Abstract
Microautoradiography combined with fluorescence in situ hybridization (MAR-FISH) was used to screen for potential polyphosphate-accumulating organisms (PAO) in a full-scale enhanced biological phosphorus removal (EBPR) plant. The results showed that, in addition to uncultured Rhodocyclus-related PAO, two morphotypes hybridizing with gene probes for the gram-positive Actinobacteria were also actively involved in uptake of orthophosphate (Pi). Clone library analysis and further investigations by MAR-FISH using two new oligonucleotide probes revealed that both morphotypes, cocci in clusters of tetrads and short rods in clumps, were relatively closely related to the genus Tetrasphaera within the family Intrasporangiaceae of the Actinobacteria (93 to 98% similarity in their 16S rRNA genes). FISH analysis of the community biomass in the treatment plant investigated showed that the short rods (targeted by probe Actino-658) were the most abundant (12% of all Bacteria hybridizing with general bacterial probes), while the cocci in tetrads (targeted by probe Actino-221) made up 7%. Both morphotypes took up Pi aerobically only if, in a previous anaerobic phase, they had taken up organic matter from wastewater or a mixture of amino acids. They could not take up short-chain fatty acids (e.g., acetate), glucose, or ethanol under anaerobic or aerobic conditions. The storage compound produced during the anaerobic period was not polyhydroxyalkanoates, as for Rhodocyclus-related PAO, and its identity is still unknown. Growth and uptake of Pi took place in the presence of oxygen and nitrate but not nitrite, indicating a lack of denitrifying ability. A survey of the occurrence of these actinobacterial PAO in 10 full-scale EBPR plants revealed that both morphotypes were widely present, and in several plants more abundant than the Rhodocyclus-related PAO, thus playing a very important role in the EBPR process.
In the wastewater treatment industry, enhanced biological phosphorus removal (EBPR) has been widely used to remove orthophosphate (Pi) from wastewater to protect the receiving water bodies against eutrophication. In EBPR processes, the principle is to enrich microorganisms that can accumulate excessive amounts of intracellular polyphosphate [poly(P)] by using sequential anaerobic-aerobic and/or anaerobic-denitrifying conditions (37). The understanding of the biochemical pathways of these poly(P)-accumulating organisms (PAO) is based mainly on analysis of chemical transformations in enriched laboratory-scale EBPR systems, because no pure cultures are available (25, 34, 37). The generally accepted hypothesis (25, 34) proposes that PAO take up organic matter (usually assumed to be acetate) during the anaerobic period by using poly(P) as an energy source and sequester the acetate taken up into polyhydroxyalkanoates (PHA). The reducing power to form PHA is provided by hydrolysis of an intracellular glycogen pool through the glycolytic pathway. The PHA are used as energy and carbon sources in the following aerobic or denitrifying period for growth and to refresh the poly(P) and glycogen pools (25, 34).
Many studies of microbial populations enriched in laboratory-scale EBPR systems (8, 13, 19) and a few in full-scale EBPR plants (39) indicate that uncultured bacteria closely related to the genus Rhodocyclus in the Betaproteobacteria are important PAO and that their enriched cultures generally behave as biochemical models predict (8, 13, 15, 19). Recently, we investigated some important physiological aspects of the Rhodocyclus-related PAO (RPAO) present in three full-scale EBPR plants by using fluorescence in situ hybridization (FISH) combined with microautoradiography (MAR). The results (15) showed that RPAO were able to take up short-chain fatty acids, including acetate, pyruvate, and propionate, but not formic acid, glucose, and ethanol, under aerobic and anaerobic conditions. The glycolytic pathway, but not the tricarboxylic acid cycle, was essential to their anaerobic substrate uptake. RPAO were able to grow and take up Pi with oxygen, nitrate, or nitrite present as an electron acceptor, strongly suggesting that they are able to denitrify while taking up Pi. Recent FISH investigations of their occurrence in full-scale EBPR plants suggest that these bacteria are generally abundant and thus important PAO (8, 15, 39).
Some bacteria belonging to the gram-positive Actinobacteria have also been suggested to be potential PAO because some isolates have demonstrated aerobic Pi uptake ability after taking up organic substrates under anaerobic conditions (34). These bacteria include Microlunatus phosphovorus (32) and Tetrasphaera elongata (strain ASP12) (29). However, their aerobic Pi uptake depends on their anaerobic uptake of glucose (for M. phosphovorus) or an amino acid mixture (Casamino Acids) (for T. elongata) instead of short-chain fatty acids, as for the RPAO. Moreover, none of them formed PHA after the anaerobic substrate uptake, thus behaving differently from what had been predicted for the putative PAO. Therefore, they were not thought to be important players in full-scale EBPR processes. Intracellular poly(P) storage has been found in many other Actinobacteria, including Tetrasphaera australiensis and Tetrasphaera japonica (23), T. elongata (strain Lp2) (11), Tessaracoccus bendigoensis (24), and the filamentous “Candidatus Nostocoida limicola” (6) and “Candidatus Microthrix parvicella” (28). However, as far as we are aware, no studies have confirmed their function as PAO in full-scale EBPR systems.
In this paper, we present the identification and ecophysiological characterization of a group of uncultured Actinobacteria closely related to Tetrasphaera and other unidentified Actinobacteria within the family Intrasporangiaceae actively involved in P removal in a full-scale EBPR plant by using a suite of molecular techniques and microautoradiography. Their distribution and abundance in some municipal and industrial activated-sludge plants were investigated by FISH using new oligonucleotide probes. The possible relationship between these actinobacterial PAO (APAO) and RPAO and their roles in EBPR systems are discussed.
MATERIALS AND METHODS
Sampling and plant description.
Activated-sludge samples were obtained from Skagen Wastewater Treatment Plant (WWTP), Skagen, Denmark. Skagen WWTP has a Biodenipho configuration (35), in which influent is mixed with the activated sludge in an anaerobic tank before entering a tank with alternating denitrifying and nitrifying conditions. Skagen WWTP mainly treats industrial wastewater from the fishing industry in an amount corresponding to a population of 280,000 persons. Activated-sludge samples collected in the oxic (nitrifying) tanks, as well as influent wastewater, were sampled between May 2003 and July 2004.
MAR-FISH.
A MAR-FISH method slightly modified from those described by Lee et al. (17) and Nielsen et al. (27) was used, as detailed in Kong et al. (15). Briefly, biomass samples were incubated with a radioactively labeled compound under different well-defined electron acceptor and electron donor conditions before fixation with freshly prepared paraformaldehyde (final concentration, 4%) in a phosphorus-buffered saline at 4°C. All incubations were carried out on a shaking disk (Kikalabortechnik, Albertslund, Denmark) at 250 rpm and kept at 20 ± 1°C. The sources and specifications of all the radioactive chemicals except the amino acid mixture used in this study have been previously described (15). The labeled amino acid mixture (1.0 mCi/ml) (ICN Biochemicals, Inc., Bie og Berntsen, Denmark) consisted of l-[2,3-3H]alanine (0.08 mCi/ml), l-[4,5-3H]arginine (0.07), l-[2,3-3H]aspartic acid (0.08), l-[G-3H]glutamic acid (0.125), l-[2-3H]glycine (0.04), l-[2,5-3H]histidine (0.015), l-[4,5-3H]isoleucine (0.05), l-[4,5-3H]leucine (0.14), l-[4,5-3H]lysine (0.06), l-[ring-2,4-3H]phenylalanine (0.08), l-[2,3,4,5-3H]proline (0.05), l-[G-3H]serine (0.04), l-[G-3H]threonine (0.05), l-[3,5-3H]tyrosine (0.04), and l-[2,3-3H]valine (0.08).
