Abstract
Criteria for the design of gene-specific and group-specific oligonucleotide probes were established experimentally via an oligonucleotide array that contained perfect match (PM) and mismatch probes (50-mers and 70-mers) based upon four genes. The effects of probe-target identity, continuous stretch, mismatch position, and hybridization free energy on specificity were tested. Little hybridization was observed at a probe-target identity of ≤85% for both 50-mer and 70-mer probes. PM signal intensities (33 to 48%) were detected at a probe-target identity of 94% for 50-mer oligonucleotides and 43 to 55% for 70-mer probes at a probe-target identity of 96%. When the effects of sequence identity and continuous stretch were considered independently, a stretch probe (>15 bases) contributed an additional 9% of the PM signal intensity compared to a nonstretch probe (≤15 bases) at the same identity level. Cross-hybridization increased as the length of continuous stretch increased. A 35-base stretch for 50-mer probes or a 50-base stretch for 70-mer probes had approximately 55% of the PM signal. Little cross-hybridization was observed for probes with a minimal binding free energy greater than −30 kcal/mol for 50-mer probes or −40 kcal/mol for 70-mer probes. Based on the experimental results, a set of criteria are suggested for the design of gene-specific and group-specific oligonucleotide probes, and the experimentally established criteria should provide valuable information for new software and algorithms for microarray-based studies.
Microarrays are one of the most powerful technologies currently available for genomic research (6, 7, 9, 12, 17, 19, 23, 28, 32, 34), and various formats and probe types have been developed. Two types of microarrays, DNA arrays and oligonucleotide arrays, are commonly used (29). Oligonucleotide arrays have increased in use because of several advantages, including better specificity, easy construction, and cost efficiency (21, 29). In previous studies that used short oligonucleotide probes, multiple oligonucleotide probe pairs (perfect match and a single-mismatch control) per gene were necessary to detect differential gene expression under different physiological conditions (13, 32). Recent studies indicated that a single 50-mer to 70-mer oligonucleotide per gene could produce comparable hybridization signals obtained with DNA arrays under different experimental conditions (11, 26; Z. He et al., unpublished data). However, a recent study that compared three different microarrays for the same gene set resulted in different sets of genes (15). To achieve specific hybridization, the major challenges are to establish probe design criteria and identify optimal probes for each gene or a group of genes in a sequence database (e.g., whole genomes) in a standardized manner.
Initially, for 50-mer oligonucleotides, Kane et al. (11) suggested that an oligonucleotide probe showing >75% identity with nontargets might cause cross-hybridization. Kane et al. (11) also showed that a 50-mer probe, which had a 15-base, 20-base, or 35-base stretch with nontargets, had approximately 1%, 4%, or 50% of the target signal intensity, respectively. Similar results were observed by Hughes et al. (8) for 60-mer oligonucleotides. Based on sequence identity and/or continuous stretch criteria, a few probe design programs, such as OligoArray (24), OligoWiz (16), and OligoPicker (31), have been developed. In OligoArray 2.0, the oligonucleotide specificity is computed by binding free energy (25). In addition, other factors that influence the specificity and sensitivity of oligonucleotide arrays, such as secondary structures and Tm, have been considered in OligoArray (24), OligoArray 2.0 (25), and OligoPicker (31).
However, many aspects regarding oligonucleotide probe design remain unclear. First, parameters that affect probe specificity have not been extensively investigated in the following aspects: (i) the effects of sequence identity and continuous stretch on cross-hybridization have not been tested separately; (ii) for long oligonucleotide probes, mismatch position has not been studied rigorously or implemented in any available probe design programs; and (iii) the relationship between theoretical free energy and experimental hybridization signal intensity has not been examined extensively. Second, most of the criteria and the respective threshold values have not been determined experimentally. For example, OligoPicker uses a 15-base stretch and a BLAST score of 30.0 as cutoffs for 70-mer oligonucleotide selection (31), and OligoArray 2.0 determines probe specificity for oligonucleotides with varying lengths by predicting secondary structures and computing the thermodynamics of probe hybridization with targets (25). Although Kane et al. (11) suggested that 75 to 85% sequence identity and 15-base continuous stretch should apply to 50-mer oligonucleotide probe design, similar experiments have not been done with 70-mer oligonucleotides, which are also used in microarray-based studies (e.g., whole-genome microarrays). Third, the recognized criteria have not been comprehensively compared in any study. A very stringent single criterion may miss truly specific probes and limit the gene coverage. In contrast, the relaxation of a single criterion may produce a significant number of nonspecific probes and decrease the quality of microarrays (X. Li et al., unpublished data). Therefore, the consideration of multiple criteria is essential to eliminate probe candidates with possible cross-hybridization and maintain specific oligonucleotide probes.
