Abstract
Lactose-limited fermentations of 49 dairy Streptococcus thermophilus strains revealed four distinct fermentation profiles with respect to galactose consumption after lactose depletion. All the strains excreted galactose into the medium during growth on lactose, except for strain IMDOST40, which also displayed extremely high galactokinase (GalK) activity. Among this strain collection eight galactose-positive phenotypes sensu stricto were found and their fermentation characteristics and Leloir enzyme activities were measured. As the gal promoter seems to play an important role in the galactose phenotype, the galR-galK intergenic region was sequenced for all strains yielding eight different nucleotide sequences (NS1 to NS8). The gal promoter played an important role in the Gal-positive phenotype but did not determine it exclusively. Although GalT and GalE activities were detected for all Gal-positive strains, GalK activity could only be detected for two out of eight Gal-positive strains. This finding suggests that the other six S. thermophilus strains metabolize galactose via an alternative route. For each type of fermentation profile obtained, a representative strain was chosen and four complete Leloir gene clusters were sequenced. It turned out that Gal-positive strains contained more amino acid differences within their gal genes than Gal-negative strains. Finally, the biodiversity regarding lactose-galactose utilization among the different S. thermophilus strains used in this study was shown by RAPD-PCR. Five Gal-positive strains that contain nucleotide sequence NS2 in their galR-galK intergenic region were closely related.
Streptococcus thermophilus is one of the most widely used lactic acid bacteria in the dairy industry, well known as a starter culture component in yoghurt fermentation and cheesemaking. In S. thermophilus, lactose is transported into the cell by a lactose permease (LacS), which operates as a lactose-galactose antiporter or as a galactoside-proton symport system (6). Although lactose is efficiently transported into the cell and subsequently hydrolyzed by an intracellular β-galactosidase, many strains of S. thermophilus used in the dairy industry ferment only the glucose moiety of lactose, while the galactose moiety is excreted into the medium in equimolar amounts with the lactose uptake (11, 12, 22). The exchange reaction, which is highly favored with an excess of galactosides on either side of the cell membrane, leads to a galactose-negative phenotype of S. thermophilus in milk (18). Although phospho-β-galactosidase activity was found in some S. thermophilus strains, which suggests alternative pathways for lactose transport and galactose fermentation, the Leloir pathway seems to be the most ubiquitous route for galactose catabolism in S. thermophilus (8, 9, 12, 21). This pathway consists of a regulator (GalR), a galactokinase (GalK), a galactose 1-phosphate uridylyltransferase (GalT), a UDP-glucose 4-epimerase (GalE), and a mutarotase (GalM) which are part of the galRKTEM gene cluster, and allows the conversion of galactose into glucose 1-phosphate that can be directed to glycolysis by phosphoglucomutase or alternatively used in anabolic pathways (2). The galR-galK intergenic region contains the promoter of the galR gene and of the gal operon. The galR gene is divergently transcribed and the gal operon consists of galK, galT, and galE. A polycistronic mRNA transcript galKTE has been reported in two S. thermophilus strains, namely CNRZ 302 and SMQ-301 (28, 30). The galM gene has its own promoter (30).
Most dairy strains of S. thermophilus are Gal negative. Mutations to a galactose-positive phenotype do not result in constitutive expression of the gal genes, which strongly suggests that S. thermophilus was Gal positive and became Gal negative in the recent past (30). Currently, the Gal-negative phenotype has been ascribed to a defect in the induction mechanism of GalK, which seems to be the rate-limiting enzyme of the Leloir pathway (2). However, under appropriate selective conditions, such as limited lactose and excess galactose concentrations, Gal-positive derivatives of S. thermophilus can be obtained which ferment galactose and possess Leloir enzyme activities (11, 14, 22). It has been shown that the Gal-negative S. thermophilus strain CNRZ 302 contains structurally intact genes for the Leloir pathway enzymes, but that they are weakly transcribed, while independently isolated Gal-positive mutants contain up mutations in the gal promoter (30). The function of GalR has been determined as a transcriptional activator of both the gal and lac operons, which negatively regulates its own expression (30). Recently, a S. thermophilus strain has been reported in which the poor translation of the galK gene seems to be responsible for the Gal-negative phenotype (28).
As the complete utilization of galactose is a desirable property in various industrial dairy fermentations, a better understanding of the origin of the Gal-negative phenotype of S. thermophilus could provide new strategies to obtain stable galactose-fermenting strains (7). Although a lot of progress has been made in elucidating galactose catabolism in S. thermophilus, most of the work focused on the molecular level and was hence carried out on a limited number of strains. Further investigation of the galactose phenotype of S. thermophilus is recommended from an industrial point of view. Gal-positive strains can be useful if applied in dairy products where galactose accumulation in the milk or curd can cause product defects, such as growth of undesirable heterofermentative lactic acid bacteria and cheese browning during baking (16, 17, 26). Lower levels of galactose in fermented dairy products can be beneficial for human health as well, as too high galactose consumption can lead to accumulation of toxic galactitol in human tissue cells (10). Finally, a fully functional Leloir pathway may also lead to a higher exopolysaccharide (EPS) production, since it has been suggested that the EPS precursors, the activated nucleotide sugars whose low level might be a potential bottleneck in EPS production, are formed from the galactose moiety of lactose (13).
In this paper, the galactose phenotype and genotype of 49 S. thermophilus strains from dairy origin has been studied to gain more insight into the origin of the Gal-negative phenotype. In particular, we have investigated if the Gal-positive phenotype is indeed, exclusively determined by mutations in its gal promoter.
MATERIALS AND METHODS
Bacterial strains.
The 49 S. thermophilus strains used in this study are listed in Table 1. The strains were stored at −80°C on Protect Beads (Technical Service Consultants Ltd., Lancashire, United Kingdom) or in de Man Rogosa Sharpe (MRS) medium (Oxoid, Basingstoke, United Kingdom) containing 25% (vol/vol) glycerol (1).
TABLE 1.