FISH probing of MAR-incubated biomass, coating with emulsion, exposure, and development of the hybridized FISH slides before being examined microscopically were carried out as described before (15). Each MAR experiment was repeated at least three times. Controls for non-radioactively induced silver grain formation (chemography) were always included (15), and no MAR-positive cells were ever observed with any of the radioactive chemicals used in this study.
Potential PAO were screened as follows. Fresh biomass samples were collected by centrifugation (3,373 × g) and resuspended in filtered (0.2-μm-pore-size) oxygen-free influent (by evacuating and refilling the headspace in a bottle closed with a chlorobutyl stopper with ultrapure nitrogen three times) to the original biomass concentration (4 to 5 g biomass [dry weight]/l). The sample was incubated anaerobically for 2 h before the biomass was collected again by centrifugation and washed three times with filtered effluent before being used to study the 33Pi uptake, as described by Kong et al. (15). Using the same criteria, bacteria taking up an amount of 33Pi that was clearly MAR positive after 3 days of exposure at 4°C were assumed to be PAO with an active Pi uptake.
Investigation of bacterial uptake of organic substrates under different electron acceptor conditions was carried out as previously described (15). In amino acid uptake tests, each of the amino acids was added to a final concentration of 1.0 mM, and the labeled amino acid mixture was added at 10 μCi (per ml) calculated on the basis of l-[2,5-3H]histidine. In order to distinguish bacterial storage from growth, preincubations were carried out with unlabeled oleic acid for 6 h before the addition of labeled oleic acid. In these incubations, the biomass concentration used was 0.2 g/liter; oleic acid was first added to a final concentration of 1.0 mM, and an equal amount was added again after 6 h, together with the labeled oleic acid.
FISH.
Sludge samples for probing gram-positive and gram-negative bacteria were fixed with 50% ethanol (30) and 4% paraformaldehyde (2) in phosphate-buffered saline, respectively. FISH was carried out according to the method of Amann (2). The oligonucleotide probes EUBmix (equimolar concentrations of EUB338, EUB338II, and EUB338III), ALF 968, BET 42a, GAM 42a, HGC69a, actino-1011, and PAO651, labeled with Cy3 or FLUOS [5(6)-carboxyfluorescein-N-hydroxy-succinimide ester], were used for FISH. Their specificities, hybridization requirements, and reference information are all described in probeBase (20). If necessary, lysozyme (0.5 g/liter in 100 mM Tris [pH 7.5] and 5 mM EDTA) and mutanolysin (5,000 U/ml in phosphate buffer) (9) were used to treat the biomass to increase the permeability of the cells.
Clone library construction, clone screening, and sequencing.
Community DNA was extracted from fresh sludge by using the FastDNA kit with the FastPrep instrument according to the protocol recommended by the provider (Qbiogene, Carlsbad, CA). PCR using primer actino-1011 (19) and bacterial primer 27F (16) as the primer pair was performed with a PCR cycler as follows: initial denaturation at 94°C for 5 min, followed by 30 cycles of denaturation (45 s at 94°C), annealing (45 s at 55°C), and extension (1 min at 72°C) before a final extension at 72°C for 5 min. After confirmation on a 0.8% agarose gel, the PCR product was ligated into the pCRII-TOPO vector provided in the TOPO TA cloning kit according to the manufacturer's instructions (Invitrogen, Groningen, The Netherlands). Clones with the correct 16S rRNA gene inserts were confirmed using PCR amplification with the vector-specific primer set M13F and M13R before being further screened by denaturing gradient gel electrophoresis (DGGE). PCR products obtained by applying the primers 341F-GC (26) and 907R (16) were used to run DGGE gels. DGGE was performed using the D-Gene System (Bio-Rad, Herlev, Denmark) according to the manufacturer's guidelines, using polyacrylamide gels with a denaturing gradient of 30 to 70%. The gels were run for 15 h at 100 V in 1× Tris-acetate-EDTA buffer at 60°C before being stained with 100 ng/μl SYBR Gold (Molecular Probes, Taastrup, Denmark). Plasmids showing different patterns were chosen to be sequenced.
PCR amplification for sequencing was performed on diluted plasmids using the M13 primer set. Sequence reactions were carried out using 0.5 μl primers (10 pmol μl−1), 5 μl DYEnamic ET dye terminator sequence kit (Amersham Biosciences, Hillerød, Denmark), and 2 to 7 μl purified PCR product in each setup. After amplification, the reaction products were sequenced on a MegaBACE1000 DNA-sequencing system (Amersham Biosciences, Hillerød, Denmark).
Phylogenetic analysis.
The partial 16S rRNA gene sequences obtained were retrieved in ARB software (http://www.arb-home.de) and aligned. The aligned sequences were checked in the Ribosomal Database Project (http://rdp.cme.msu.edu/) for chimeric artifacts using the check_chimera tool (22) before being compared in GenBank using the BLAST program (1). Then, the sequences and their closely related sequences were aligned in ARB, and a phylogenetic tree based on the neighbor-joining method (21, 31) was built. For calculation of a consensus tree, sequences longer than 1,300 bp were processed by the maximum-likelihood method to determine the topology of the tree before the shorter sequences were added using the function provided in the ARB program.
Oligonucleotide probe design and specification.
Oligonucleotide probes Actino-221 and Actino-658 targeting the 16S rRNA sequences retrieved were designed using functions provided in the ARB software. The specificities of these probes were further confirmed by the use of the Check Probe program in the Ribosomal Database Project (21). Competitors for each probe were designed to exclude bonding to bacteria with one mismatch. The clone-FISH technique (33) was adopted to specify the optimal formamide (FA) concentrations for each probe. Briefly, clones Ska2, Ska6, and Ska18, which have zero to two mismatches with probes Actino-221 and Actino-658, were inserted into plasmid pGEMT (Promega, Mannheim, Germany), which was cloned into JM109(DE3) host cells (Promega, Mannheim, Germany) equipped with a T7 RNA polymerase. The JM109(DE3) cells with the right insertion were incubated at 37°C with IPTG (1 mM) and chloramphenicol (170 mg/liter) before being fixed in 4% paraformaldehyde for later FISH probing. The fluorescence intensities of 200 individual cells at FA concentrations of 0 to 60% (in 5% steps) were evaluated by image analysis using MetaMorph software (Universal Image Corporation, Downingtown, PA).
FISH combined with chemical staining.