In addition, the criteria for group-specific probe design have not been experimentally established. For a group of highly homologous sequences with >90% sequence identity, the selection of gene-specific probes will be difficult. In this case, multiple probes should be considered to represent a group. In addition, direct performance comparisons between commonly used 50-mer and 70-mer oligonucleotide probes for oligonucleotide array construction have not been evaluated. Therefore, in this study, we have experimentally determined the effects of probe-target identity, length of continuous stretch, free energy, and mismatch positions on microarray hybridization specificity. Based on the experimental results, a set of criteria for the design of gene-specific and group-specific 50-mer and 70-mer probes were established.
MATERIALS AND METHODS
Oligonucleotide probe preparations.
50-mer and 70-mer perfect match (PM) oligonucleotide probes were designed with a modified version of the software, PRIMEGENS (33) based on four genes (SO1679, SO1744, SO2680, and SO0848) from the Shewanella oneidensis MR-1 genome. The mismatch (MM) probes were generated with a C++ program as follows: based on the PM probes designed above n (n = 3, 5, …37), random matches were introduced for each probe to generate MM probes. Three random probes were selected at each level of mismatches. The nucleotide composition (A, T, C, or G) at each mismatched position was randomly assigned. Thus, in total, 45 MM probes were generated for each template with a length of 50 or 70 nucleotides. All designed oligonucleotides were commercially synthesized without modification by MWG Biotech, Inc. (High Point, NC). The concentration of oligonucleotide probes was adjusted to 100 pmol/μl. Detailed information about all designed oligonucleotide probes are listed in Table S1 in the supplemental material.
Microarray construction.
Oligonucleotide probes prepared in 50% dimethyl sulfoxide (Sigma Chemical Co., MO) were spotted onto SuperAmine glass slides (Telechem International, CA) using a PixSys 5500 robotic printer (Cartesian Technologies Inc., CA). Each probe had two replicates on a single slide. In total, there were 736 spots on the array. After printing, the oligonucleotide probes were fixed onto the slides by UV cross-linking (300 mJ of energy) according to the protocol of the manufacturer (Telechem International, CA).
Synthesis and preparation of artificial target templates.
Four 70-mer artificial targets (T1-SO1679, T2-SO1744, T3-SO2680, and T4-SO0848) that were complementary to the 70-mer PM probes were synthesized (Molecular Structure Facility at Michigan State University, East Lansing). The artificial oligonucleotide targets were labeled at the 5′ end with Cy5 (T1-SO1679, T2-SO1744, and T3-SO2680) or Cy3 (T4-SO0848) fluorescent dyes during synthesis.
Genomic DNA extraction, purification, and labeling.
Genomic DNA was isolated and purified from S. oneidensis MR-1 as described previously (35). The purified genomic DNA was fluorescently labeled by random priming using Klenow fragment of DNA polymerase. Mixture I (35 μl), which contained 500 ng of genomic DNA and 20 μl of random primers (Invitrogen Life Technologies, CA), was heated at 98°C for 3 to 5 min, cooled on ice, and then centrifuged. Mixture II (15 μl), which contained 1 μl of a solution consisting of 5 mM (each) dATP, dGTP, and dTTP and 2.5 mM dCTP, as well as 2 μl (80 U) of Klenow (Invitrogen Life Technologies, CA), and 0.5 μl of Cy3 or Cy5 dye (Amersham BioSciences, United Kingdom) were added to mixture I. A total of 50 μl of labeling reaction solution was incubated for 3 h at 42°C. The labeling reaction was terminated by heating at 98°C for 3 min. The tubes were removed and placed on ice. After a quick centrifugation, the sample was hydrolyzed in 50 mM NaOH at 37°C for 10 min and then neutralized with the same amount of HCl. The labeled cDNA targets were purified immediately using a QIAquick PCR purification column and concentrated in a Savant Speedvac centrifuge (Savant Instruments Inc., Holbrook, NY).
Microarray hybridization, washing, and scanning.