Biokinetic parameters of Streptococcus thermophilus strains grown in M17 medium containing 0.5% (wt/vol) lactose
| Strain | Source (original reference)a | μmax (h−1)b | YX/Sb | YP/Sb | YGal,Lacb | Fermentation profile |
|---|---|---|---|---|---|---|
| IMDOST01 | Greek yogurt (ACA-DC 492) | 1.93 | 0.37 | 0.48 | 0.47 | B |
| IMDOST02 | Greek yogurt (ACA-DC 491) | 1.82 | 0.36 | 0.56 | 0.42 | B |
| IMDOST03 | Greek yogurt (ACA-DC 490) | 1.88 | 0.33 | 0.56 | 0.44 | B |
| IMDOST04 | Unknown (5FT) | 1.82 | 0.35 | 0.50 | 0.44 | B |
| IMDOST05 | Greek yogurt (ACA-DC 611) | 1.58 | 0.34 | 0.56 | 0.41 | B |
| IMDOST06 | Unknown [(t) 359] | 1.52 | 0.31 | 0.46 | 0.45 | B |
| IMDOST07 | Greek yogurt (ACA-DC 511) | 1.71 | 0.37 | 0.51 | 0.51 | C |
| IMDOST08 | Greek yogurt (ACA-DC 595.1) | 1.46 | 0.33 | 0.62 | 0.38 | C |
| IMDOST09 | Greek yogurt (ACA-DC 615) | 1.73 | 0.38 | 0.66 | 0.42 | C |
| IMDOST10 | Greek yogurt (ACA-DC 619) | 1.74 | 0.36 | 0.44 | 0.44 | B |
| IMDOST11 | Industrial yogurt starter (UN 1YRF) | 1.68 | 0.41 | 0.61 | 0.33 | C |
| IMDOST12 | Industrial yogurt starter (UN 7FT) | 1.44 | 0.32 | 0.48 | 0.45 | A |
| IMDOST13 | Greek yogurt (ACA-DC 602.1) | 1.72 | 0.32 | 0.49 | 0.43 | B |
| IMDOST14 | Industrial yogurt starter (UN 4L) | 2.02 | 0.36 | 0.45 | 0.43 | B |
| IMDOST15 | Greek yogurt (ACA-DC 487) | 1.31 | 0.31 | 0.39 | 0.44 | B |
| IMDOST16 | Greek yogurt (ACA-DC 486) | 1.62 | 0.32 | 0.40 | 0.45 | B |
| IMDOST17 | Greek yogurt (ACA-DC 482) | 1.62 | 0.33 | 0.46 | 0.44 | B |
| IMDOST18 | Greek yogurt (ACA-DC 481) | 1.85 | 0.36 | 0.45 | 0.42 | B |
| IMDOST19 | Industrial yogurt starter (UN 5FR) | 1.63 | 0.34 | 0.45 | 0.42 | B |
| IMDOST21 | Greek yogurt (ACA-DC 489) | 1.89 | 0.51 | 0.58 | 0.43 | B |
| IMDOST22 | Greek yogurt (ACA-DC 488) | 1.58 | 0.40 | 0.68 | 0.41 | C |
| IMDOST23 | Greek yogurt (ACA-DC 623.1) | 1.58 | 0.48 | 0.67 | 0.34 | C |
| IMDOST24 | Industrial yogurt starter (Yoplait C417) | 1.61 | 0.46 | 0.67 | 0.34 | C |
| IMDOST25 | Industrial yogurt starter (UN 7FT) | 1.74 | 0.39 | 0.52 | 0.47 | B |
| IMDOST26 | Unknown (510) | 1.41 | 0.41 | 0.49 | 0.44 | B |
| IMDOST27 | Industrial yogurt starter (UN 6FC) | 1.64 | 0.37 | 0.46 | 0.42 | B |
| IMDOST28 | Unknown (NR) | 1.78 | 0.34 | 0.46 | 0.44 | A |
| IMDOST29 | Industrial yogurt starter (UN ATX) | 1.73 | 0.36 | 0.37 | 0.42 | B |
| IMDOST30 | Unknown (4) | 1.66 | 0.41 | 0.47 | 0.49 | B |
| IMDOST31 | Yoghurt (NCFB 2393) | 1.80 | 0.39 | 0.52 | 0.41 | B |
| IMDOST32 | Greek yogurt (ACA-DC 613) | 1.96 | 0.34 | 0.44 | 0.39 | B |
| IMDOST33 | Unknown (Gal+) | 1.96 | 0.38 | 0.49 | 0.44 | B |
| IMDOST34 | Greek yogurt (ACA-DC 638) | 1.53 | 0.39 | 0.59 | 0.37 | B |
| IMDOST36 | Industrial yogurt starter (LY03) | 1.66 | 0.34 | 0.42 | 0.51 | B |
| IMDOST37 | Industrial yogurt starter (Sfi20) | 1.75 | 0.37 | 0.46 | 0.42 | B |
| IMDOST38 | Industrial yogurt starter (BTC) | 1.55 | 0.32 | 0.47 | 0.45 | B |
| IMDOST39 | Greek yogurt (ACA-DC 480) | 1.75 | 0.33 | 0.44 | 0.41 | B |
| IMDOST40 | Industrial yogurt starter (EU20) | 1.76 | 0.80 | 0.95 | 0 | D |
| IMDOST41 | Industrial yogurt starter (STCH101) | 2.19 | 0.40 | 0.50 | 0.50 | A |
| IMDOST42 | Romanian yogurt (ACA-DC ST111) | 1.38 | 0.34 | 0.36 | 0.47 | A |
| MB1655 | Pasteurized milk (NCFB 575) | 2.00 | 0.46 | 0.46 | 0.36 | B |
| MB1656 | Gruyère cheese starter (NCFB 2564) | 1.06 | 0.28 | 0.46 | 0.45 | B |
| MB1657 | Unknown (NCFB 489) | 1.59 | 0.38 | 0.53 | 0.40 | A |
| MB1658 | Unknown (LMG 13564) | 1.76 | 0.46 | 0.49 | 0.46 | A |
| MB1661 | Unknown | 2.01 | 0.47 | 0.44 | 0.38 | B |
| MB1663 | Unknown (NCFB 2075) | 1.65 | 0.42 | 0.47 | 0.40 | B |
| MB1664 | Unknown (NCFB 1242) | 1.85 | 0.45 | 0.50 | 0.37 | A |
| MB1665 | Unknown | 1.91 | 0.46 | 0.58 | 0.31 | A |
| MB1667 | Pasteurized milk (NCFB 574) | 1.77 | 0.41 | 0.58 | 0.51 | A |
ACA-DC, Collection of the Laboratory of Dairy Research, Agricultural University of Athens, Athens, Greece, LMG, BCCMTM/LMG Bacteria Culture Collection, Laboratory of Microbiology, Ghent University, Ghent, Belgium; NCFB, National Collection of Food Bacteria, Reading, United Kingdom; UN, Université de Nancy, Nancy, France.
μmax, maximum specific growth rate (h−1); YX/S, cell yield coefficient (OD620 · g of substrate−1); YP/S, product yield coefficient (g of product · g of substrate−1); YGal,Lac, galactose-lactose exchange coefficient (g of galactose · g of lactose−1).
The authenticity of the strains was checked by a 259-bp PCR amplification product of the 16S-23S rRNA gene spacer region using the species-specific primer pair ThI and ThII (23). The PCR amplification profile was used as described by Tilsala-Timisjärvi et al. (23).