Neisser staining (for polyphosphate), Nile blue staining, and Sudan black staining (for lipidic inclusions) were carried out as previously described (18). When carried out in combination with FISH, FISH was performed prior to identifying the cells that were of interest, followed by chemical staining. The staining reaction of the probe-defined cell was examined after relocation on the glass slide.
Microscopy.
FISH images of bacterial cells responding positively to the various probes used were captured with an epifluorescence microscope as described before (15). For quantitative FISH analysis, at least 12 microscopic fields (magnification, ×1,000) were analyzed. Within each field, cells hybridized to a given probe were expressed as a percentage of the total area of bacteria hybridized by the EUBmix using the functions provided in MetaVue (Universal Image Corporation, Downingtown, PA).
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences obtained in this study have been deposited in the GenBank database under accession numbers AY710271 to AY710289.
RESULTS
Screening for PAO in Skagen WWTP.
MAR-FISH was used to identify potential PAO in the industrial full-scale EBPR plant Skagen WWTP. Activated sludge was first incubated under anaerobic conditions with organic substrates so that potential PAO could fill their intracellular storage pools. Subsequently, the sludge was incubated with 33Pi, and bacteria able to take up 33Pi and accumulate poly(P) with oxygen, nitrate, or nitrite present as an electron acceptor were regarded as potential PAO. FISH analysis with various oligonucleotide probes was applied to identify these MAR-positive bacteria. The results showed that when acetate was used as the organic substrate during the 2-h anaerobic preincubation, many RPAO hybridizing with probe PAO651 subsequently took up 33Pi in the presence of oxygen, nitrate, or nitrite. When wastewater was used as an organic substrate during the anaerobic period, both RPAO and many Actinobacteria hybridizing with probe HGC69a were able to take up 33Pi in the presence of oxygen or nitrate, but not nitrite. Further FISH probing showed that these APAO also hybridized with probe actino-1011 designed for uncultured Tetrasphaera-related bacteria with poly(P) storage ability (19). Three different morphotypes hybridized with both actino-1011 and HGC69a in the sludge. These included small cocci in clusters of tetrads, short rods in clumps, and some filamentous bacteria. Only some of the cocci in tetrads and the short rods were able to take up 33Pi under the conditions tested, while most of the cocci in tetrads and short rods and all the filaments were positive with the Neisser stain, suggesting poly(P) storage ability. During the entire experimental period, the cocci in clusters of tetrads and the short rods were much more abundant than the filamentous morphotype.
Clone library analysis.
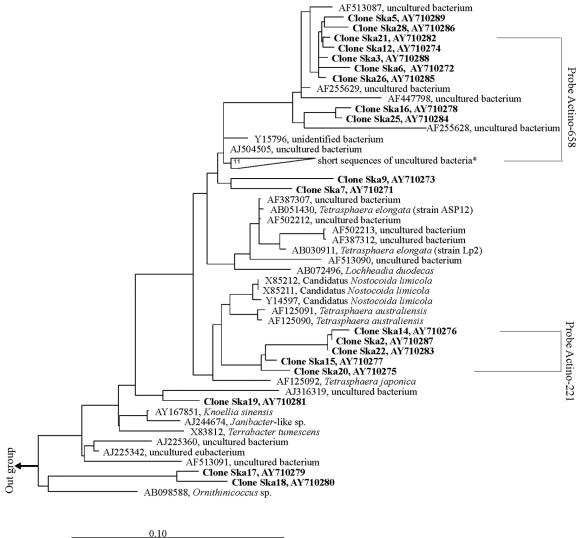
To further investigate the phylogeny of these APAO, a clone library was constructed based on the community 16S rRNA gene amplified by using the unlabeled probe actino-1011 as the reverse primer and the bacterial primer 27F as the forward primer. Forty clones with correct DNA inserts were selected and screened using DGGE. Nineteen operational taxonomic units were identified and sequenced. The same number of clones (AY710271 to AY710289) with 950 to 970 base pairs and sharing 88.3 to 99.9% similarity were successfully obtained. Their phylogenetic relationship with other closely related bacteria is shown in the phylogenetic tree (Fig. 1). Clones Ska3, Ska5, Ska6, Ska12, Ska16, Ska21, Ska25, Ska26, and Ska28 share 97.9 to 99.9% similarity, and they are closely (97.4 to 99.1% similarity) affiliated with four previously reported full sequences (>1,400 bp)—AF513087 (38), AF255629, AF255628 (19), and Y15796 (7, 10)—and with 13 partial sequences (200 to 699 bp) (7, 10) of uncultured bacteria. All these sequences were obtained from EBPR systems. Clones Ska7 and Ska9 are also related to this group, but with lower similarity (<96% similarity). Clones Ska2, Ska14, Ska15, Ska20, and Ska22, sharing 99.1 to 99.9% similarity, form a separate cluster, which is related to the 16S rRNA gene sequences of T. australiensis (96.3 to 97.6% similarity) and T. japonica (95.3 to 96.3% similarity), both isolated from activated sludge. Clone Ska19 is affiliated with a sequence (AJ316319) of an uncultured bacterium from a mural painting environment. Clones Ska18 and Ska17 are related (95.2%) to sequences of Ornithinicoccus sp. found in composting sludge.
FIG. 1.
Phylogenetic tree based on the neighbor-joining method showing the phylogenetic affiliation of Tetrasphaera-related bacteria. Longer sequences (>1,300 bp) were used to build the tree before shorter sequences were inserted using the function provided in the ARB software. Boldface names indicate the sequences obtained from this study. The tree was rooted with Microlunatus phosphovorus (D26169). The scale bar corresponds to one substitution per 10 nucleotides. *, short sequences (200 to 250 bp), including AJ225376 to AJ225379, AJ225370, AJ225364, AJ225356, AJ225355, AJ225353, AJ225351, and AJ225341.
Design and testing of oligonucleotide probes.