Hybridization was conducted in triplicate, and each slide contained two replicate spots of each probe so that six data points for each probe were obtained. The microarrays were hybridized at 45°C overnight in the presence of 50% formamide. The labeled cDNAs were resuspended in 20 to 25 μl of hybridization solution that contained 50% formamide, 1 mM dithiothreitol, 3× saline-sodium citrate, 0.3% sodium dodecyl sulfate, and 0.8 μg/μl of herring sperm DNA (Invitrogen Life Technologies, CA). The sample was incubated at 98°C for 5 min, centrifuged to collect condensation, and kept at 50 to 60°C. The sample was immediately applied to the microarray slide, and hybridization was carried out in a waterproof Corning hybridization chamber (Corning Life Science, NY) submerged in a 45°C water bath in the dark for 16 h. After hybridization, microarray slides were washed according to the protocol of the manufacturer (Telechem International, CA). Microarrays were scanned with a ScanArray 5000 microarray analysis system (Packard BioChip Technologies, MA). Normally, 95 to 100% of laser power and 70 to 80% photomultiplier tube efficiency were selected.
Data analysis and normalization.
Scanned images were analyzed using the software ImaGene 5.5 (Biodiscovery Inc., CA). Prior to normalization, negative spots and poor-quality spots were flagged by ImaGene and then removed in Excel. The signal-to-noise ratio (SNR) was also computed for each spot to discriminate true signals from noise. The SNR ratio was calculated as follows: SNR = (signal mean − background mean)/(background standard deviation). A commonly accepted criterion for the minimum signal (threshold) that can be accurately quantified is an SNR of >3.0 (30).
Stretch length and free energy calculation.
The BLAST program (5) was used to identify regions with high probe-target identities. A simple Perl program extracts the BLAST output and calculates stretch length using 2.0 bit scores as 1 base perfect match. If more than one stretch was identified in a probe, the longest one was used. A C++ program was used to calculate binding free energy values based on a nearest-neighbor model using established thermodynamic parameters (1-4, 10, 14, 18, 20, 27).
MMPD calculation.
The distance of the mismatch in the middle position (mid) is set as zero. For each mismatch at position i, each distance (di) value is calculated using the formula di = [(mid − i) · (mid − i)]1/2. The average distance (Davg) of a probe to a particular nontarget is the sum of each individual di value divided by the number (n) of mismatches, Davg = (Σdi)/n. The maximal mismatch position distance (MMPD) value is obtained by taking the maximal value of all Davg values. The calculation of all MMPD values was performed by a Perl program.
RESULTS
Effects of probe-target identity on signal intensity.
Probe-target identity is a crucial factor that determines the specificity of microarray probes, particularly oligonucleotide arrays. To investigate the relationship between probe-target identity and signal intensity, artificial oligonucleotide targets with 0 to 37 mismatches were used. Four artificial targets were mixed equally in different concentrations to determine the optimal concentrations to achieve specific signals and good sensitivity. The experimental results indicated that l0.0 pg or 2.0 pg of synthetically labeled target was needed to achieve appropriate specificity and sensitivity for 50-mer or 70-mer probes, respectively. The experimentally determined optimal concentrations of targets were used in the later studies.