Fermentation conditions.
Fermentations were done in 1-liter laboratory fermentors with a working volume of 0.5 liters. The fermentor inoculum was prepared in three steps. To obtain fresh cultures, the bacteria were propagated twice (12 h at 42°C) in a medium identical to the one used for the fermentations. The inoculation volume was always 1% (vol/vol). After another inoculation (1%, vol/vol) and 12 h of incubation at 42°C, 5 ml (1%, vol/vol) of this preculture was used to inoculate the fermentor. All experiments were carried out in M17 medium supplemented with 0.5% (wt/vol) lactose, unless stated otherwise. The fermentor was sterilized in an autoclave at 121°C for 20 min. Lactose and galactose were sterilized separately. After sterilization they were added aseptically to the fermentor. The temperature was held at 42 ± 0.5°C and the pH was uncontrolled. The fermentors were agitated at 100 rpm with a magnetic stirrer to keep the broth homogeneous.
Analyses.
During the first 8.5 h of fermentation, 5-ml samples were aseptically withdrawn from the fermentor every 30 min to determine optical density, and lactose, galactose, glucose, and lactic acid concentrations. The optical density at 620 nm (OD620) was used to monitor cell growth after appropriate dilution of the samples. Samples for substrate and product determination were microcentrifuged at 14,000 × g for 5 min, filtered through 0.2-μm-pore-size filters (Advantec MFS, Dublin, CA) and stored at −20°C until further analysis. Lactose, galactose, and lactate were separated on a cation-exchange column (Aminex HPX-87H; Bio-Rad Laboratories Ltd., Hemel Hempstead, United Kingdom) and quantified using a refractive index detector (RID 6A; Shimadzu Co., Kyoto, Japan). The mobile phase was 5 mM H2SO4 at a flow rate of 0.6 ml min−1.
The maximum specific growth rate μmax (h−1) was calculated as the maximum slope from the linearized values of the optical density (OD620) as a function of fermentation time (in hours). The cell yield coefficient YX/S (OD620 · g of substrate−1) and the product yield coefficient YP/S (g of product. g of substrate−1) both based on substrate consumption, were calculated as the slope from the OD620 and product yield, as a function of substrate (lactose or galactose) consumption during the exponential growth phase, respectively. The galactose-lactose exchange coefficient YGal,Lac (g of galactose · g of lactose−1) was calculated as the slope of the galactose excretion as a function of lactose uptake before lactose depletion.
Preparation of cell extracts.
Samples for enzyme activity measurements were taken during exponential growth. For a selected number of strains, samples were taken at two points, namely before and after lactose depletion. The cells were cooled on ice and centrifuged at 5,500 × g for 5 min, and the pellet was stored at −80°C until further use. All centrifugation steps were carried out in a cooled centrifuge at 4°C, and samples were kept on ice during the preparation. For preparation of the extracts, cells were washed twice in 10 ml ice-cold 50 mM potassium phosphate buffer (pH 7.0) and disrupted with zirconium glass beads in a Bead Beater (Biospec Products, Bartlesville, OK) by three 20-s treatments at 4,600 rpm with intervals of 1 min on ice to cool between treatments. The cell debris was removed by centrifugation at 20,000 × g for 20 min. Extracts were stored on ice and enzyme assays were performed within 4 h.
Enzyme assays.
Protein concentrations were determined by the micro BCA method (Pierce, Rockford, IL). The measurements were done in triplicate and standard deviations were below 4%. All enzyme assays were performed at 37°C with a Cobas Mira Plus autoanalyzer (Roche Diagnostics, Mannheim, Germany) by monitoring the absorbance at 340 nm. The GalK assay mixture contained 100 mM triethanolamine (TEA) buffer (pH 7.8), 5 mM MgCl2, 2 mM phosphoenolpyruvate, 2 mM ATP, 0.4 mM NADH, 4 U of lactate dehydrogenase ml−1, and 3 U of pyruvate kinase ml−1. The assay was started by adding 10 mM galactose. To correct for NADH oxidase activity the same assay was carried out, except for the use of water instead of 10 mM galactose as starting reagent. Subtraction of the values obtained with water from the values obtained with galactose gave the correct experimental value. In case GalK activity could not be detected, at least two new cell extracts were made from independent fermentation samples to confirm the validity of the enzyme activity measurements. Specific GalK activity values below 0.02 U mg−1 were reported as not detected because their corresponding standard deviations were as high as the measured values, and hence GalK activity was below the detectable limit. GalT activity was measured in 100 mM TEA buffer (pH 7.8), 10 mM MgCl2, 1 mM NADP+, 0.25 mM glucose 1,6-diphosphate (GDP), 5 U of glucose 6-phosphate dehydrogenase (G6PDH) ml−1, and 3 U of phosphoglucomutase ml−1. The assay was started by adding 2 mM UDP-glucose and 4 mM galactose 1-phosphate. GalE activity was measured in 100 mM Tris-HCl buffer (pH 8.5), 10 mM MgCl2, 1 mM NAD+, and 0.15 U of UDP-glucose dehydrogenase ml−1, and the assay was started with 0.8 mM UDP-galactose. All results were expressed in units (U) per mg of total protein, and are an average of triplicate measurements with standard deviation. The t test and one-way analysis of variance (ANOVA) were performed on the enzyme activity data using Statistica 6.1 (Statsoft Inc., Tulsa, OK). Significant differences were assessed by Duncan's post-hoc test. A value of P < 0.05 was considered to be statistically different.
Galactose phenotype.
To identify galactose-positive strains among the 49 S. thermophilus strains tested, every strain was first propagated twice for 12 h in 10 ml M17 medium containing 0.5% (wt/vol) lactose to yield a well grown inoculum that was subsequently used to inoculate (1%, vol/vol) 10 ml M17 medium containing 0.5% (wt/vol) galactose as the sole energy source. High-pressure liquid chromatography (HPLC) analysis confirmed that no residual lactose was transferred during inoculation. After another 12 h of incubation at 42°C, the optical density, pH, galactose consumption, and lactate production were measured and used as characteristics to assign the phenotype.
DNA isolation.