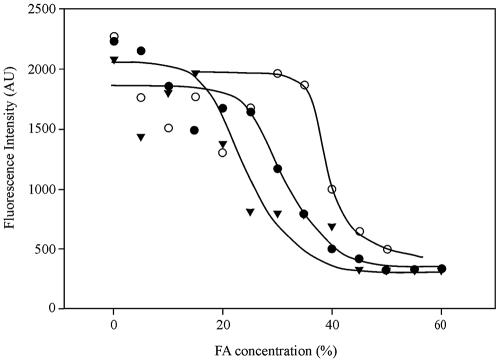
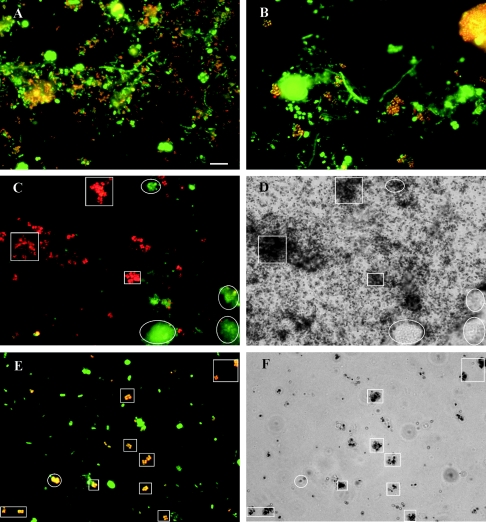
Two oligonucleotide probes, Actino-658 and Actino-221, targeting most, but not all, of the clones retrieved in this study (Fig. 1) were designed to further clarify the phylogenies of different morphotypes hybridizing by probe actino-1011. The sequences, target regions, and specificities of all probes and competitors are listed in Table 1. The optimal FA concentrations for FISH analysis were found by using the clone-FISH technique (33), since no pure culture is available which perfectly matches any of the probes. The optimal FA concentration was approximately 40% for probe Actino-658 and 30% for probe Actino-221. Figure 2 gives an example of the dissociation curve of probe Actino-658 with the host cells JM109(DE3) at different FA concentrations. The dissociation curves obtained agreed with those measured by biomass FISH probing (data not shown). In the presence of the competitors, probe Actino-658 hybridized with the short rods (Fig. 3A) (0.3 to 0.5 μm thick and 0.8 to 1.0 μm long), also fluorescing with probe actino-1011. Probe Actino-221 hybridized with all the cocci (0.5 to 1.0 μm) in tetrads (Fig. 3B), also fluorescing with probe actino-1011. No pretreatment was necessary for FISH probing using probe Actino-658, while probe Actino-221 needed lysozyme treatment for 15 min at room temperature. Further treatment with mutanolysin did not improve the signal.
TABLE 1.
Oligonucleotide probes and competitors designed for detection of actinobacterial PAO
| Name | Abbreviation | Target group | Sequence (5′-3′) | % Formamide |
|---|---|---|---|---|
| S-S-Actino-221-a-A-18 | Actino-221 | Actinobacterial PAO | CGCAGGTCCATCCCAGAC | 30 |
| Actino-221 comp1 | c1Actino-221 | CGCAGGTCCATCCCATAC | ||
| Actino-221 comp2 | c2Actino-221 | CGCAGGTCCATCCCAGAG | ||
| S-S-Actino-658-a-A-18 | Actino-658 | Actinobacterial PAO | TCCGGTCTCCCCTACCAT | 40 |
| Actino-658comp1 | c1Actino-658 | TCCGGTCTCCCCTACCAC | ||
| Actino-658comp2 | c2Actino-658 | ATTCCAGTCTCCCCTACCAT |
FIG. 2.
Dissociation curves of probe Actino-658 with clones Ska6 (zero mismatches; ○), Ska2 (one mismatch; •), and Ska18 (two mismatches; ▾). Each point represents the average fluorescence intensity (AU [arbitrary units]) of 200 individual cells.
FIG. 3.
FISH and MAR images of activated sludge with APAO. (A and B) FISH images showing bacteria hybridized with the bacterial probes EUBmix (green) and the rod-APAO probe Actino-658 (A) or the coccus-APAO probe Actino-221 (B) (red). Yellow microcolonies and cells (overlay of red and green) are the rod-APAO (A) or the coccus-APAO (B). (C) FISH image showing the coccus-APAO hybridized with probe Actino-221 (red) and RPAO hybridized with probe PAO651 (green). (D) Bright-field MAR image showing that most coccus-APAO (shown with squares) and a few RPAO (shown with ellipses) seen in panel C take up 33Pi aerobically after an anaerobic preincubation with Casamino Acids. (E and F) FISH image (E) and MAR image (F) show the coccus-APAO (yellow) taking up labeled oleic acid anaerobically. The coccus-APAO, indicated with squares (E), have positive MAR signals (F). An example of only slightly MAR-positive coccus-APAO is indicated by a circle (E and F). Bar = 10 μm.
Ecophysiology determined by MAR-FISH and chemical staining.
The uptake of 33Pi (Table 2) and various organic substrates by bacteria hybridizing with probe Actino-221 or Actino-658 were further characterized by using MAR-FISH. For both probe-defined populations, it was found that the aerobic uptake of 33Pi strongly depended on the type of organic substrate that was used during the 2-h anaerobic preincubation. When raw wastewater was added, 10 to 90% (values varied in different experiments with different wastewater samples) of the Actino-221-positive cocci in tetrads and almost all (>95%) the Actino-658-positive short rods took up 33Pi. Their poly(P) storage was confirmed by using FISH combined with Neisser stain, which clearly showed intracellular Neisser-positive granules. In control experiments without any added external organic substrate in the anaerobic preincubation, only a few of the Actino-658-positive short rods and none of the Actino-221-positive cocci were able to take up 33Pi in the aerobic period, indicating that both morphotypes were able to take up organic substrates anaerobically and store them as energy sources, which subsequently were used to take up 33Pi under aerobic conditions. Thus, the physiological traits shown here meet the criteria for PAO, so for convenience, the two gene probe-defined morphotypes are here called coccus-APAO and rod-APAO.
TABLE 2.
Effects of different organic substrates during anaerobic preincubation on the uptake of 33Pi under different electron acceptor conditions by the coccus- and rod-APAOa
| Organism | Incubation conditions
|
Uptake by APAO with organic substrates used in anaerobic preincubationc
|
|||||
|---|---|---|---|---|---|---|---|
| EAb | Time (h) | Wastewater | Casamino Acids | Oleic acid | Acetate | Not added | |
| Coccus-APAO | O2 | 3 | 10-90 | 95-100 | 0 | 0 | 0 |
| NO3− | 3 | 10-90 | 95-100 | 0 | 0 | 0 | |
| NO2− | 3 | 0 | 0 | 0 | 0 | ND | |
| Not added | 3 | 0 | 0 | 0 | 0 | ND | |
| O2 (+WW)d | 3 | 0 | ND | ND | ND | ND | |
| Rod-APAO | O2 | 3 | 95-100 | 95-100 | ND | 0-5 | 0-5 |
| NO3− | 3 | 95-100 | 95-100 | 0 | 0 | 0-5 | |
| NO2− | 3 | 0 | 0 | ND | 0 | ND | |
| Not added | 3 | 0 | 0 | ND | 0 | ND | |
| O2 (+WW) | 3 | 0 | ND | ND | ND | ND | |
Incubation conditions and results based on MAR investigations. Incubation with 33Pi was carried out after 2 h of anaerobic incubation with different organic substrates.
EA, electron acceptors.
Sludge concentration used in all anaerobic preincubations was 1 g (suspended solids)/liter. Acetate, oleic acid, or Casamino Acids were added at a final concentration of 1.5 mM, 1.0 mM, and 125 mg/liter, respectively. Ranges of APAO taking up 33Pi are shown as averages of percentages obtained by counting 50 APAO cells in at least three independent experiments. ND, not determined.
WW, wastewater.
The possible effect of complex organic substrates other than wastewater during anaerobic preincubation on the subsequent 33Pi uptake by the APAO was also investigated (Table 2). Anaerobic incubation with Casamino Acids made most of the two morphotypes of APAO (>95%) able to take up 33Pi under aerobic conditions. Figure 3C and D show examples of the coccus-APAO taking up 33Pi. This indicates that Casamino Acids were taken up and stored anaerobically as a so-far-unknown compound (see below) by the APAO, suggesting that amino acids are favorable organic substrates. To further confirm this, a labeled amino acid mixture with an amino acid composition similar to that of Casamino Acids was added under anaerobic and aerobic conditions. Both the coccus-APAO and the rod-APAO were able to take up the labeled amino acids under both electron acceptor conditions.