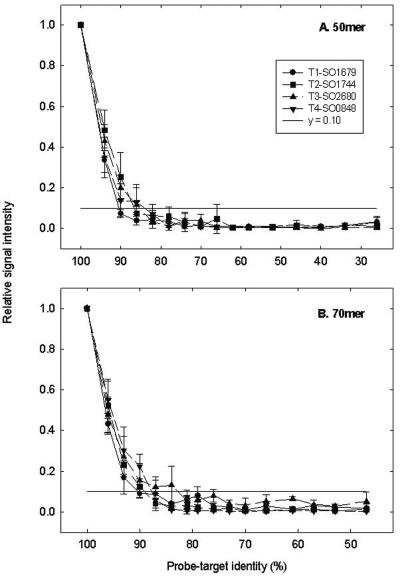
The effects of probe-target identity on relative signal intensity were assessed (Fig. 1). Relative signal intensity was calculated with the signal intensity of each PM probe as 1.0, and all MM probe signals were compared to that of the PM probe. In general, little hybridization was observed for both 50-mer and 70-mer probes with less than 85% identity to the respective targets, whereas the signal intensity increased substantially for probes that had more than 90% identity to the respective targets (Fig. 1). Approximately 33 to 48% of the PM probe signal intensities were detected when probe-target identity increased to 94% for 50-mer oligonucleotides (Fig. 1A). Similarly, 70-mer probes that showed 96% similarities to the targets detected between 43 and 55% of the PM probe signal intensities (Fig. 1B). In addition, the effect of sequence identity on signal intensity appeared to be sequence dependent. For example, for 70-mer probes, little hybridization (<10% of the PM probe signal intensity) was observed for target gene SO1679 at a probe-target identity of 90%, whereas at the same identity relatively strong signals (approximately 25% of the PM probe signal intensity) were observed for target gene SO0848 (Fig. 1B). Similar results were obtained by Rhee et al. (22) with 50-mer oligonucleotide arrays for genes involved in the biodegradation of organic contaminants. The data suggested that GC content or Tm could not explain the observed phenomenon. For instance, the GC content and Tm for the PM and MM probes from SO0848 were not significantly different from other probes. These results suggested that a gene-specific probe should have an identity of <85% to nontargets and that a group-specific probe should have an identity of >95% within a group and an identity of <85% outside the group under the conditions examined.
FIG. 1.
Relationships between probe-target identity and relative signal intensity. Four artificial oligonucleotides (T1-SO1679, T2-SO1744, T3-SO2680, and T4-SO0848) containing 10 or 2.0 pg of Cy dye (2.5 or 0.5 pg for each target) were hybridized with 50-mer (A) and 70-mer (B) oligonucleotide arrays, respectively, at 45°C in the presence of 50% formamide. Data are presented as the average and standard deviation from six replicate data points. The horizontal line (y = 0.10) shows that less than 10% of the PM signal was observed when identity was <85%.
Due to the complicated nature of surface hybridization, one would expect low levels of cross-hybridization for the MM probes. A central question is how to distinguish true hybridization signals from nonspecific background noise. One common approach is to determine signal-to-noise ratios. In general, the hybridization of a probe with an SNR less than 3.0 was treated as background noise (30). To translate this to the signal intensity of the MM probes relative to the PM probes, the relationships of relative signal intensities to SNR values were further analyzed for the MM probes with an SNR less than 3.0. On average, 2.5% ± 2.9%, 7.5% ± 2.7%, and 9.5% ± 3.2% of the PM probe signal was detected for the 50-mer MM probes at an SNR between 0 and 3.0, 1.0 and 3.0, and 2.0 and 3.0, respectively. Similarly, an average of 3.5% ± 3.8%, 8.0% ± 4.4%, and 9.0% ± 3.3% of the PM probe signal was observed for the 70-mer MM probes at an SNR between 0 and 3.0, 1.0 and 3.0, and 2.0 and 3.0, respectively (data not shown). The data also suggested that a probe with up to 16% of the PM probe signal had an SNR of <3.0, or that a probe with as low as 8% of the PM signal had an SNR of >3.0. Therefore, in this study, if an MM probe had less than approximately 10% of the PM probe signal, the hybridization signal of the MM probe was considered to be background noise. This value was used in subsequent analyses for the establishment of probe design criteria.
Relationship between a continuous stretch and signal intensity.
Probes with a 15-base or shorter stretch to nontargets were treated as nonstretch probes, and probes with a 16-base or longer stretch were treated as stretch probes. All designed probes with more than 90% probe-target identity had long stretches, and all probes with less than 74% probe-target identity did not have long stretches. Therefore, only probes with a probe-target identity between 90% and 76% for 70-mer probes or between 90% and 70% for 50-mer probes were selected for the determination of the effect of long stretches on signal intensity (see Fig. S1 in the supplemental material).
