Abstract
Concentration of water samples is a prerequisite for the detection of the low virus levels that are present in water and may present a public health hazard. The aim of this study was to develop a rapid, standardized molecular method for the detection of enteroviruses in large-volume surface water samples, using a concentration method suitable for the detection of infectious viruses as well as virus RNA. Concentration of water was achieved by a conventional filter adsorption-elution method and ultrafiltration, resulting in a 10,000-fold concentration of the sample. Isolation of virus RNA by a silica-based RNA extraction method was compared with the nonmagnetic and magnetic NucliSens RNA isolation methods. By using the silica-based RNA extraction method in two out of five samples, enterovirus RNA was detected, whereas four out of five samples were positive following RNA isolation with magnetic silica beads. Moreover, estimated RNA levels increased at least 100 to 500 times. Furthermore, we compared enterovirus detection by an in-house reverse transcription (RT)-PCR with a novel commercially available real-time nucleic acid sequence-based amplification (NASBA) assay. We found that the rapid real-time NASBA assay was slightly less sensitive than our in-house RT-PCR. The advantages, however, of a commercial real-time NASBA assay, like the presence of an internal control RNA, standardization, and enormous decrease in turnaround time, makes it an attractive alternative to RT-PCR.
Contamination of surface waters with enteric viruses through the disposal of human wastewater is a concern for public health, especially if these surface waters are used as recreational waters but also if sources are utilized for the production of drinking water. Waterborne outbreaks of enteric viruses have been repeatedly reported (1, 2, 12, 17, 20). Although noroviruses are the most common agents involved in waterborne outbreaks causing gastroenteritis, enteroviruses can cause a wide variety of symptoms in a healthy host. Many enterovirus infections result in mild febrile illness, but they are also capable of causing a wide range of serious illnesses, including aseptic meningitis, myocarditis, and poliomyelitis (26), which emphasize that research on enteroviruses in different water sources is important to be able to assess public health risks.
In water, viruses are usually present in low numbers. To be able to detect these low numbers, several hundred liters of surface water need to be analyzed. A variety of concentration methods is available, many of them consisting of two successive steps. Frequently used methods to concentrate water samples are either electronegative or electropositive membrane filtration or filtration using glass powder or glass wool. A second concentration step can be performed by ultrafiltration (UF) or organic flocculation. Viruses remain infectious in the resulting concentrates but are less suitable to isolate viral RNA. Despite repeated efforts to improve RNA isolation from water samples, no standard method is available (5, 15, 23). Two-phase separation using Dextran T40 and PEG 6000 is a commonly used method to isolate RNA (3, 16, 23, 27) but is, however, not suitable to detect culturable viruses. A more detailed overview of these methods has been described by Wyn-Jones and Sellwood (32).
Enterovirus concentrations in surface waters are often determined by cell culture using Buffalo green monkey (BGM) cells. These cells are widely used in monolayer plaque assays for the routine detection of infectious enteroviruses in water (8). Unfortunately, not all enterovirus types are detected in BGM cells, especially the pathogenic coxsackievirus types A (26), which are difficult to detect; only coxsackievirus A7, A9, and A16 can infect and multiply in BGM cells, whereas other coxsackie A viruses (A1 through A6, A8, A10 through A15, A17 through A22, and A24) are not detected (8). Furthermore, cell culture is an expensive and time-consuming method, implicating the need for the detection of viruses by molecular methods such as conventional reverse transcription (RT)-PCR (11, 29) or nucleic acid sequence-based amplification (NASBA), which isothermally amplifies RNA (6). NASBA detection of viral RNA in water has been described for hepatitis A virus in artificially contaminated sewage water (7, 18). Although mainly described as end point assays, amplification products of both techniques can be detected by real-time methods (14). The major advantage of real-time detection is the ability to quantify amplification products, which is very important to be able to estimate the public health risks of low levels of enteric viruses in surface water. Furthermore, detection and hands-on time are decreased enormously. Real-time RT-PCR is frequently used for detection of enterovirus in clinical samples (19, 25, 28) but has less frequently been described for environmental samples (9). An enterovirus NASBA using end point detection has been described for clinical samples (10, 13, 21). At present, a real-time commercial enterovirus NASBA assay is available.
The aim of this study was to develop a rapid, standardized method for the detection of enteroviruses in large-volume surface water samples. We used a concentration method which was appropriate for the detection of infectious viruses as well as for the detection of viral RNA. We compared three different RNA extraction protocols and found the best results with a new isolation method using magnetic silica beads. Furthermore, we compared enterovirus detection by an in-house RT-PCR with a novel commercially available real-time NASBA assay. These molecular data were compared with the numbers of infectious enterovirus particles detected by cell culture.
MATERIALS AND METHODS
Cell cultures.
The continuous BGM cell line derived from African green monkey kidney cells was used. The cell line was maintained in 75-cm2 and 162-cm2 tissue culture-treated flasks (Corning BV, Schiphol-Rijk, The Netherlands) on medium 199 supplemented with Hanks' solution and l-glutamine (Cambrex, Verviers, Belgium). Growth medium was supplemented with 0.09% sodium bicarbonate, 10% fetal bovine serum (Life Technologies, Breda, The Netherlands), and antibiotics (penicillin at 100 IU/ml, streptomycin at 100 μg/ml). Three days after seeding of cells, the medium was replaced by medium 199 (as mentioned above) as maintenance medium and the same concentration of antibiotics.