Total genomic DNA was isolated from all S. thermophilus strains as described by Flamm et al. (5). Briefly, bacterial cells from 2 ml of an overnight culture, grown in M17 medium (Oxoid), were pelleted by microcentrifugation (13,000 × g, 2 min), washed in 1 ml of 15 mM sodium citrate buffer (pH 7.0) containing 150 mM NaCl, suspended in 100 μl of lysozyme solution (10 mM sodium phosphate buffer [pH 7.0]; 20% [wt/vol] sucrose, 4 mg of lysozyme ml−1 [Boehringer, Mannheim, Germany]), and incubated at 37°C for 45 min. To these suspensions, 200 μl of TE buffer (50 mM Tris-HCl [pH 8.0], 20 mM EDTA), 100 μl of Sarkosyl solution (5% [wt/vol] Sarkosyl in TE buffer; Boehringer), and 100 μl of proteinase K solution (25 mg ml−1 in TE buffer; Boehringer) were added and incubated at 37°C for 1 h. Cell lysates were extracted once with phenol and twice with chloroform. Precipitation of nucleic acids was done with sodium acetate (final concentration, 0.3 M) and two volumes of pure ethanol. The DNA pellet was washed twice with 80% (vol/vol) ethanol and finally dissolved in TE buffer. The DNA solution was incubated at 37°C for 1 h in the presence of a diluted RNase solution. The concentration of the DNA was determined spectrophotometrically at 260 nm.
Presence of the gal genes by PCR.
To verify the presence of the individual genes of the Leloir pathway and their relative position within the gene cluster, five primer pairs were designed that amplify a specific target in each gene. Primer pairs galR FW1-galR REV1, galK FW2-galK REV2, and galT FW3-galT REV3 were based on the sequence with accession number U61402, and have as target galR, galK, and galT, respectively (Table 2). Primer pairs galE FW4-galE REV4 and galM FW5-galM REV5 were based on the sequence with accession number M38175, and have as target galE and galM, respectively (Table 2). By combining forward (FW) and reverse (REV) primers of adjacent targets, the relative organization of the genes within the cluster was confirmed.
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| galR FW1 | 5′-TCC TAT TCA GGT TTA TGG TT-3′ |
| galR REV1 | 5′-CGG AAC TAC TGT ATG GAA AA-3′ |
| galK FW2 | 5′-TGG TGT AGA AGC AGA TCA TA-3′ |
| galK REV2 | 5′-TCC AGG CGT TCC AAT TTA AT-3′ |
| galT FW3 | 5′-TAT GAG GAA TTG GAC CGT AT-3′ |
| galT REV3 | 5′-AAA CGA ACA ACA CGG TGA TT-3′ |
| galE FW4 | 5′-ATT AGG TGG AGC TGG TTA TA-3′ |
| galE REV4 | 5′-TAA CGA AGA GGG ACA TAC TT-3′ |
| galM FW5 | 5′-ACT TGC TAC CTC GAA TTG AA-3′ |
| galM REV5 | 5′-GAG ACT TCA TCT GAC CTT TA-3′ |
| galRK FW6 | 5′-ATC CGA TTT CAT CAG CGA TA-3′ |
| galRK REV6 | 5′-CGT AAG TAC CTA GGG TAA TA-3′ |
DNA sequencing.
All sequencing was performed with the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction kit v2.0 and an ABI PRISM 310 Genetic Analyzer (PE Biosystems, Warrington, United Kingdom) according to the manufacturer's protocol. All templates for sequencing purposes were amplified with a proofreading polymerase. As template for sequencing of the galR-galK intergenic regions purified PCR product galR FW1-galK REV2 (1,722 bp) was used for all strains. All PCR templates were purified using the High Pure PCR Product Purification Kit (Boehringer). Primers galRK FW6 or galRK REV6 were used in the cycle sequencing reaction (Table 2). Sequence analysis was carried out with GeneCompar software (Applied Maths, Sint-Martens-Latem, Belgium). Sequencing of the complete Leloir gene clusters, both sense and antisense, was done with purified galR FW1-galM REV5 (6,210 bp) as template.
Genetic typing by RAPD-PCR.
Template DNA for Randomly Amplified Polymorphic DNA (RAPD) analysis was made by a dilution of the genomic DNA in water to a final concentration of 25 ng μl−1. The RAPD analysis was performed with the commercially available Ready-To-Go RAPD analysis kit (Amersham Biosciences AB, Uppsala, Sweden) according to the manufacturer's instructions. One microliter of the DNA dilution and 25 pmol of primer XD9 (5′-GAAGTCGTCC-3′; Isogen Bioscience BV, Maarssen, The Netherlands) were used in the PCR. RAPD-PCR amplification with this primer yields distinctive and reproducible patterns and allows the identification of S. thermophilus (15). The PCR was performed with a thermocycler (model 9700; Perkin-Elmer, Shelton, CT) by using 45 cycles of 1 min at 94°C, 1 min at 33°C, and 2 min at 72°C. The initial denaturation was performed at 94°C for 5 min, and the final extension was done at 72°C for 7 min. The ramping speed between annealing temperature and elongation temperature was reduced to 70% of the normal speed. A mixture of the DNA molecular mass markers X and IX (Boehringer) in a 1:1 ratio was used as a size standard. The DNA bands were visualized under UV illumination and the gel image was captured using a Geldoc 1000 system with Molecular Analyst software version 1.0 (Bio-Rad Laboratories Ltd.). Conversion, normalization, and further analysis of the patterns were carried out with the Gel Compar II version 3.0 software (Applied Maths). Similarity coefficients for pairs of tracks were calculated by using Pearson's product-moment correlation coefficient, and strains were grouped by using the unweighted pair group method with arithmetic averages.
Nucleotide sequences accession numbers.
The eight different nucleotide sequences found in the galR-galK intergenic region have been deposited in GenBank under accession no. AY721595 through AY721602. The nucleotide sequences of the complete Leloir gene clusters of S. thermophilus strains IMDOST22, IMDOST36, IMDOST40, and IMDOST42 have been deposited in GenBank under accession no. AY704366, AY704365, AY704367, and AY704368, respectively.
RESULTS
The galactose phenotype. (i) Lactose fermentations.
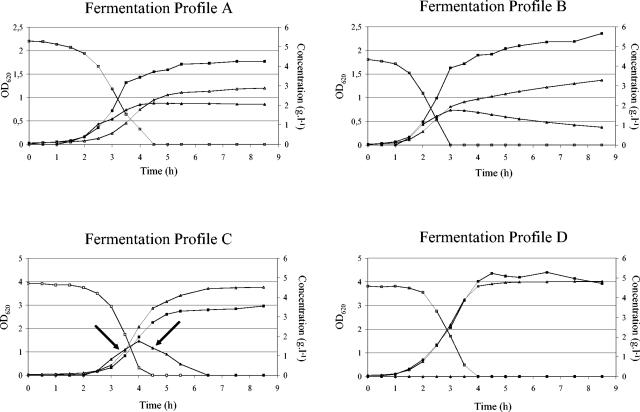
Among the 49 S. thermophilus strains studied, four typical lactose-limited fermentation profiles based on the fate of galactose were observed, referred to as A, B, C, and D, and the strains were grouped accordingly (Fig. 1; Table 1). This low sugar concentration (0.5% [wt/vol] lactose) was necessary to follow the fate of galactose after lactose depletion. In a medium containing higher lactose concentrations, the acidified medium could already inhibit the growth of S. thermophilus and prevent the subsequent metabolism of the galactose portion of lactose. In these fermentations, the pH after lactose depletion was typically around 5.6 and the final pH varied between pH 5.6 and pH 4.8. All the strains, except for strain IMDOST40, excreted galactose into the medium during growth on lactose, indicating the activity of the lactose-galactose antiport uptake system. In case of a perfect stoichiometry, the theoretical galactose-lactose exchange coefficient YGal,Lac should be 0.5 g of galactose. g of lactose−1. However, the galactose excretion was for most strains less than stoichiometrically expected (Table 1). After lactose depletion, repression was relieved and most of the strains started consumption of the galactose that was excreted in the medium during growth on lactose (Fig. 1).