The hybridization intensity for the nonstretch probes increased slightly with the increase of probe-target identity (see Fig. S1 in the supplemental material). For example, 9% and 2% of the PM signal intensities were observed at probe-target identities of 90% and 70%, respectively, for 50-mer probes (see Fig. S1A). In a similar fashion, 4% and 2% of the PM signal intensities were observed at probe-target similarities of 90% and 76%, respectively, for 70-mer probes (see Fig. S1B). For stretch probes, the signal intensity increased as the probe-target identity increased, but the relationship was not linear (see Fig. S1). The highest cross-hybridization signal was observed at a probe-target identity of 78% for 50-mer probes, and the stretch probes had 22% of the PM signal compared to 5% for the nonstretch probes at the same sequence identity (see Fig. S1A). Similarly, the highest cross-hybridization was observed at a sequence identity of 84% for 70-mer probes, and the stretch probes had 17% of the PM signal compared to 4% for the nonstretch probes at the same sequence identity (see Fig. S1B). For the stretch probes, the relationship between signal and identity may also depend on stretch characteristics (the length and the position of a stretch) and other parameters (free energy and MMPD). The results indicated that the stretch probes might be responsible for an additional 9% (3 to 17%) of the PM probe signal intensity compared to the nonstretch probes at the same sequence identity. Significant hybridization was not observed even at a probe-target identity of 90% when a probe did not have a 16-base or longer stretch to the nontarget templates (see Fig. S1 in the supplemental material).
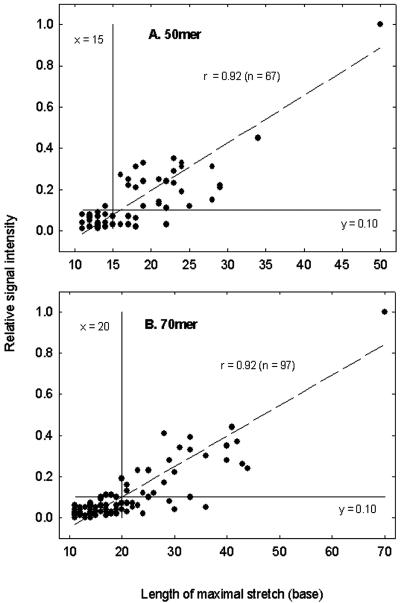
To investigate the effects of stretch length on signal intensity, more-than-10-base stretches of a probe with the four artificial targets were calculated and plotted against relative signal intensity, and a linear correlation between stretch length and relative signal intensity was observed with a P value of <0.001 for both 50-mer and 70-mer probes (Fig. 2). For 50-mer probes, little signal intensity (<8% of the PM probe signal) was observed when a probe had a 15-base or shorter stretch. The signal intensity increased when the stretch length was increased. For example, 30% or 55% of the PM probe signal intensity was expected when a probe had a 25- or 35-base stretch, respectively (Fig. 2A). For 70-mer probes, about 3% or 10% of the PM probe signal was detected when a probe had a 15- or 20-base stretch, respectively. When stretch length increased to 35 or 50 bases, the signal intensity reached 32% or 55% of the PM probe signal intensities, respectively (Fig. 2B).
FIG. 2.
Relationships between the maximal length of stretch and the relative signal intensity for 50-mer (A) and 70-mer (B) oligonucleotide probes. Only more-than-10-base stretches were considered. The vertical line (x = 15 or 20) and horizontal line (y = 0.10) show that less than 10% of the PM signal was observed when stretch length was <15 or 20 bases.
Relationship between mismatch position and signal intensity.
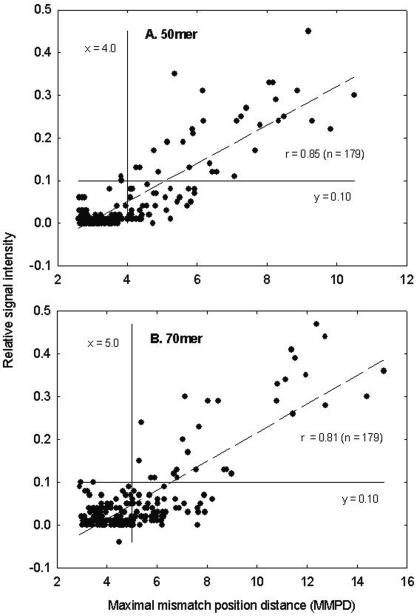
When the 50-mer probes were used to examine the relationship between the position of mismatches and relative signal intensity, the MM probes had an average MMPD value of 4.09. As expected, the signal intensity increased as the MMPD increased. Little signal intensity (less than 3% of the PM probe signal) was observed when the MMPD was less than 3.5, but the signal intensity was approximately 10% of the PM probe signal when the MMPD value increased to 5.0. The majority of data points were well fitted to the line (P < 0.001), and this result indicated that hybridization signal intensity was closely correlated to MMPD (Fig. 3A). Similar results were observed for 70-mer probes. For example, the MM probes had an average MMPD value of 5.37. Approximately 1.5% and 8% of the PM signals were observed when the MMPD values were 4.0 and 6.0, respectively (Fig. 3B).