Sampling and concentration by UF.
Ten liters of raw and treated sewage water and large volumes of river water (approximately 600 liters) were collected and concentrated by a conventional filter adsorption-elution method (30). Magnesium chloride was added to the water sample to a final concentration of 0.05 M to enable the formation of a virus-magnesium complex. By reducing the pH to 3.8 with 0.5 M HCl, these complexes adsorb to a negatively charged cartridge filter (1.2 μm nominal; Millipore, Etten-Leur, The Netherlands). Viruses were eluted from the filter with elution buffer (pH 9.0) containing 3% beef extract (Difco Laboratories, Detroit, MI) and 0.05 M Tris (Bisolve, Valkenswaard, The Netherlands). The typical eluate volume of 10 liters of raw sewage water is approximately 650 ml and approximately 1,800 ml for volumes of 600 liters of water. The eluate was neutralized with a concentrated acetic acid buffer (pH 5.0), resulting in a final eluate with a pH of approximately 7.4. Two-thirds of the eluates were further concentrated to approximately 40 ml by UF using a cellulose-acetate filter (nominal molecular weight limit, 10,000) under high pressure (three bars). The ultrafilter was rinsed with 3% beef extract (pH 9.0). This final UF concentrate was stored at −70°C until further use. The remaining one-third of the eluates was concentrated by two-phase separation as described previously (23).
Detection of enterovirus by monolayer plaque assay.
Infectious viruses were detected by use of a monolayer plaque assay (22). UF concentrate was quickly thawed at 37°C. Antibiotics (final concentrations of penicillin G at 576 μg/ml, streptomycin sulfate at 4,476 U/ml, Amphotericin B at 72 μg/ml, kanamycin monosulphate at 2.9 mg/ml, and neomycin at 576 μg/ml) were added to inactivate bacteria in the concentrate. The suspension was incubated for 1 h at room temperature, after which it was inoculated onto BGM kidney cells. BGM cells were grown to confluent monolayers in 75-cm2 plastic flasks. Before inoculation, culture medium was removed and the sample was added to the flasks. The cultures were incubated at room temperature for 2 h to allow virus adsorption to the cells. The cells were overlaid with medium 199 supplemented with Earle's salts (Life Technologies, Breda, The Netherlands) and 10% fetal bovine serum (Life Technologies), 0.9% Bacto Agar (Difco, Amsterdam, The Netherlands), 0.2% bicarbonate, 100 IU penicillin, and 100 μg/ml streptomycin (Life Technologies). After 9 days of incubation in an inverted position at 37°C, the cells were stained with 0.03% neutral red (Sigma-Aldrich, Zwijndrecht, The Netherlands) in 0.9% agar and incubated for 4 to 6 h at 37°C in the dark. After 24 h, visible plaques were counted and the virus concentration in the original water sample was calculated from the analyzed volume and the virus count.
Two-phase separation (2PHS) and RNA isolation (method I).
Prior to the isolation of viral RNA, the eluate of the conventional filter adsorption-elution method was further concentrated by a modified protocol of a two-phase separation method (27) described by Lodder et al. (23). Briefly, 650 ml eluate, 1% (wt/vol) dextran T40 (Pharmacia, Roosendaal, The Netherlands), 10% (wt/vol) PEG 6000 (Merck, Amsterdam, The Netherlands), 0.2 M NaCl, and 10 mM phosphate buffer (pH 7.2) were mixed for 1 h at 4°C. The suspension was then transferred to a separation funnel and left overnight at 4°C. After separation, the bottom phase and the interphase were harvested. Further purification was done by spin-column chromatography using Sephadex G200 (ICN, Zoetermeer, The Netherlands) and by ultrafiltration in a Centricon 100 microconcentrator with a 100,000-molecular-weight cutoff (Amicon, Dronten, The Netherlands). The average retentate volumes of 1 to 5 ml were subjected to RNA extraction using guanidinium (iso)thiocyanate (GITC)-silica according to the method of Boom et al. (4).
RNA isolation from UF concentrate using NucliSens isolation reagents (method II).
For NucliSens-based RNA isolation, 12.5 μl to 6 ml UF concentrate was used. At least two volumes of NucliSens lysis buffer (bioMérieux, Boxtel, The Netherlands) was added, followed by incubation for 10 min at room temperature. Thereafter, 4,000 copies of internal control (IC) RNA, supplied with the NucliSens EasyQ enterovirus assay, were added. IC RNA consists of RNA derived from a cloned poliovirus 5′ untranslated region in which a segment of 24 bp was replaced by a fragment of 20 bp from the genome of potato leafroll virus. Either 50 μl of NucliSens nonmagnetic or 50 μl of NucliSens magnetic silica particle suspension (bioMérieux, Boxtel, The Netherlands) was added, followed by 10 min of incubation at room temperature to allow nucleic acid binding. Silica particles were extensively washed with different wash buffers according to the instructions of the manufacturer. The NucliSens miniMAG instrument (bioMérieux, Boxtel, The Netherlands) was used to collect and wash the magnetic silica particles. Nucleic acids were recovered from the particles during a 5-min incubation period at 60°C, using 50 μl elution buffer.