FIG. 1.
Four typical batch fermentation profiles as found among 49 S. thermophilus strains when grown in M17 medium with 0.5% (wt/vol) lactose at 42°C. S. thermophilus IMDOST04, S. thermophilus IMDOST10, S. thermophilus IMDOST07, and S. thermophilus IMDOST40 are representative strains for fermentation profiles A, B, C, and D, respectively. The black arrows in fermentation profile C indicate the different sampling points for enzyme activity measurements. Symbols: ▴, galactose; ▵, lactic acid; ▪, OD620; □, lactose.
Nine strains (18.4%) displayed fermentation profile A and consumed none of the excreted galactose within 8.5 h of fermentation. The majority of the strains (32; 65.3%) displayed fermentation profile B, and were only able to consume part of the excreted galactose within 8.5 h of fermentation. Strains belonging to this group consumed the excreted galactose at various speed and to various extent, but never to completion. The inability of the members of group A and B to use all of the excreted galactose was also persistent after prolonged incubation up to 24 h. Seven strains (14.3%) displayed a fermentation profile C and consumed all of the excreted galactose within 8.5 h of fermentation. Strain IMDOST40, the only strain (2.0%) with a fermentation profile D, did not excrete galactose during growth on lactose and consumed the glucose and galactose moieties of lactose simultaneously. The biokinetic parameters of all the strains grown in M17 medium containing 0.5% (wt/vol) lactose are represented in Table 1.
(ii) Lactose repression of the gal genes.
For strain IMDOST07 and IMDOST22, strains that both displayed a fermentation profile C, enzyme activities were measured at two points during fermentation, namely before (t = 3.5 h) and after (t = 4.5 h) lactose depletion (Fig. 1). After lactose repression was relieved, a twofold increase of the specific activity of GalK and GalT was observed while a two- to threefold decrease of the specific activity of GalE was measured (Table 3).
TABLE 3.
Lactose repression of the gal genes
| Strain | Lactose depletion | Specific enzyme activitya (U · mg of protein−1)
|
||
|---|---|---|---|---|
| GalK | GalT | GAIE | ||
| IMDOST07 | Before | 0.10 ± 0.02A | 6.0 ± 0.1A | 0.62 ± 0.00B |
| After | 0.19 ± 0.03B | 11.8 ± 0.1B | 0.22 ± 0.00A | |
| IMDOST22 | Before | Not detected | 3.7 ± 0.0A | 0.52 ± 0.01B |
| After | Not detected | 7.5 ± 0.1B | 0.28 ± 0.00A | |
Means ± standard deviations. Mean values for each strain in each column with a common superscript did not differ at α = 0.05.
(iii) Galactose fermentations.
Among the S. thermophilus strains examined, eight Gal-positive strains were found when grown in M17 medium containing 0.5% (wt/vol) galactose as the sole energy source. All the Gal-positive strains exhibited a fermentation profile C or D when grown on lactose, while the strains exhibiting fermentation profile A or B were all Gal negative. These Gal-positive strains were also grown in M17 medium supplemented with 0.5% (wt/vol) galactose as the sole energy source and their biokinetic parameters are represented in Table 4.
TABLE 4.
Biokinetic parameters and specific activities of the enzymes of the Leloir operon for Gal-positive Streptococcus thermophilus strains grown in M17 medium containing 0.5% (wt/vol) galactose
| Straina | μmax (h−1)b | YX/Sb | YP/Sb | galR-galK region | GalK (U · mg−1)c | GalT (U · mg−1)c | GalE (U · mg−1)c |
|---|---|---|---|---|---|---|---|
| IMDOST07 | 0.99 | 0.58 | 0.87 | NS1 | 0.41 ± 0.02A | 18.7 ± 0.2E | 0.33 ± 0.01B |
| IMDOST08 | 1.49 | 0.48 | 0.90 | NS2 | Not detected | 10.3 ± 0.1C | 0.44 ± 0.01C |
| IMDOST09 | 1.51 | 0.51 | 0.90 | NS2 | Not detected | 10.2 ± 0.1C | 0.83 ± 0.01H |
| IMDOST11 | 0.73 | 0.54 | 0.89 | NS2 | Not detected | 9.3 ± 0.3B | 0.53 ± 0.001E |
| IMDOST22 | 0.65 | 0.57 | 0.88 | NS2 | Not detected | 9.3 ± 0.4B | 0.68 ± 0.01G |
| IMDOST23 | 1.02 | 0.66 | 0.87 | NS2 | Not detected | 9.4 ± 0.4B | 0.50 ± 0.01D |
| IMDOST24 | 0.72 | 0.59 | 0.96 | NS6 | Not detected | 13.0 ± 0.6D | 0.62 ± 0.01F |
| IMDOST40 | 1.71 | 0.66 | 0.98 | NS3 | 1.04 ± 0.05B | 2.7 ± 0.2A | 0.27 ± 0.01A |
The source and the original reference are given in Table 1.
μmax, maximum specific growth rate (h−1); YX/S, cell yield coefficient (OD620 · g of substrate−1); YP/S, product yield coefficient (g of product · g of substrate−1)
Means ± standard deviations. Mean values in each column with a common superscript did not differ at α = 0.05.
(iv) Activities of the gal enzymes.
For the Gal-positive strains, activities of the gal enzymes were measured during mid-exponential growth phase (Table 4). The specific GalT activity ranged from 2.7 U mg−1 to 18.7 U mg−1 with a mean value of 10.4 U mg−1, while the specific GalE activity ranged from 0.27 U mg−1 to 0.83 U mg−1 with a mean value of 0.52 U mg−1. Strain IMDOST40 showed low specific activity for GalT and GalE, 2.7 U mg−1 and 0.27 U mg−1, respectively, but extremely high specific activity for GalK (1.04 U mg−1). The specific activity of GalK could only be detected for strains IMDOST07 and IMDOST40, giving 0.41 U mg−1 and 1.04 U mg−1, respectively. For the other Gal-positive strains normal GalT and GalE activities were measured, but no GalK activity was apparently present.
