. Appl. Microbiol. 93:345-352. [DOI] [PubMed] [Google Scholar]
- 49.Tate, M. E., P. J. Murphy, W. P. Roberts, and A. Kerr. 1979. Adenine N substituent of agrocin 84 determines its bacteriocin-like specificity. Nature 280:697-698. [DOI] [PubMed] [Google Scholar]
- 50.Tudor-Nelson, S. M. 1995. An analysis of antagonism in tomato race-three strains of Xanthomonas campestris pv. vesicatoria. M.S. thesis. University of Florida, Gainesville.
- 51.Tudor-Nelson, S. M. 1999. Molecular characterization of bacteriocin-like activity in tomato race-three strains of Xanthomonas campestris pv. vesicatoria. Ph.D. dissertation. University of Florida, Gainesville.
- 52.Tudor-Nelson, S. M., G. V. Minsavage, R. E. Stall, and J. B. Jones. 2003. Bacteriocin-like substances from tomato race 3 strains of Xanthomonas campestris pv. vesicatoria. Phytopathology 93:1415-1421. [DOI] [PubMed] [Google Scholar]
- 53.Vanneste, J. L., Yu, J., and S. V. Beer. 1992. Role of antibiotic production by Erwinia herbicola Eh252 in biological control of Erwinia amylovora. J. Bacteriol. 174:2785-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vidaver, A. K. 1983. Bacteriocins: the lure and the reality. Plant Dis. 67:471-475. [Google Scholar]
- 55.Wang, J. F., J. B. Jones, J. W. Scott, and R. E. Stall. 1990. A new race of the tomato group of strains of Xanthomonas campestris pv. vesicatoria. Phytopathology 80:1070. [Google Scholar]
- 56.Xu, G. W., and C. F. Gonzalez. 1991. Plasmid, genomic, and bacteriocin diversity in U.S. strains of Xanthomonas campestris pv. oryzae. Phytopathology 81:628-631. [Google Scholar]