Enterovirus RT-PCR.
The highly conserved 5′ untranslated region of enteroviruses was used as the target for amplification. The RT-PCR was performed as described earlier (29). The specificity of the detected enteroviruses was confirmed by the hybridization of RT-PCR products as described previously.
Enterovirus NASBA.
Enteroviruses were detected using the NucliSens Basic kit and the NucliSens EasyQ enterovirus reagents (bioMérieux, Boxtel, The Netherlands) mainly as described by Landry et al. (21) with modifications for real-time detection of the amplification products. A sequence-specific enterovirus molecular beacon was used for the detection of wild-type enterovirus, whereas the IC molecular beacon was used to detect an enterovirus-specific internal control RNA. The entero-beacon contains a 6-carboxyfluorescein (FAM) fluorophore at the 5′ end, whereas the IC beacon contains a carboxyrhodamine fluorophore. The NucliSens EasyQ analyzer was used for the real-time detection of NASBA amplicons (bioMérieux, Boxtel, The Netherlands).
Estimation of enterovirus concentrations in water.
Estimation of the numbers of viral genomes present in water by RT-PCR (PCR-detectable units [PDU]) and NASBA (NASBA-detectable units [NDU]) was performed on 10-fold serially diluted RNA samples (end point dilution). Virus concentrations in the undiluted samples were estimated as the most probable numbers by the use of the presence or absence of virus genomes in the 10-fold RNA dilutions on replicate samples under the assumption that negative samples do not contain viral RNA. Application of the Poisson distribution was justified by the assumption that the infectious virus particles or viral RNA was dispersed randomly in the sample. The maximum-likelihood method was used to estimate the number of virus particles in the undiluted sample (24). A negative binominal model gives the best fit for the distribution of virus particles in the original and diluted samples. The 95% confidence interval was estimated for each virus concentration.
RESULTS
Viral RNA extraction from UF-concentrated water samples.
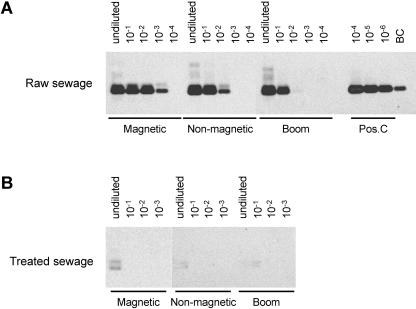
A standardized and time-efficient concentration and detection protocol for viruses in water is required for detection by molecular and cell culture methods. To this end, we analyzed whether UF concentrates, which are commonly used for the detection of infectious enterovirus by cell culture, could be subjected to viral RNA extraction. Raw and treaded sewage types were analyzed because of their high virus concentrations. Three different RNA extraction methods were compared, all based on the use of silica beads (4) (Fig. 1). The method (Boom) using GITC-silica was compared with two commercially available isolation kits, the NucliSens isolation kit (nonmagnetic) and the NucliSens magnetic extraction kit (Magnetic). Tenfold serial dilutions of the RNA samples were analyzed by RT-PCR and Southern blot hybridization of the RT-PCR products. As shown in Fig. 1A, RNA isolated from concentrated raw sewage with magnetic silica beads was amplified most efficiently, which is shown by positive signals up to a 1,000-fold dilution of RNA. Nonmagnetic silica isolated RNA was detected in 100-fold diluted RNA, whereas the Boom-isolated RNA was detected only in undiluted and 10-fold-diluted RNA, indicating that the Boom method resulted in the least efficient isolation of RNA from concentrated raw sewage. The strongest signal in the analysis of RNA isolated from treated sewage was generated with undiluted RNA isolated by the magnetic isolation procedure (Fig. 1B).
FIG. 1.
RNA was extracted from concentrated raw (A) and treated (B) sewage by a conventional filter adsorption-elution method and ultrafiltration. Three different RNA extraction methods were compared, GITC-silica method (Boom), the NucliSens isolation kit (Non-magnetic), and the NucliSens magnetic extraction kit (Magnetic). Tenfold serial dilutions of the RNA samples were analyzed by RT-PCR and Southern blot hybridization. Poliovirus RNA was amplified as a positive control (Pos.C), and a previously amplified positive RNA was used as a blot control (BC).
Although one RT-PCR product was expected, two bands are visible in Fig. 1. Both bands hybridized with an enterovirus-specific probe, indicating that both were enterovirus-specific PCR products. The presence of the doublet can be explained by the use of a nondenaturing 2% agarose gel for the detection by gel electrophoresis, resulting in migration differences of the highly structured amplified 5′ untranslated region of the enterovirus RNA.
Thus, UF concentrates commonly used to detect enterovirus by cell culture are applicable to detect RNA as well. Isolation with the NucliSens magnetic extraction reagents resulted in the most efficient detection of enterovirus RNA.
Extraction efficiency of RNA isolation with the miniMAG system.