The abilities of APAO to take up formic acid, acetate, propionate, butyric acid, pyruvate, lactate, ethanol, glucose, oleic acid, aspartic acid, glutamic acid, leucine, glycine, thymidine, or mixed amino acids were also investigated under anaerobic or aerobic conditions. None of the substrates tested could be taken up by the rod-APAO. The coccus-APAO could take up only oleic acid (Fig. 3E and F), but surprisingly, after anaerobic uptake, no subsequent uptake of 33Pi took place (Table 2). The presence of the glycolysis inhibitor iodoacetate, which acts on glyceraldehyde-3-phosphate dehydrogenase (5), added at a final concentration of 200 mg/liter (15) did not inhibit the coccus-APAO's uptake of oleic acid or the APAO's (both the coccus- and the rod-APAO) uptake of Casamino Acids (results inferred from their ability to take up Pi aerobically after anaerobic incubation with Casamino Acids in the presence of iodoacetate) under anaerobic conditions. This indicates that hydrolysis of glycogen was not necessary for the uptake of oleic acid (for the coccus-APAO only) and Casamino Acids. In agreement with the survey (see above), APAO could not take up acetate, so addition of acetate during the anaerobic preincubation could not promote any aerobic 33Pi uptake in the APAO.
A series of experiments were conducted in order to investigate whether nitrate or nitrite could be used as an electron acceptor by these APAO for growth or to promote uptake of 33Pi after anaerobic uptake of substrate (Table 1). Under anaerobic conditions, most (>95%) of the coccus-APAO were able to take up oleic acid during a 3-h incubation, but they could not continue to do so after 6 h (after a 6-h preincubation with unlabeled oleic acid). This shows that the coccus-APAO were not able to grow on oleic acid under anaerobic conditions but were only able to store a certain amount of it as a so-far-unknown compound. However, in the presence of nitrate or oxygen, but not nitrite, they continued to take up oleic acid after 6 h. This strongly indicates that the coccus-APAO were not able to denitrify but were probably able to reduce nitrate to nitrite and thereby get energy for growth. This was also supported by the observation that coccus-APAO could take up 33Pi in the presence of nitrate, but not nitrite, after anaerobic uptake of Casamino Acids or raw wastewater. The rod-APAO's ability to reduce nitrate or nitrite is still not clear, although their 33Pi uptake in the presence of nitrate, but not nitrite (Table 2), seems to indicate that they behaved like the coccus-APAO, unable to denitrify but able to grow in the presence of oxygen or nitrate.
The potential of APAO to store PHA during anaerobic substrate uptake was further investigated. When Nile blue or Sudan black staining was combined with FISH, none of the APAO were positively stained after anaerobic incubation with wastewater, Casamino Acids, or oleic acid, indicating a lack of capability to store PHA.
Distribution of PAO in full-scale EBPR plants.
The distributions of APAO and RPAO were investigated in 10 full-scale, well-working EBPR plants with nitrogen removal (nitrification and denitrification). The bacteria were identified and quantified by FISH probing using the probes Actino-221, Actino-658, and PAO651. APAO were present (3 to 35% of all Bacteria) in all treatment plants investigated (Table 3). The coccus-APAO were the most abundant and seemed to be most common in treatment plants treating only, or mainly, industrial wastewater, although they were equally present in the plants treating mainly domestic wastewater. These plants typically receive a 10 to 30% organic load from various industries. Up to one-third of all Bacteria were coccus-APAO in Dan Shellfish WWTP, treating wastewater from the production of various shellfish. Rod-APAO were present in significant numbers in both some industrial and some domestic plants, but not all. The RPAO were mainly dominant in the domestic plants (9 to 17%) and were hardly present in the industrial plants (<3%), except Skagen WWTP (17% ± 6%).
TABLE 3.
Abundances of APAO and RPAO in full-scale EBPR plants as determined by FISHa
| Plant name | Wastewater characteristics | Abundanceb
|
||
|---|---|---|---|---|
| Coccus-APAO | Rod-APAO | RPAO | ||
| Egaa WWTP | Domestic | 6 ± 3 | 10 ± 2 | 17 ± 6 |
| Aalborg East WWTP | Domestic | 3 ± 2 | <1 | 16 ± 11 |
| Aalborg West WWTP | Domestic | 5 ± 3 | <1 | 15 ± 10 |
| Naestved WWTP | Domestic | 9 ± 5 | <1 | 9 ± 5 |
| Usseroed WWTP | Domestic | 3 ± 1 | 5 ± 2 | 3 ± 1 |
| Vedbaek WWTP | Domestic | 5 ± 2 | 4 ± 1 | 1 ± 1 |
| Assens WWTP | Mainly industrial (dairy) | 15 ± 5 | 14 ± 3 | 2 ± 1 |
| Rodkaersbro WWTP | Industrial (dairy) | 17 ± 9 | <1 | <1 |
| Skagen WWTP | Industrial (fish) | 7 ± 3 | 12 ± 5 | 19 ± 3 |
| Dan Shellfish WWTP | Industrial (mussels) | 30 ± 8 | 5 ± 2 | 2 ± 1 |
The oligonucleotide probes used were Actino-221 (coccus-APAO), Actino-658 (rod-APAO), and PAO657 (RPAO).
Abundances are expressed as percentages (mean ± standard error) of all Bacteria (EUBmix).
If the fraction of APAO (including the coccus- and the rod-APAO) is compared to the fraction of RPAO, it appears that APAO are also important PAO in several municipal EBPR plants treating domestic wastewater. The ratio between the total APAO and RPAO, as estimated from their mean percentages, was low in Aalborg West WWTP (0.3 to 0.4) and Aalborg East WWTP (0.2 to 0.3), around 1 in Egaa WWTP (0.9) and Naestved WWTP (1.1), and high or very high in Usseroed WWTP (2.7) and Vedbaek WWTP (9.0). With the information about plant operation and wastewater characteristics available, it was not possible to explain these differences.
DISCUSSION
Some Actinobacteria have been suggested to be potential PAO, based on laboratory-scale reactor studies and on physiological investigations of pure cultures (11, 19, 29), but this is the first time they have been identified and found in significant amounts and exhibiting PAO behavior in full-scale EBPR systems. The polyphosphate-accumulating capability was evaluated with MAR-FISH and FISH combined with Neisser staining, and the APAO found in this study demonstrated the ability to take up Pi and accumulate poly(P) using oxygen or nitrate as an electron acceptor after anaerobic uptake of organic substrates. However, their physiology differs significantly from that proposed for the putative PAO (34), e.g., RPAO (15), as will be discussed below.
Identification and design of oligonucleotide probes for APAO.