FIG. 3.
Relationships between the MMPD and the relative signal intensity for 50-mer (A) and 70-mer (B) oligonucleotide probes. The vertical line (x = 4.0 or 5.0) and horizontal line (y = 0.10) show that less than 10% of the PM signal was observed when MMPD was <4.0 or 5.0.
Relationships between free energy and signal intensity.
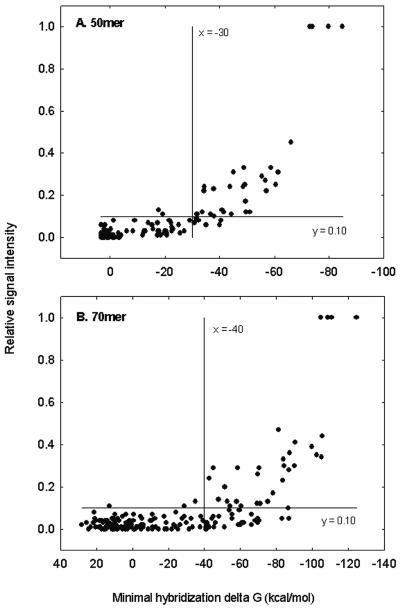
The relationship between the calculated minimal free energy and relative signal intensity is shown in Fig. 4. The binding free energy of a 50-mer PM probe with the respective target was less than −70 kcal/mol, and hybridization signal intensities decreased as binding free energy increased (absolute values decreased). Approximately 35% of the PM signal was observed when free energy was −60 kcal/mol, and cross-hybridization was almost eliminated when the binding free energy was greater than −30 kcal/mol (Fig. 4A). A 70-mer PM probe had a binding free energy value of less than −100 kcal/mol, and approximately 45% of the PM signal intensity might be observed when the free energy value was −90 kcal/mol; little (<5% of the PM probe signal) cross-hybridization was obtained when the binding free energy value was greater than −40 kcal/mol (Fig. 4B).
FIG. 4.
Relationships between the minimal hybridization free energy and the relative signal intensity for 50-mer (A) and 70-mer (B) oligonucleotide probes. The vertical line (x = −30 or x = −40) and horizontal line (y = 0.10) show that less than 10% of the PM signal was observed when binding free energy was less than −30 or −40 kcal/mol.
Comparison of theoretical prediction and hybridization results.
Further comparisons showed that each single criterion (identity, stretch, free energy, or MMPD) was able to identify some common specific probes and could also remove some additional nonspecific probes (data not shown), and these results suggested that the combination of different criteria was needed. To understand how different combinations of criteria affected the probe design outcome, the hybridization data for the MM probes were further examined (see Table S2 in the supplemental material). The threshold values of identity, stretch, free energy, and MMPD were set to 85%, 15 bases, −30 kcal/mol, and 4.0 for 50-mer probes, respectively, and 85%, 20 bases, −40 kcal/mol, and 5.0 for 70-mer probes, respectively. In our experiments, 32 (50-mer) and 36 (70-mer) out of 180 MM probes were experimentally detected to be nonspecific based on SNR ratios. Our results demonstrated that it was difficult to exclude all nonspecific probes based on a single criterion (see Table S2). For example, for 50-mer probes, the best condition was C (free energy of −30 kcal/mol). Based on this criterion, 39 out of 180 MM probes should be excluded, and among the 39 predicted nonspecific probes, 29 were consistent with the experimental results. These data suggested a prediction rate of 74%, and 3 of 32 experimentally proven nonspecific probes were not eliminated by the criterion (misinclusion rate of 9%). In addition, 10 theoretically excluded probes actually did not have significant cross-hybridization signals (misexclusion rate of 26%). The criterion MMPD (D) predicted most nonspecific probes (49) but had the highest mis-excluded rate (45%), the lowest prediction rate (57%), and a mis-included rate of 12% (see Table S2 in the supplemental material).