To assess the efficiency of RNA isolation by magnetic silica beads, three large-volume water samples (02-8, 01-6, 00-1) from different rivers in The Netherlands were UF concentrated. Enterovirus concentrations as determined by cell culture were 1.4, 0.65, and 14.9 PFU/liter, respectively. Different volumes of UF concentrates were analyzed, ranging from 12.5 μl to 6 ml. Four thousand copies of an enterovirus-specific IC RNA were added, which were coamplified with the viral RNA using the same set of primers. The presence of wild-type and IC RNA was analyzed by real-time NASBA. Amplification products were detected by hybridization using two different molecular beacon probes, one specific for the wild-type enterovirus and the other specific for the IC product. IC RNA was not detected when RNA was extracted from concentrate volumes of greater than 200 μl (data not shown), indicating that the use of larger volumes results in either a less efficient RNA extraction or the presence of inhibitors. Thus, for these samples that contain a large amount of background nucleic acids (3% beef extract), we recommend that RNA be isolated with magnetic silica beads from samples of 200 μl or less. Analysis of larger volumes will consequently underestimate the virus titer.
Comparison of two RNA isolation methods.
The conventional RNA isolation following two-phase separation, spin column gel chromatography, and ultrafiltration (method I: 2PHS plus Boom) (23) was compared with RNA isolation by using the NucliSens magnetic extraction kit following UF concentration (method II: UF plus magnetic extraction) with respect to extraction efficiencies and the presence of inhibitors. To this end, five large-volume water samples of rivers A and B were concentrated by filtration on a negatively charged membrane and further processed to isolate RNA by method I and method II. As depicted in Table 1, the input volumes of water used for RNA isolation (extracted volume) largely differed between the two extraction methods. To compare the two RNA isolation methods, 10-fold dilutions of the five RNA samples were analyzed by RT-PCR for the presence of enterovirus RNA in triplicate. Despite the analyses of relatively small volumes with method II, in four out of five samples, enterovirus RNA was detected (02-8, 02-9, 04-1, and 04-2); in these four samples, undiluted RNAs were positive as well as three of the 10-times-diluted RNA samples (02-8, 04-1, and 04-2) (Table 1). By using method I, two out of five samples (02-8 and 02-9) gave positive results. Enterovirus RNA was detected in none of the undiluted samples, suggesting that PCR inhibitors were present. Moreover, method I appeared to be less sensitive because the analysis of 10−3-diluted RNA samples, corresponding to volumes varying between 7.61 ml and 14.0 ml, were all negative. In four out of five undiluted samples of method II, enteroviruses were detected, corresponding to volumes ranging from 6.98 ml to 31.9 ml of water sample.
TABLE 1.
Detection of enterovirus in large-volume river water samples by RT-PCR
| Sample | River | Method I (2PHS + Boom)
|
Method II (UF + magnetic extraction)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Extracted vol (liters) | Dilution | Analyzed vol | RT-PCR results
|
Extracted vol (ml) | Dilution | Analyzed vol (ml) | RT-PCR results
|
||||||
| I | II | III | I | II | III | ||||||||
| 02-8 | A | 153.6 | 100 | 14.0 liters | − | − | − | 319 | 100 | 31.9 | + | + | + |
| 10−1 | 1.4 liters | − | − | − | 10−1 | 3.19 | + | + | + | ||||
| 10−2 | 140 ml | + | − | − | 10−2 | 0.32 | − | − | + | ||||
| 10−3 | 14.0 ml | − | − | − | 10−3 | 0.03 | − | − | − | ||||
| 02-9 | A | 183.2 | 100 | 16.7 liters | − | − | − | 252 | 100 | 25.2 | + | + | + |
| 10−1 | 1.67 liters | − | + | + | 10−1 | 2.52 | − | − | − | ||||
| 10−2 | 167 ml | − | − | + | 10−2 | 0.25 | − | − | − | ||||
| 10−3 | 16.7 ml | − | − | − | 10−3 | 0.03 | − | − | − | ||||
| 03-2 | A | 140.3 | 100 | 12.8 liters | − | − | − | 288 | 100 | 28.8 | − | − | − |
| 10−1 | 1.28 liters | − | − | − | 10−1 | 2.88 | − | − | − | ||||
| 10−2 | 128 ml | − | − | − | 10−2 | 0.29 | − | − | − | ||||
| 10−3 | 12.8 ml | − | − | − | 10−3 | 0.03 | − | − | − | ||||
| 04-1 | B | 106.6 | 100 | 7.6 liters | − | − | − | 69.8 | 100 | 6.98 | + | + | + |
| 10−1 | 761 ml | − | − | − | 10−1 | 0.70 | + | + | − | ||||
| 10−2 | 76.1 ml | − | − | − | 10−2 | 0.07 | − | − | − | ||||
| 10−3 | 7.61 ml | − | − | − | 10−3 | 0.01 | − | − | − | ||||
| 04-2 | B | 202.9 | 100 | 14.5 liters | − | − | − | 224 | 100 | 22.4 | + | + | + |
| 10−1 | 1.45 liters | − | − | − | 10−1 | 2.24 | + | − | − | ||||
| 10−2 | 145 ml | − | − | − | 10−2 | 0.22 | − | − | − | ||||
| 10−3 | 14.5 ml | − | − | − | 10−3 | 0.02 | − | − | − | ||||
Virus concentrations were estimated semiquantitatively on the 10-fold serially diluted RNA samples by calculating the mean concentration of enterovirus RNA in the initial water samples and the 95% confidence interval. As shown in Table 2 (samples marked with footnote a), method II resulted in estimated virus concentrations which were at least 100 times higher (02-9; 91.5 versus 0.88 detectable units [PDU]/liter) or even 500 times higher (02-8; 1,340 versus 2.6 PDU/liter) than those by method I. In two samples (04-1 and 04-2), no virus was detected by method I, whereas viral RNA was detected by method II (1,317 and 189 PDU/liter, respectively). The latter two samples were from a different river than the first three. Our data suggest that in river B, more inhibitors were present which were not removed with method I but were with method II.