Most (17 of 19) of the retrieved sequences are more closely related to the genus Tetrasphaera in the family Intrasporangiaceae of the class Actinobacteria than to any other known genus. This genus currently consists of three main species, T. australiensis (cocci growing in tetrads), T. japonica (cocci growing in tetrads), and T. elongata (short rods). However, a relatively reliable identification must await the retrieval of full sequences. Of the 19 sequences, 12 were covered by the probes Actino-658 and Actino-221 designed in this study. It was not possible to design a single probe targeting the clones Ska28 and Ska5 and sequence AF51308 together with all the sequences targeted by probe Actino-658 (>3 mismatches to Ska5 and Ska28; 1 mismatch to AF51308), although they were all closely affiliated, so these three sequences were left out. The possibility that clones Ska5 and Ska18 are artifacts introduced by PCR amplification cannot be ruled out, although they do not appear to be, based on results obtained by checking with the CHECK-CHIMERA tool. In FISH probing, Actino-658 hybridized with most of the short rods, while Actino-221 hybridized with all the cocci in tetrads; both were able to take up 33Pi and store poly(P) as visualized using the probes HGC69a and actino-1011. Thus, most probably all tetrads and most rods able to take up 33Pi in the sludge were identified. Therefore, we did not attempt to further clarify the morphologies and physiologies related to the other sequences by designing other probes. It is recognized, however, that the phylogenies and physiologies (see below) of these Tetrasphaera-related Actinobacteria are diverse.
Phylogenetically, the sequences targeted by probe Actino-658 are more similar (97.4 to 99.1%) to sequences AF255629 and AF255628 (both longer than 1,400 bp) of uncultured bacteria with poly(P) storage ability found in a laboratory-scale EBPR system (19) than to sequences of available isolates of Tetrasphaera (92.9 to 94.0%). Sequence AF255629 shares only <95% similarity with sequence AF255628, and both are more distantly related (<95% similarity) to the sequences of Tetrasphaera spp., suggesting that the phylogenies of those bacteria targeting by probe Actino-658 are diverse and that they may belong to an unknown genus other than Tetrasphaera. Thus, the exact phylogeny of the rod-APAO has to be further clarified.
As mentioned above, the sequences of the coccus-APAO are more closely affiliated with those of T. australiensis and T. japonica (95.3 to 97.6% similarity) than the rod-APAO, and they are all cocci typically growing in tetrads. However, the fact that the coccus-APAO hybridized with probe actino-1011, which has four mismatches with T. australiensis, suggests that they most probably are not T. australiensis. Similarly, when the oligonucleotide probe Tet63 (14), specifically targeting T. japonica, was applied to the Skagen WWTP biomass, no positive signal was observed (data not shown), indicating that the coccus-APAO are not T. japonica. It is still unknown whether they belong to a new species of the genus Tetrasphaera or to another, unknown genus. Also some physiological differences between the coccus-APAO and T. australiensis and T. japonica as revealed by using MAR-FISH (see below) support the difference in their phylogenies. A better determination of their phylogenetic affiliation requires longer sequences than the 950 to 970 bp obtained in this study. We tried to construct a clone library using bacterial universal primers 27F and 1492R to obtain nearly full-length sequences for a better identification of the APAO, but only a few clones affiliated with Actinobacteria could be identified by sequencing 80 clones, and they were all distantly related to Tetrasphaera (data not shown). Tetrad-forming bacteria can be hard to break in DNA extraction procedures, as reported by Beer et al. (4), which could be one explanation.
The clone-FISH technique is a useful tool for specifying the hybridization conditions for oligonucleotide probes targeting bacteria for which no pure cultures with a perfect match (or one or two mismatches) are available (33). The sequences retrieved in this study have different numbers of mismatches (zero, one, and two) to probes Actino-658 and Actino-221, and clone-FISH was successfully used to specify the FA concentrations of these probes. Some differences in the fluorescence intensities of individual host cells hybridized by a probe at a certain FA concentration were found, so the fluorescence intensity for at least 200 cells was examined at each FA concentration.
Besides the two morphotypes hybridizing with Actino-221 and Actino-568, the filamentous morphotype hybridizing with probe actino-1011 also had intracellular poly(P) granules, as determined by Neisser staining. This is very similar to “Ca. Nostocoida limicola” reported from activated sludge (6). However, as no 33Pi uptake could be detected under the conditions tested, and as this morphotype was not very common, no further studies were carried out.
Comparison of the physiology of APAO to those of isolated Tetrasphaera spp.
The APAO found in this study are in some physiological and morphological aspects similar to their closest isolated relatives in the genus Tetrasphaera. The morphology of the rod-APAO was very similar to that of T. elongata strain ASP12 (29) and strain LP2 (11), all being short rods of similar sizes. They all contain intracellular poly(P) granules and have nitrate-reducing ability (to nitrite). Moreover, rod-APAO and strain ASP12 consume Casamino Acids anaerobically and take up Pi aerobically. Furthermore, the rod-APAO and strain ASP12 grow only on complex media. Strain LP2 is different, as it can utilize various organic substrates, including short-chain fatty acids and glucose.
The morphology of coccus-APAO is very similar to those of T. australiensis and T. japonica (23), all being typical cocci growing in clusters of tetrads. Physiologically, they seem to use only a restricted range of specific organic substrates. They all grow on complex media, such as Casamino Acids, indicating that they can use certain amino acids. Furthermore, the coccus-APAO utilized oleic acid, while T. japonica grows on pyruvate and T. australiensis on lysine, sucrose, and xylose, as revealed by the Biolog GP analysis systems (23). Moreover, the coccus-APAO were most likely able to reduce nitrate to nitrite, while T. australiensis and T. japonica cannot. All these observations indicate that, potentially, there are a number of Actinobacteria with rather diverse physiologies that could act as PAO in activated-sludge systems. This also shows that it is difficult to predict the behavior of the APAO found in this study in real treatment plants on the basis of studies of pure cultures of close relatives (bacteria related to the genus Tetrasphaera).
Physiology of APAO compared to EBPR biochemical models.
The APAO described in this study differ significantly in their physiologies from the putative PAO, the RPAO. Consequently, there is also a substantial difference between the physiology of the APAO and that predicted by the general biochemical models used to explain the behavior of PAO (25, 34, 37). A great difference is the range of organic substrate to be taken up, where APAO utilize certain amino acids while the RPAO consume acetate, propionate, some amino acids, and other simple compounds (15). The identities of the amino acids consumed by the APAO are still unclear, as the labeled amino acid mixture used consisted of 15 different amino acids. Of these, only aspartic acid, glutamic acid, glycine, and leucine were tested individually in MAR-FISH, and none of these could be taken up by the APAO, while glutamic acid could be utilized by RPAO. In anaerobic Pi release experiments using sludge from Skagen WWTP, different Pi release profiles were observed by adding Casamino Acids or acetate to the sludge, and more Pi was released when both Casamino Acids and acetate were added under anaerobic conditions (data not shown), confirming that both APAO and RPAO were actively involved in the EBPR process in the plant. Furthermore, in contrast to RPAO, the APAO seemed not to form PHA during anaerobic uptake of organic substrates, as no PHA could be detected by chemical staining using Nile blue and Sudan black. This is in agreement with observations of all the Tetrasphaera isolates (11, 23). Furthermore, the glycolytic pathway was unimportant for anaerobic substrate uptake in the APAO, while RPAO need to degrade internal glycogen by the glycolytic pathway to provide reducing power for PHA formation (25). This was observed in the finding that the presence of iodoacetate, which inhibits glycolysis (15), did not affect the anaerobic uptake of oleic acid (for coccus-APAO only) or Casamino Acids. Thus, it is unknown how the bacteria store energy from anaerobic substrate uptake of organic substrate to take up Pi under aerobic conditions.