The galactose genotype.
The authenticity of the 49 dairy strains of S. thermophilus used in this study (Table 1) was confirmed by PCR using a species-specific primer pair (ThI and ThII) based on the 16S-23S rRNA spacer region which is very conserved among various LAB (23).
To verify that the 49 S. thermophilus strains examined had the genetic potential to catabolize galactose via the Leloir pathway, the presence and the genetic organization of the Leloir genes were checked by PCR. All strains contained the structural Leloir genes in the same genetic order, namely, galRKTEM.
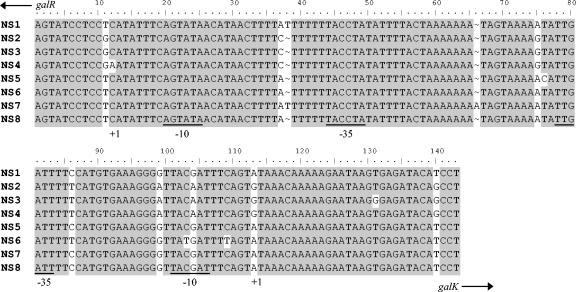
To investigate the possible link between weak transcription of the genes of the Leloir operon and the Gal-negative phenotype, the galR-galK intergenic regions containing the galR and gal promoter were sequenced. Among the 49 strains only eight different nucleotide sequences were found in this intergenic region. These eight different nucleotide sequences are further referred to as NS1 to NS8 and their alignment is shown in Fig. 2. The number of differences found between the nucleotide sequences NS1 to NS8 ranged from one to ten. These differences were distributed over 15 distinct positions in the 142-bp region (Fig. 2). Most strains (28; 57.1%) possessed a nucleotide sequence identical to NS1, which is also found in the Gal-negative S. thermophilus strain CNRZ 302 (accession number U61402). Eight strains (16.3%) possessed a nucleotide sequence identical to NS2, three strains (6.1%) possessed a nucleotide sequence identical to NS5, and six strains (12.3%) possessed a nucleotide sequence identical to NS8. NS3, NS4, NS6, and NS7 were only found in a single strain each (2.0% each) (Fig. 3, Table 4). Strains with NS2, NS3, NS4, or NS6 in the galR-galK intergenic region contained one nucleotide difference in the −10 region of the gal promoter, (TACAAT) or (TATGAT), respectively, compared with the other nucleotide sequences (TACGAT), bringing the former sequences closer to consensus (TATAAT) (Fig. 2). All the Gal-positive strains, except for IMDOST07, had only one difference in the −10 region of the gal promoter, which was therefore closer to consensus. Five out of eight Gal-positive strains had nucleotide sequence NS2 in their galR-galK intergenic region and strain IMDOST40 had nucleotide sequence NS3. NS2 and NS3 differed only in 1 nucleotide, namely NS3 had a T-to-G substitution in its Shine-Dalgarno (SD) sequence.
FIG. 2.
Alignment of the eight different nucleotide sequences (NS) as found in the galR-galK intergenic region of 49 wild-type S. thermophilus strains. The intergenic region contains the promoter sequences for the galR and the gal genes. The promoter regions are defined according to Vaughan et al. (30). The −10 and −35 regions are underlined, and the transcriptional start sites are indicated at +1.
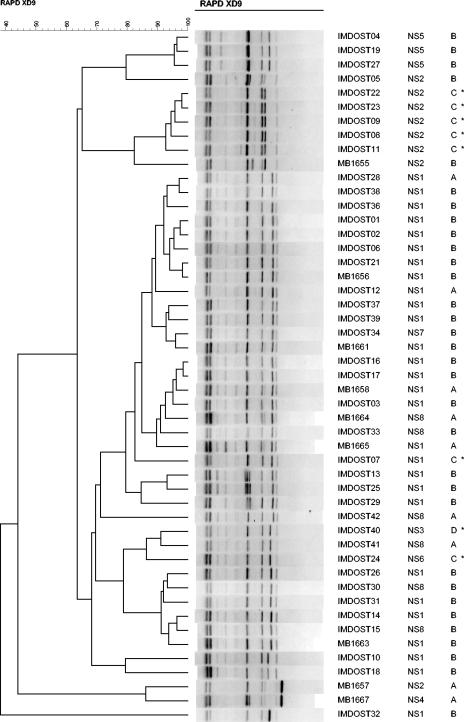
FIG. 3.
Genetic relationship among the 49 S. thermophilus strains as estimated by clustering analysis of RAPD patterns obtained with primer XD9 (15). The dendrogram was generated by the unweighted pair group method. The three columns on the right give the strain name, the nucleotide sequence (NS) of the galR-galK intergenic region, and the type of fermentation profile. Galactose-positive strains are indicated with an asterisk.
The complete Leloir gene clusters were sequenced for four strains, namely IMDOST22, IMDOST36, IMDOST40, and IMDOST42. These strains displayed a type C, type B, type D, and type A fermentation profile, respectively. Among these strains only IMDOST22 and IMDOST40 displayed the Gal-positive phenotype. After translation of the coding region of the four Leloir gene clusters, both Gal-positive strains seemed to have more amino acid differences within the amino acid sequence of GalK, GalT, GalE, and GalM, compared with the Gal-negative strains (Table 5). Both Gal-positive strains contained the same deletion in galR, which resulted in a frameshift affecting the amino acid sequence of GalR after position 58.
TABLE 5.
Number of differences in the amino acid sequence of the translated Leloir genes of IMDOST22, IMDOST36, IMDOST40, and IMDOST42, with regard to the galactose-negative S. thermophilus strains CNRZ 302 and A147a
RAPD-PCR typing of the strains.
The RAPD data shows the biodiversity regarding lactose-galactose utilization among the S. thermophilus strains studied. All the Gal-positive strains that had the nucleotide sequence NS2 in their galR-galK intergenic region were closely related (Fig. 3), as they grouped together in one RAPD subcluster. A similar observation was made for Gal-negative strains that had the nucleotide sequence NS5 in their galR-galK intergenic region (Fig. 3).