TABLE 2.
Estimated enterovirus concentrations in river samples as determined by cell culture, RT-PCR, and NASBA
| Sample | River | PFU/liter in cell culture | Method I (2PHS + Boom) RT-PCR at PDU/liter (p2.5-p97.5)b | Method II (UF + Magnetic extraction)
|
|
|---|---|---|---|---|---|
| RT-PCR at PDU/liter (p2.5-p97.5)b | NASBA at NDU/liter (p2.5-p97.5)b | ||||
| 02-8a | A | 1.4 | 2.6 (0.15-11.6) | 1,340 (308-5,165) | 9,548 (1,249-72,843) |
| 02-9a | A | 1.35 | 0.88 (0.21-2.5) | 91.5 (19.5-358) | 388 (76.0-1,304) |
| 03-2a | A | 0 | 0 (0-0.05) | 0 (0-20.0) | 0 (0-28.9) |
| 04-1a | B | 0.26 | 0 (0-0.08) | 1,317 (282-4,380) | 0 (0-82.9) |
| 04-2a | B | 0.17 | 0 (0-0.04) | 189 (43.6-726) | 32.9 (5.4-104) |
| 01-1 | C | 0.31 | 1,390 (244-5,753) | 826 (107-5,978) | |
| 01-2 | C | 0.27 | 168 (26.6-762) | 178 (20.2-824) | |
| 01-3 | C | 0.80 | 97.3 (13.7-502) | 435 (77.6-1,479) | |
| 01-4 | C | 0.90 | 144 (22.7-649) | 152 (17.2-702) | |
| 01-5 | C | 0.19 | 0 (0-31.5) | 0 (0-57.8) | |
| 01-6 | C | 0.65 | 55.0 (7.7-284) | 706 (91.6-5,110) | |
| 01-7 | C | 0.35 | 68.1 (11.0-224) | 346 (39.2-1,602) | |
| 03-3 | A | 0.12 | 30.5 (1.7-141) | 0 (0-48.9) | |
| 04-3 | D | 0.56 | 241 (33.8-1,243) | 0 (0-95.5) | |
| 04-4 | D | 0 | 0 (0-26.0) | 0 (0-26.2) | |
Samples described in Table 1.
95% confidence intervals are given in parentheses.
Thus, the UF concentrate from large-volume surface water samples can be used to isolate RNA with magnetic silica beads. In spite of the much smaller sample volumes that were analyzed using the rapid method II, enteroviruses were detected in more samples by RT-PCR than with method I (Table 2). Consequently, virus concentrations were estimated to be at least 100 times higher with the NucliSens magnetic extraction kit (method II) than those obtained by the Boom protocol (method I).
Comparison of RT-PCR and real-time NASBA.
Detection of enterovirus RNA by RT-PCR was compared with real-time NASBA. To this end, NucliSens magnetic extraction was used to isolate the RNA of 10 additional water samples from rivers A, C, and D and the presence of enterovirus RNA was determined by our in-house RT-PCR assay. The presence of enterovirus RNA in all samples was analyzed by the real-time enterovirus NASBA assay as well, using the same amount of input RNA. Virus concentrations were estimated semiquantitatively on 10-fold serially diluted RNA samples by both molecular methods and were compared with virus titers as determined by the BGM monolayer plaque assay. Table 2 shows that 12 out of 15 samples (80%) were positive by RT-PCR; two out of three (13%) negatives (03-2, 01-5, and 04-4) were also negative by cell culture (03-2 and 04-4). Nine of the 15 samples (60%) were positive when detected by NASBA. Three of the six negative samples were also negative by RT-PCR (03-2, 01-5, and 04-4). The other three samples (04-1, 03-3, and 04-3) contained 0.26, 0.12, and 0.56 PFU/liter culturable enteroviruses, respectively, suggesting that our in-house RT-PCR assay might be slightly more sensitive than NASBA. On the other hand, in four samples (02-8, 01-3, 01-6, and 01-7), the detected virus concentrations by NASBA were at least five times higher than the concentrations detected by RT-PCR, suggesting the opposite. In one sample (04-1), a high quantitation was obtained with RT-PCR (1,317 PDU/liter) whereas no enterovirus RNA was detected by NASBA. Average enterovirus concentrations in the 15 analyzed surface waters as determined by cell culture were 0.49 PFU/liter, whereas estimated concentrations determined by RT-PCR and NASBA were 342 PDU/liter and 841 NDU/liter, respectively.