It was surprising that the coccus-APAO were able to take up oleic acid anaerobically and store it as an unknown compound(s) that was used to grow in the following aerobic phase but could not use the storage compound(s) as an energy source for taking up 33Pi under aerobic conditions, as with Casamino Acids. However, an almost similar physiology has been described for “Ca. Microthrix parvicella”, a filamentous member of the Actinobacteria that grows in EBPR systems, where it can take up oleic acid under anaerobic and aerobic conditions and use it as an energy and carbon source under aerobic conditions (28). Like APAO, “Ca. Microthrix parvicella” is probably able to take up Pi (based on the presence of Neisser-positive granules), but it has so far not been possible experimentally to induce 33Pi uptake based on uptake of oleic acid (3). Thus, this physiology seems very similar to that of the coccus-APAO, except that intracellular granules positive with Nile blue and Sudan black indicate the presence of PHA or lipid in “Ca. Microthrix parvicella”. The identities of intracellular (storage) compounds likely formed by APAO after anaerobic substrate uptake are still unknown and may depend on the substrate taken up, thus providing different abilities for uptake of Pi. These effects of different organic substrates on PAO metabolism (growth with or without Pi uptake) have never been reported before. Whether other PAO, e.g., RPAO, have the same physiological traits is still unknown. Further research using pure cultures of Tetrasphaera spp., e.g., T. australiensis and T. elongata (strain AS12), which share some physiological traits with APAO, may help to clarify this.
Another difference between RPAO and APAO was the ability to denitrify. In contrast to RPAO, the APAO could not denitrify and use nitrite as an electron acceptor for uptake of Pi. However, APAO could take up Pi in the presence of nitrate, so it is likely that they could reduce nitrate to nitrite. This is in accordance with some of the pure cultures of Tetrasphaera (11, 23).
The efficiency of Pi uptake on a single-cell level appeared very similar for APAO and RPAO based on MAR signals of 33Pi uptake. However, when wastewater was used as a substrate during the anaerobic preincubation, only a fraction of APAO took up 33Pi in different experiments (10 to 95%) during the experimental period, probably reflecting variations in the composition of the wastewater, as most of them (>95%) could take up 33Pi after anaerobic preincubation with Casamino Acids. An inactive fraction was also found for RPAO (15), indicating that growth conditions were often suboptimal for some PAO. It also shows that it is difficult to use their abundance, as quantified by FISH, to accurately estimate EBPR activity in a treatment plant.
The results obtained here also emphasize that it is impossible to explain the behavior of the EBPR processes in full-scale plants using the usual biochemical or mathematical models (12, 34). The models are all based on uptake of acetate and metabolism of PHA and glycogen, so they cannot explain the processes going on in EBPR plants with a predominance of APAO. Our findings in this study may help to explain the unreliability of EBPR processes (34). Also, common tests for monitoring potential EBPR activity in treatment plants may give erroneous results; usually acetate is added to simulate anaerobic release of Pi (36), which will not work in plants with a predominance of APAO.
Role of APAO in full-scale EBPR systems.
The widespread distribution and the high number of APAO in full-scale EBPR systems, all with nitrogen removal (nitrification and denitrification), show that the probe-defined APAO and RPAO can be considered the most important PAO known today in full-scale EBPR plants. Furthermore, it is important to stress that the two groups together constituted up to 38% of all bacteria present (in Skagen WWTP), and thus were very important for the entire ecosystem in terms of transformation of organic matter, P, and N and for floc formation. The numbers of APAO were highest in the four plants treating industrial wastewater (17 to 35%), and it is interesting that one plant contained only APAO and no RPAO at all (Roedkjaersbro). Our results here suggest that APAO are mainly amino acid consumers during anaerobic uptake of substrate, while RPAO are broader consumers, using acetate, propionate, and a few other compounds (15), so the availability of proper organic substrates may be a key factor in the selection of the different species. Industrial wastewater often contains high levels of proteins, and the APAO may benefit from that. The ecological relationship between the coccus-APAO and the rod-APAO is still not clear. We could see hardly any difference in the physiologies of the two types (except for uptake of oleic acid), so other, so-far-unresolved factors must regulate their dominance. More information about plant operation and wastewater characteristics may help resolve in detail the differences that control the selection of the three PAO.
Acknowledgments
The Danish Technical Research Council supported this study under the framework program “Activity and Diversity in Complex Microbial Systems.”
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I. 1995. In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes, p. 1-15. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecological manual. Kluwer Academic Publications, London, United Kingdom.
- 3.Andreasen, K., and P. H. Nielsen. 2000. Growth of Microthrix parvicella in nutrient removal activated sludge plants: studies of in situ physiology. Water Res. 34:1559-1569. [Google Scholar]
- 4.Beer, M., Y. H. Kong, and R. J. Seviour. 2004. Are some putative glycogen accumulating organisms (GAO) in anaerobic:aerobic activated sludge systems members of the α-Proteobacteria? Microbiology 150:2267-2275. [DOI] [PubMed] [Google Scholar]
- 5.Bickis, I. J., and J. H. Quastel. 1965. Effects of metabolic inhibitors on energy metabolism of Ehrlich ascites carcinoma cells. Nature 205:44-46. [DOI] [PubMed] [Google Scholar]
- 6.Blackall, L. L., E. M. Seviour, D. Bradford, S. Rossetti, V. Tandoi, and R. J. Seviour. 2000. ‘Candidatus Nostocoida limicola’, a filamentous bacterium from activated sludge. Int. J. Syst. Evol. Microbiol. 50:703-709. [DOI] [PubMed] [Google Scholar]
- 7.Christensson, M., L. L. Blackall, and T. Welander. 1998. Metabolic transformations and characterisation of the sludge community in an enhanced biological phosphorus removal system. Appl. Microbiol. Biotechnol. 49:226-234. [Google Scholar]
- 8.Crocetti, G. R., P. Hugenholtz, P. L. Bond, A. Schuler, J. Keller, D. Jenkins, and L. L. Blackall. 2000. Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation. Appl. Environ. Microbiol. 66:1175-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erhart, R., D. Bradford, R. J. Seviour, R. Amann, and L. L. Blackall. 1997. Development and use of fluorescent in situ hybridization probes for the detection and identification of “Microthrix parvicella” in activated sludge. Syst. Appl. Microbiol. 20:310-318. [Google Scholar]
- 10.Eschenhagen, M., M. Schuppler, and I. Roske. 2003. Molecular characterization of the microbial community structure in two activated sludge systems for the advanced treatment of domestic effluents. Water Res. 37:3224-3232. [DOI] [PubMed] [Google Scholar]
- 11.Hanada, S., W. T. Liu, T. Shintani, Y. Kamagata, and K. Nakamura. 2002. Tetrasphaera elongata sp. nov., a polyphosphate-accumulating bacterium isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 52:883-887. [DOI] [PubMed] [Google Scholar]
- 12.Henze, M., W. Gujer, T. Mino, T. Matsuo, M. C. Wentzel, G. V. R. Marais, and M. C. M. Van Loosdrecht. 1999. Activated sludge model no. 2d, ASM2d. Water Sci. Technol. 39:165-182. [Google Scholar]
- 13.Hesselmann, R. P. X., C. Werlen, D. Hahn, J. R. van der Meer, and A. J. B. Zehnder. 1999. Enrichment, phylogenetic analysis and detection of a bacterium that performs enhanced biological phosphate removal in activated sludge. Syst. Appl. Microbiol. 22:454-465. [DOI] [PubMed] [Google Scholar]
- 14.Kong, Y. H., M. Beer, R. J. Seviour, K. C. Lindrea, and G. N. Rees. 2001. Structure and functional analysis of the microbial community in an aerobic:anaerobic sequencing batch reactor (SBR) with no phosphorus removal. Syst. Appl. Microbiol. 24:597-609. [DOI] [PubMed] [Google Scholar]
- 15.Kong, Y. H., L. J. Nielsen, and H. P. Nielsen. 2004. Microautoradiographic study of Rhodocyclus-related polyphosphate-accumulating bacteria in full-scale EBPR plants. Appl. Environ. Microbiol. 70:5383-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 113-175. In E. Stackebrandt and M. Goodfellow. (ed.), Nucleic acid techniques in bacterial systematics. Wiley Publications, Chichester, United Kingdom.