DISCUSSION
Most strains of the species S. thermophilus display a Gal-negative phenotype that is currently ascribed to the weak induction of the gal operon (2, 30). Four different fermentation profiles in M17 medium with 0.5% (wt/vol) lactose were found among the strains studied. Except for strain IMDOST40, galactose was always excreted into the medium during growth on lactose, indicating the activity of the lactose-galactose antiport uptake system. This corresponds with earlier reports stating that galactose accumulation, even when a lactose-limited medium is used, is independent of the galactose phenotype (11, 20, 22). However, in most fermentations galactose accumulation occurred in somewhat lower than stoichiometric amounts with regard to lactose utilization. This is probably due to the low amount of initial lactose used during the fermentations, since equimolar amounts are commonly found when an excess of lactose is present (25). Galactose excretion by LacS is probably both a consequence of the preferred mode of action (antiport) of the permease LacS in vivo, and of the repression of the gal operon during growth on lactose (6). The repressed status of the gal operon during growth on lactose has been confirmed for strain IMDOST07 and strain IMDOST22 by comparing enzyme activities at two stages during exponential growth, namely before and after lactose depletion. A twofold increase of the specific activities of GalK and GalT was measured, although specific GalE activity was two- to threefold lower after lactose depletion. It might be that the epimerase GalE is not only needed to recycle the UDP-galactose formed during the conversion of galactose 1-phosphate into glucose 1-phosphate by GalT, but also in other reactions where UDP-glucose and UDP-galactose conversions are involved, for instance cell wall biosynthesis and EPS production (4). The decrease in activity can then be explained by the different states of the growth of the cell. The first sample was taken in the mid-exponential phase and the second sample in the late exponential phase.
During growth on lactose there was no galactose accumulation detected in the medium for strain IMDOST40. This could be explained in two ways: first, the glucose and galactose moiety of lactose were consumed simultaneously, or, secondly, the excretion of galactose and its consecutive uptake occurred at the same speed resulting in no net galactose excretion. Growing this strain in M17 medium containing both 0.5% (wt/vol) lactose and 0.5% (wt/vol) galactose showed that the consumption of extracellular galactose only started when lactose was depleted, which rules out the latter assumption. Consequently, in this strain LacS has to function as a lactose-proton symporter instead of a galactoside antiporter. As LacS functions as a symport system, the intracellular galactose concentration is probably low. This could be a consequence of a defect in the catabolite repression causing the gal genes to be sufficiently transcribed to allow a fully functional Leloir pathway. As the precise mechanism by which the Leloir enzymes are repressed has not been determined, explaining this phenomenon becomes more difficult.
Another explanation might be that the rate-limiting enzyme of the Leloir pathway, GalK, even when repressed, is sufficiently present to allow fast galactose catabolism. This explanation is supported by the finding that strain IMDOST40 contained a one nucleotide difference bringing its putative GalK SD sequence (AAGGGAGA) closer to consensus, which may result in a more efficient translation of the gene. During growth on galactose, specific GalK activity of strain IMDOST40 was indeed higher than for other strains. The optimal mRNA sequence for ribosome binding seems to be AAAGGAGG for strain S. thermophilus SMQ-301 (28). Due to this additional difference in strain IMDOST40, the SD sequence is closer to optimal and the free energy of the mRNA(SD)-rRNA 16S complex formation is probably more favorable (24).
Among the Gal-positive strains four different nucleotide sequences (NS1, NS2, NS3, and NS6) were found in the galR-galK intergenic region. However, most Gal-positive strains contained nucleotide sequence NS2 that contained many differences in the gal promoter compared with nucleotide sequence NS1. The latter nucleotide sequence is most commonly found in the galR-galK intergenic region of Gal-negative strains. RAPD analysis revealed that all the Gal-positive strains that had the nucleotide sequence NS2 in their galR-galK intergenic region were closely related. This finding supports the hypothesis that S. thermophilus was Gal positive and became Gal negative in the recent past. One class of Gal-positive mutants described by Vaughan et al. contained a G-to-A substitution in the −10 box which turned out to be a promoter up mutation (30). Interesting to note is that strains possessing NS2, NS3, or NS4 in their galR-galK intergenic region, still possessed the original A instead of G in the −10 box. In this study, most of the strains that still possessed this A also displayed the Gal-positive phenotype. Recently, van den Bogaard et al. (29) also identified two naturally occurring Gal-positive strains that still possessed the original A in the −10 box. The Gal-negative strains IMDOST05, MB1655, and MB1657 possessed an identical gal promoter sequence as six of the Gal-positive strains, indicating that the Gal-positive phenotype is not exclusively determined by the gal promoter sequence. The higher number of amino acid differences found within the translated coding region of the Leloir genes, compared with the Gal-negative strains support this idea (Table 5). This is also consistent with the finding that Gal-negative S. thermophilus strains often contain significant amounts of GalT and GalE activities (3, 19, 22, 28), and a polycistronic galKTE mRNA transcript has been reported before (19, 28).
However, GalT and GalE activity were found for all the strains tested, while GalK activity could only be detected for two out of eight Gal-positive strains. This finding suggests that these other strains possibly metabolized galactose via an alternative route.
This study showed that most wild-type S. thermophilus strains started to consume galactose after lactose repression is relieved, regardless their galactose phenotype. However, Gal-positive strains had a tendency to consume galactose faster than Gal-negative strains and to completion. IMDOST40 was the only strain that was capable of metabolizing the glucose and the galactose moieties of lactose simultaneously, a feature that is commonly found in S. salivarius (27). Although the gal promoter played an important role in the Gal-positive phenotype, it did not determine the Gal-positive phenotype exclusively. Indeed, both Gal-positive strains had also more amino acid differences in the amino acid sequence of the Leloir enzymes compared with the Gal-negative strains.
Acknowledgments
We acknowledge financial support from the Research Council of the Vrije Universiteit Brussel (VUB), the Belgian Ministry of Agriculture, the Fund for Scientific Research-Flanders, and the European Commission (grant FAIR-CT98-4267). F.d.V. was recipient of a research fellowship at VUB and a Marie Curie Host Fellowship (QLK3-CT-2001-60077) at the Department of Applied Microbiology at Lund University.