In enterovirus-containing surface waters with virus concentrations lower than 0.6 PFU/liter, enteroviruses were not always detected by molecular methods whereas they were by cell culture. This indicates that the cell culture detection limit is lower than that of the molecular methods. Enterovirus concentrations lower than 0.6 PFU/liter are detected more consistently by RT-PCR than by NASBA.
DISCUSSION
This paper describes a rapid method for the detection of enteroviruses in large-volume surface water samples, using a concentration method which is appropriate for the detection of infectious viruses as well as for the detection of viral RNA. Up to 600 liters of surface water was concentrated by a conventional negative membrane adsorption-elution method followed by a second concentration step using ultrafiltration. In previous studies, this concentrate was analyzed to detect culturable viruses by a BGM monolayer plaque assay (8). We demonstrated that such large-volume surface water concentrates can also be subjected to viral RNA isolation using the NucliSens magnetic extraction kit. This is a substantial improvement compared to other described RNA extraction methods using concentration methods which are not applicable for cell culture. Furthermore, no additional purification steps have to be performed to remove inhibitors, making it a rapid method to isolate RNA from water.
Enteroviruses in surface waters are mainly determined by cell culture, using a BGM monolayer plaque assay (8). This very sensitive, quantitative method, however, is unable to detect certain enterovirus types, especially many of the coxsackie A virus types (8). These enteroviruses can be detected by molecular methods if general primers are used for amplification. We compared two different molecular detection methods, an in-house RT-PCR versus a commercial real-time NASBA assay. Detection of viruses in water by RT-PCR has been described before (11, 29), whereas detection by NASBA has been described only for hepatitis A virus in artificially contaminated sewage water (18). NASBA has several potential advantages over RT-PCR, including the fact that NASBA was developed specifically for RNA target amplification, making it suitable for the detection of viruses in water, which are mainly RNA viruses. In addition, real-time NASBA is a one-step closed amplification process, which makes it less prone to cross-contamination and reduces the hands-on time. According to Heim and Schumann, a one-step enterovirus NASBA achieved about the same sensitivity as a three-step nested-RT-PCR (13). Taking these possible advantages of NASBA into consideration, we compared the detection of enteroviruses in water by RT-PCR with NASBA. In the highly conserved 5′ untranslated region, a fragment of 229 nucleotides is amplified by NASBA (21) and a fragment of 196 nucleotides by RT-PCR (29). As shown in Table 2, only 1 of the 15 samples that were positive by cell culture was negative by RT-PCR. On the other hand, NASBA analysis of the same RNA samples resulted in four negative results that were positive by cell culture. These data suggest that our in-house RT-PCR assay is slightly more sensitive than NASBA. In four samples, however, virus concentrations determined by NASBA were at least five times higher than concentrations that were found by RT-PCR, suggesting the opposite. These discrepancies in detection between NASBA and RT-PCR can be caused by the differences in methods, but also the use of different primers and probes might cause the differences in detection. This is argued, however, by sequence alignments of RT-PCR and NASBA primer and probes with enterovirus sequences, which give almost identical results with regard to the enterovirus types that can be detected. Typing of enteroviruses in the NASBA-negative samples might answer the question of whether the concentration of enterovirus RNA is too low to detect or whether enterovirus types which cannot be detected are present. In addition, the commercial real-time NASBA assay contains a homologous internal control RNA competing for the same primers as the wild-type RNA, potentially affecting sensitivity, whereas our in-house RT-PCR assay has no internal control.
Although enterovirus detection by real-time NASBA might not be as sensitive as RT-PCR, it does have several advantages. NASBA analysis is performed with the NucliSens Basic kit and NucliSens EasyQ Enterovirus reagents, which are commercial kits ensuring a constant quality. The RT-PCR is an in-house method and thus is less standardized, which consequently might give more variable results. Another major advantage is the availability of an internal control RNA, which will distinguish a truly negative result from a false-negative result because of the presence of inhibitors. In our false-negative samples, internal control RNA signals were present, indicating that samples were not negative because of the presence of inhibitors but because of the sensitivity of the assay. Furthermore, hands-on time is decreased because no separate RT step or labor-intensive Southern blotting is needed. In addition, total turnaround times from amplification to result for the RT-PCR and NASBA assays differ dramatically. Our in-house RT-PCR assay typically takes 12 h, resulting in a 2-day procedure, while the commercial real-time NASBA assay takes only 4 h.
To be able to compare the quantitative aspects of RT-PCR and NASBA to those of cell culture, virus concentrations (PCR- or NASBA-detectable units) were estimated as most probable numbers using the presence or absence of virus RNA in 10-fold RNA dilutions. As shown in Table 2, RNA isolation using magnetic silica beads highly increases the detectable amounts of PCR-detectable units. Thus, although estimated virus concentrations might be higher because viral genomes are detected and not infectious viruses, the RNA extraction method also appears to be critical for virus estimation. Our data clearly demonstrated that virus concentrations as determined by 2PHS concentration and RT-PCR underestimate the concentrations at least 100 times, which consequently will effect assessments of the public health risk, like the evaluations of environmental exposure to pathogenic microorganisms and drinking water treatment criteria. Although only infectious viruses have potential public health effects, additional information obtained by RT-PCR on the presence of nonculturable enteroviruses might be beneficial to the estimation of these health risks.