- 17.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K. H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindrea, K. C., E. M. Secviour, R. J. Seviour, L. L. Blackall, and J. A. Soddell. 1999. Practical methods for the examination and characterization of activated sludge, p. 257-300. In R. J. Seviour and L. L. Blackall (ed.), The microbiology of activated sludge. Kluyver Academic Publications, Dordrecht, The Netherlands.
- 19.Liu, W. T., A. T. Nielsen, J. H. Wu, C. S. Tsai, Y. Matsuo, and S. Molin. 2001. In situ identification of polyphosphate- and polyhydroxyalkanoate-accumulating traits for microbial populations in a biological phosphorus removal process. Environ. Microbiol. 3:110-122. [DOI] [PubMed] [Google Scholar]
- 20.Loy, A., M. Horn, and M. Wagner. 2003. probeBase: an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 28:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maszenan, A. M., R. J. Seviour, B. K. C. Patel, P. Schumann, J. Burghardt, Y. Tokiwa, and H. M. Stratton. 2000. Three isolates of novel polyphosphate-accumulating Gram-positive cocci, obtained from activated sludge, belong to a new genus, Tetrasphaera gen. nov., and description of two new species, Tetrasphaera japonica sp. nov. and Tetrasphaera australiensis sp. nov. Int. J. Syst. Evol. Microbiol. 50:593-603. [DOI] [PubMed] [Google Scholar]
- 24.Maszenan, A. M., R. J. Seviour, B. K. C. Patel, P. Schumann, and G. N. Rees. 1999. Tessaracoccus bendigoensis gen. nov., sp. nov., a Gram-positive coccus occurring in regular packages or tetrads, isolated from activated sludge biomass. Int. J. Syst. Bacteriol. 49:459-468. [DOI] [PubMed] [Google Scholar]
- 25.Mino, T., M. C. M. Van Loosdrecht, and J. J. Heijnen. 1998. Microbiology and biochemistry of the enhanced biological phosphate removal process. Water Res. 32:3193-3207. [Google Scholar]
- 26.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel-electrophoresis analysis of polymerase chain reaction-amplified genes-coding for 16S ribosomal RNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen, J. L., D. Christensen, M. Kloppenborg, and P. H. Nielsen. 2003. Quantification of cell-specific substrate uptake by probe-defined bacteria under in situ conditions by microautoradiography and fluorescence in situ hybridization. Environ. Microbiol. 5:202-211. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen, P. H., P. Roslev, T. E. Dueholm, and J. L. Nielsen. 2002. Microthrix parvicella, a specialized lipid consumer in anaerobic-aerobic activated sludge plants. Water Sci. Technol. 46:73-80. [PubMed] [Google Scholar]
- 29.Onda, S., and S. Takii. 2002. Isolation and characterization of a Gram-positive polyphosphate-accumulating bacterium. J. Gen. Appl. Microbiol. 48:125-133. [DOI] [PubMed] [Google Scholar]
- 30.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K. H. Schleifer. 1994. In situ probing of gram-positive bacteria with high DNA G+C content using 23S-ribosomal-RNA-targeted oligonucleotides. Microbiology 140:2849-2858. [DOI] [PubMed] [Google Scholar]
- 31.Saitou, N., and M. Nei. 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 32.Santos, M. M., P. C. Lemos, M. A. M. Reis, and H. Santos. 1999. Glucose metabolism and kinetics of phosphorus removal by the fermentative bacterium Microlunatus phosphovorus. Appl. Environ. Microbiol. 65:3920-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schramm, A., B. M. Fuchs, J. L. Nielsen, M. Tonolla, and D. A. Stahl. 2002. Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ. Microbiol. 4:713-720. [DOI] [PubMed] [Google Scholar]
- 34.Seviour, R. J., T. Mino, and M. Onuki. 2003. The microbiology of biological phosphorus removal in activated sludge systems. FEMS Microbiol. Rev. 27:99-127. [DOI] [PubMed] [Google Scholar]
- 35.Seviour, R. K., C. Lindrea, P. C. Griffiths, and L. L. Blackall. 1999. The activated sludge process, p. 44-75, In R. J. Seviour and L. L. Blackall (ed.), The microbiology of activated sludge. Kluyver Academic Publications, Dodrecht, The Netherlands.
- 36.Tykesson, E., H. Aspegren, M. Henze, P. H. Nielsen, and J. L. Jansen. 2002. Use of phosphorus release batch tests for modelling an EBPR pilot plant. Water Sci. Technol. 45:99-106. [PubMed] [Google Scholar]
- 37.van Loosdrecht, M. C. M., C. M. Hooijmans, D. Brdjanovic, and J. J. Heijnen. 1997. Biological phosphate removal processes. Appl. Microbiol. Biotechnol. 48:289-296. [Google Scholar]
- 38.Wagner, A. M., and E. T. Cloete. 2002. 16S rRNA sequence analysis of bacteria present in foaming activated sludge. Syst. Appl. Microbiol. 25:434-439. [DOI] [PubMed] [Google Scholar]
- 39.Zilles, J. L., J. Peccia, M. W. Kim, C. H. Hung, and D. R. Noguera. 2002. Involvement of Rhodocyclus-related organisms in phosphorus removal in full-scale wastewater treatment plants. Appl. Environ. Microbiol. 68:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]