REFERENCES
- 1.de Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 2.de Vos, W. M. 1996. Metabolic engineering of sugar catabolism in lactic acid bacteria. Antonie Leeuwenhoek 70:223-242. [DOI] [PubMed] [Google Scholar]
- 3.Degeest, B., and L. de Vuyst. 2000. Correlation of activities of the enzymes α-phosphoglucomutase, UDP-galactose 4-epimerase, and UDP-glucose pyrophosphorylase with exopolysaccharide biosynthesis by Streptococcus thermophilus LY03. Appl. Environ. Microbiol. 66:3519-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escalante, A., J. Villegas, C. Wacher, M. Garcia-Garibay, and A. Farres. 2002. Activity of the enzymes involved in the synthesis of exopolysaccharide precursors in an overproducing mutant copy strain of Streptococcus thermophilus. FEMS Microbiol. Lett. 209:289-293. [DOI] [PubMed] [Google Scholar]
- 5.Flamm, R. K., D. J. Hinrichs, and M. F. Thomashow. 1984. Introduction of pAMβ1 into Listeria monocytogenes by conjugation and homology between native L. monocytogenes plasmids. Infect. Immun. 44:157-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foucaud, C., and B. Poolman. 1992. Lactose transport system of Streptococcus thermophilus. Functional reconstitution of the protein and characterization of the kinetic mechanism of transport. J. Biol. Chem. 267:22087-22094. [PubMed] [Google Scholar]
- 7.Grossiord, B. P., E. E. Vaughan, E. J. Luesink, and W. M. de Vos. 1998. Genetics of galactose utilisation via the Leloir pathway in lactic acid bacteria. Le Lait 78:77-84. [Google Scholar]
- 8.Hemme, D., M. Nardi, and D. Jette. 1980. Beta-galactosidases et phosphobeta-galactosidases de Streptococcus thermophilus. Le Lait 60:595-618. [Google Scholar]
- 9.Hemme, D., D. Wahl, and M. Nardi. 1980. Variation de l'équipement enzymatique de Streptococcus thermophilus. Le Lait 60:111-130. [Google Scholar]
- 10.Hirasuka, Y., and G. Li. 1992. Alcohol and eye diseases: a review of epidemiologic studies. J. Stud. Alcohol 62:372-402. [DOI] [PubMed] [Google Scholar]
- 11.Hutkins, R., H. A. Morris, and L. L. McKay. 1985. Galactokinase activity in Streptococcus thermophilus. Appl. Environ. Microbiol. 50:777-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hutkins, R. W., and H. A. Morris. 1987. Carbohydrate metabolism by Streptococcus thermophilus: a review. J. Food Prot. 50:876-884. [DOI] [PubMed] [Google Scholar]
- 13.Levander, F., and P. Rådström. 2001. Requirement for phosphoglucomutase in exopolysaccharide biosynthesis in glucose- and lactose-utilizing Streptococcus thermophilus. Appl. Environ. Microbiol. 67:2734-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levander, F., M. Svensson, and P. Rådström. 2002. Enhanced exopolysaccharide production by metabolic engineering of Streptococcus thermophilus. Appl. Environ. Microbiol. 68:784-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moschetti, G., G. Blaiotta, F. Villani, S. Coppola, and E. Parente. 2001. Comparison of statistical methods for identification of Streptococcus thermophilus, Enterococcus faecalis, and Enterococcus faecium from randomly amplified polymorphic DNA patterns. Appl. Environ. Microbiol. 67:2156-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukherjee, K. K. 1994. Isolation of galactose-fermenting thermophilic cultures and their use in the manufacture of low browning Mozzarella cheese. J. Dairy Sci. 77:2839-2849. [Google Scholar]
- 17.O'Leary, V. S., and J. H. Woychik. 1976. Utilization of lactose, glucose, and galactose by a mixed culture of Streptococcus thermophilus and Lactobacillus bulgaricus in milk treated with lactase enzyme. Appl. Environ. Microbiol. 32:89-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poolman, B. 1993. Energy transduction in lactic acid bacteria. FEMS Microbiol. Rev. 12:125-147. [DOI] [PubMed] [Google Scholar]
- 19.Poolman, B., T. J. Royer, S. E. Mainzer, and B. F. Schmidt. 1990. Carbohydrate utilization in Streptococcus thermophilus: characterization of the genes for aldose 1-epimerase (mutarotase) and UDP-glucose 4-epimerase. J. Bacteriol. 172:4037-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somkuti, G. A., and D. H. Steinberg. 1979. Adaptability of Streptococcus thermophilus to lactose, glucose and galactose. J. Food Prot. 42:881-882, 887. [DOI] [PubMed] [Google Scholar]
- 21.Somkuti, G. A., and D. H. Steinberg. 1979. Beta-D-galactoside galactohydrolase of Streptococcus thermophilus: induction, purification, and properties. J. Appl. Biochem. 1:357-368. [Google Scholar]
- 22.Thomas, T. D., and V. L. Crow. 1984. Selection of galactose-fermenting Streptococcus thermophilus in lactose-limited chemostat cultures. Appl. Environ. Microbiol. 48:186-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tilsala-Timisjärvi, A., and T. Alatossava. 1997. Development of oligonucleotide primers from the 16S-23S rRNA intergenic sequences for identifying different dairy and probiotic lactic acid bacteria by PCR. Int. J. Food Microbiol. 35:49-56. [DOI] [PubMed] [Google Scholar]
- 24.Tinoco, I., Jr., P. N. Borer, B. Dengler, M. D. Levin, O. C. Uhlenbeck, D. M. Crothers, and J. Bralla. 1973. Improved estimation of secondary structure in ribonucleic acids. Nat. New Biol. 246:40-41. [DOI] [PubMed] [Google Scholar]
- 25.Tinson, W., A. J. Hillier, and G. R. Jago. 1982. Metabolism of Streptococcus thermophilus. 1. Utilization of lactose, glucose, and galactose. Aust. J. Dairy Tech. 38:8-13. [Google Scholar]
- 26.Tinson, W., M. F. Ratcliff, A. J. Hillier, and G. R. Jago. 1982. Metabolism of Streptococcus thermophilus. 3. Influence on the level of bacterial metabolites in Cheddar cheese. Aust. J. Dairy Tech. 37:17-21. [Google Scholar]
- 27.Vaillancourt, K., J. D. LeMay, M. Lamoureux, M. Frenette, S. Moineau, and C. Vadeboncoeur. 2004. Characterization of a galactokinase-positive recombinant strain of Streptococcus thermophilus. Appl. Environ. Microbiol. 70:4596-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaillancourt, K., S. Moineau, M. Frenette, C. Lessard, and C. Vadeboncoeur. 2002. Galactose and lactose genes from the galactose-positive bacterium Streptococcus salivarius and the phylogenetically related galactose-negative bacterium Streptococcus thermophilus: organization, sequence, transcription, and activity of the gal gene products. J. Bacteriol. 184:785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Bogaard, P. T., P. Hols, O. P. Kuipers, M. Kleerebezem, and W. M. de Vos. 2004. Sugar utilisation and conservation of the gal-lac gene cluster in Streptococcus thermophilus. Syst. Appl. Microbiol. 27:10-17. [DOI] [PubMed] [Google Scholar]
- 30.Vaughan, E. E., P. T. van den Bogaard, P. Catzeddu, O. P. Kuipers, and W. M. de Vos. 2001. Activation of silent gal genes in the lac-gal regulon of Streptococcus thermophilus. J. Bacteriol. 183:1184-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]