Further improvements in estimations of virus concentrations can be accomplished by quantitative detection. This has been described for enterovirus in seawater by real-time RT-PCR using an external standard of serially diluted poliovirus RNA (9). The main disadvantage, however, of an external standard is the lack of an internal control safeguarding proper nucleic acid isolation and amplification as well as checking for possible inhibitors of the reaction. We experienced high variation in amplification efficiencies between different sample locations and between different sampling dates at one location. The internal control RNA can be used as an internal calibrator, as has been described for the quantitation of HIV RNA by NASBA in clinical samples (31). The internal calibrator is coisolated and coamplified with the endogenous viral nucleic acids. Both amplification products can be detected simultaneously using specific molecular beacons with specific fluorophores. Quantitation is based on the assessment of the relative RNA growth rates during the transcriptional phase of amplification (31). This would be an ideal approach to quantitate viruses in environmental samples.
Acknowledgments
This work was funded by the Environmental Inspectorate under project number 330000, Health-Related Water Microbiology.
We gratefully acknowledge Peter Teunis from the Computerization and Methodological Consultancy Unit for statistical analyses. We also thank bioMérieux for technical assistance and support.
REFERENCES
- 1.Amvrosieva, T. V., L. P. Titov, M. Mulders, T. Hovi, O. V. Dyakonova, V. I. Votyakov, Z. B. Kvacheva, V. F. Eremin, R. M. Sharko, S. V. Orlova, O. N. Kazinets, and Z. F. Bogush. 2001. Viral water contamination as the cause of aseptic meningitis outbreak in Belarus. Centr. Eur. J. Public Health 9:154-157. [PubMed] [Google Scholar]
- 2.Beller, M., A. Ellis, S. H. Lee, M. A. Drebot, S. A. Jenkerson, E. Funk, M. D. Sobsey, O. D. Simmons III, S. S. Monroe, T. Ando, J. Noel, M. Petric, J. P. Middaugh, and J. S. Spika. 1997. Outbreak of viral gastroenteritis due to a contaminated well. JAMA 278:563-568. [PubMed] [Google Scholar]
- 3.Blomqvist, S., C. Savolainen, P. Laine, P. Hirttiö, E. Lamminsalo, E. Penttilä, S. Jöks, M. Roivainen, and T. Hovi. 2004. Characterization of a highly evolved vaccine-derived poliovirus type 3 isolate from sewage in Estonia. J. Virol. 78:4876-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim van Dillen, and J. S. O. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgener, M., U. Candrian, and M. Gilgen. 2003. Comparative evaluation of four large-volume RNA extraction kits in the isolation of viral RNA from water samples. J. Virol. Methods 108:165-170. [DOI] [PubMed] [Google Scholar]
- 6.Compton, J. 1991. Nucleic acid sequence-based amplification. Nature 350:91-92. [DOI] [PubMed] [Google Scholar]
- 7.Cook, N. 2003. The use of NASBA for the detection of microbial pathogens in food and environmental samples. J. Microbiol. Methods 53:165-174. [DOI] [PubMed] [Google Scholar]
- 8.Dahling, D. R., and B. A. Wright. 1986. Optimization of the BGM cell line culture and viral assay procedures for monitoring viruses in the environment. Appl. Environ. Microbiol. 51:790-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donaldson, K. A., D. W. Griffin, and J. H. Paul. 2002. Detection, quantitation and identification of enteroviruses from surface waters and sponge tissue from the Florida Keys using real-time RT-PCR. Water Res. 36:2505-2514. [DOI] [PubMed] [Google Scholar]
- 10.Fox, J. D., S. Han, A. Samuelson, Y. Zhang, M. L. Neale, and D. Westemoreland. 2002. Development and evaluation of nucleic acid sequence based amplification (NASBA) for diagnosis of enterovirus infections using the NucliSens Basic Kit. J. Clin. Virol. 24:117-130. [DOI] [PubMed] [Google Scholar]
- 11.Gilgen, M., D. Germann, J. Luthy, and P. Hubner. 1997. Three-step isolation method for sensitive detection of enterovirus, rotavirus, hepatitis A virus, and small round structured viruses in water samples. Int. J. Food Microbiol. 37:189-199. [DOI] [PubMed] [Google Scholar]
- 12.Hafliger, D., P. Hubner, and J. Luthy. 2000. Outbreak of viral gastroenteritis due to sewage-contaminated drinking water. Int. J. Food Microbiol. 54:123-126. [DOI] [PubMed] [Google Scholar]
- 13.Heim, A., and J. Schumann. 2002. Development and evaluation of a nucleic acid sequence based amplification (NASBA) protocol for the detection of enterovirus RNA in cerebrospinal fluid samples. J. Virol. Methods 103:101-107. [DOI] [PubMed] [Google Scholar]
- 14.Hibbitts, S., A. Rahman, R. John, D. Westmoreland, and J. D. Fox. 2003. Development and evaluation of NucliSens basic kit NASBA for diagnosis of parainfluenza virus infection with ‘end-point’ and ‘real-time’ detection. J. Virol. Methods 108:145-155. [DOI] [PubMed] [Google Scholar]
- 15.Hot, D., O. Legeay, J. Jacques, C. Gantzer, Y. Caudrelier, K. Guyard, M. Lange, and L. Andreoletti. 2003. Detection of somatic phages, infectious enteroviruses and enterovirus genomes as indicators of human enteric viral pollution in surface water. Water Res. 37:4703-4710. [DOI] [PubMed] [Google Scholar]
- 16.Hovi, T., M. Stenvik, H. Partanen, and A. Kangas. 2001. Poliovirus surveillance by examining sewage specimens. Quantitative recovery of virus after introduction into sewerage at remote upstream location. Epidemiol. Infect. 127:101-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikeda, R. M., S. F. Kondracki, P. D. Drabkin, G. S. Birkhead, and D. L. Morse. 1993. Pleurodynia among football players at a high school. An outbreak associated with coxsackievirus B1. JAMA 270:2205-2206. [PubMed] [Google Scholar]
- 18.Jean, J., B. Blais, A. Darveau, and I. Fliss. 2001. Detection of hepatitis A virus by the nucleic acid sequence-based amplification technique and comparison with reverse transcription-PCR. Appl. Environ. Microbiol. 67:5593-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kares, S., M. Lonnrot, P. Vuorinen, S. Oikarinen, S. Taurianen, and H. Hyoty. 2004. Real-time PCR for rapid diagnosis of entero- and rhinovirus infections using LightCycler. J. Clin. Virol. 29:99-104. [DOI] [PubMed] [Google Scholar]
- 20.Kee, F., G. McElroy, D. Stewart, P. Coyle, and J. Watson. 1994. A community outbreak of echovirus infection associated with an outdoor swimming pool. J. Public Health Med. 16:145-148. [DOI] [PubMed] [Google Scholar]
- 21.Landry, M. L., R. Garner, and D. Ferfuson. 2003. Rapid enterovirus RNA detection in clinical specimens by using nucleic acid sequence-based amplification. J. Clin. Microbiol. 41:346-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lodder, W. J., and A. M. de Roda Husman. 2005. Presence of noroviruses and other enteric viruses in sewage surface waters in The Netherlands. Appl. Environ. Microbiol. 71:1453-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lodder, W. J., J. Vinje, R. van de Heide, A. M. de Roda Husman, E. J. T. M. Leenen, and M. P. G. Koopmans. 1999. Molecular detection of Norwalk-like caliciviruses in sewage. Appl. Environ. Microbiol. 65:5624-5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mood, A. M., F. A. Graybill, and D. C. Boes. 1974. Introduction to the theory of statistics, 3rd ed. McGraw-Hill Book Company, New York, N.Y.
- 25.Nijhuis, M., N. van Maarseveen, R. Schuurman, S. Verkuijlen, M. de Vos, K. Kendriksen, and A. M. van Loon. 2002. Rapid and sensitive routine detection of all members of the genus Enterovirus in different clinical specimens by real-time PCR. J. Clin. Microbiol. 40:3666-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pallansch, M. A., and R. P. Roos. 2001. Enteroviruses: Polioviruses, Coxsackieviruses, Echoviruses and newer enteroviruses. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. [Online.] Lippincott Willliams & Wilkins, Philadelphia, Pa.
- 27.Pöyry, T., M. Stenvik, and T. Hovi. 1988. Viruses in sewage waters during and after a poliomyelitis outbreak and subsequent nationwide oral poliovirus vaccination campaign in Finland. Appl. Environ. Microbiol. 54:371-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rabenau, H. F., A. M. K. Clarici, G. Muhlbauer, A. Berger, A. Vince, S. Muller, E. Daghofer, B. I. Danther, E. Marth, and H. H. Kessler. 2002. Rapid detection of enterovirus infection by automated RNA extraction and real-time fluorescence PCR. J. Clin. Virol. 25:155-164. [DOI] [PubMed] [Google Scholar]
- 29.Schwab, K. J., R. De Leon, and M. D. Sobsey. 1995. Concentration and purification of beef extract mock eluated from water samples for the detection of enteroviruses, hepatitis A virus, and Norwalk virus by reverse trancription-PCR. Appl. Environ. Microbiol. 61:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Olphen, M., J. G. Kapsenberg, E. van de Baan, and W. A. Kroon. 1984. Removal of enteric viruses from surface water at eight waterworks in The Netherlands. Appl. Environ. Microbiol. 47:927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weusten, J. J., P. A. Wouters, M. C. van Zuijlen, and P. A. van de Wiel. 2002. Stochastic processes defining sensitivity and variability of internally calibrated quantitative NASBA-based viral load assays. Nucleic Acids Res. 30:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyn-Jones, A. P., and J. Sellwood. 2001. Enteric viruses in the aquatic environment. J. Appl. Microbiol. 91:945-962. [DOI] [PubMed] [Google Scholar]