An appropriate combination of the criteria (F, G, and H) could exclude all nonspecific probes verified by experiments. In this case, F and H had the same results for theoretically predicted nonspecific probe number (47), prediction rate (68%), and mis-included rate (0%), but H had a lower mis-excluded rate (30%) than F (38%), and these results indicated that a relaxation of one or more criteria may exclude fewer qualified candidates without an effect on specificity. The combination of criteria G had more nonspecific probes (57) predicted, lower prediction rate (56%), and higher mis-excluded rate (44%) than F or H, although it also had all experimentally verified nonspecific probes excluded (see Table S2 in the supplemental material). It appears that the MMPD criterion did not help the probe selection in combination G but did increase the number of mis-excluded probes compared to combination F. The combination of identity and stretch criteria (E) did not exclude all experimentally verified nonspecific probes (see Table S2). The results suggested that an appropriate combination of three criteria (identity, stretch, and free energy) was able to select specific oligonucleotide probes. Similar results were observed for 70-mer probes (see Table S2).
DISCUSSION
Specificity of oligonucleotide probes.
Specificity is the most important parameter to evaluate performance of oligonucleotide arrays. The specificity of oligonucleotide arrays is commonly evaluated by the PM/MM method based on sequence identity (21). Due to the complicated hybridization dynamics of oligonucleotides and target sequences, other parameters should also be considered. In this study, the effects of four factors (sequence identity, continuous stretch, free energy, and mismatch position) on oligonucleotide specificity have been determined.
The relationship of hybridization signal intensity to probe-target identity was recently examined using artificial targets. With a 50-mer oligonucleotide microarray containing 763 probes for genes involved in nitrogen cycling and sulfate reduction, Tiquia et al. (29) showed that with hybridization conditions of 50°C, the oligonucleotide microarray hybridizations could differentiate sequences with <86% identity, whereas at 55°C, sequences with <90% identity could be differentiated. With a 50-mer oligonucleotide microarray that contained 1,662 probes for genes involved in contaminant degradation, Rhee et al. (22) showed that under hybridization conditions of 50°C and 50% formamide, the 50-mer microarray hybridization was able to differentiate sequences with <88% identities. In general, the results in this study were consistent with those observations.
Kane et al. (11) showed that a 50-mer probe that had a 15-base or longer stretch with nontargets could cause significant cross-hybridization, and we observed similar results. This feature should be included in the development and design of oligonucleotides probes. A gene-specific probe should keep a stretch as short as possible with other nontargets, and a group-specific probe should have a common stretch as long as possible within a group but as short as possible outside a group. Our results indicated that a 50-mer gene-specific probe should not have 16-base or longer stretches with nontargets, and the stretch length may be extended up to 20 bases for 70-mer probes. For group-specific probes, the length of stretches may be set as 35 bases for 50-mer probes and 50 bases for 70-mer probes so that each member in a group should have relatively uniform and strong signal intensity. By theoretical calculations, the signal intensity of each group member is able to reach approximately 55% of the PM signal for both 50-mer and 70-mer oligonucleotides under the above conditions. If more than 80% of the PM signal is expected, the common stretch length should be increased to 45 bases for 50-mer and 65 for 70-mer oligonucleotides.
Previous studies (13, 21) suggested that a short probe with a single mismatch in the middle could be more easily discriminated than a probe with a mismatch at other positions. For long oligonucleotide probes, we showed that the relative signal intensity is correlated with MMPD, and this result suggested that a probe with mismatches located closer to the middle position of nontargets should have higher specificity. In oligonucleotide probe design, the MMPD value may help choose the best probes from a probe candidate pool by minimizing MMPD values for gene-specific probes, or maximizing MMPD values within a group for group-specific probes. In addition, MMPD could be used to select gene-specific probes, and the suggested values for 50-mer and 70-mer probes are 4.0 and 5.0, respectively.
The length of stretches is dependent on mismatch positions. To achieve high specificity, stretches should be kept to a minimum and should be evenly distributed within an oligonucleotide probe, particularly when multiple mismatches are present. This does not completely agree with the MMPD criterion that requires mismatch positions to be as close to the middle of the probe as possible. Our results indicated that continuous stretches should be used as an essential criterion and that the MMPD value would be better suited for the optimization process.
Rouillard et al. (25) suggested that the computation of the oligonucleotide specificity might be more accurate using the thermodynamic parameters (binding free energy) rather than sequence identities. In this study, the minimal binding free energy was calculated, and the results indicated that binding free energy was more sensitive than sequence identity, most likely because the matches, mismatches, adjacent nucleotides, and interactions between the oligonucleotide probe and targets or nontargets were considered. Actually, at probe-target identity of 75% or greater, minimal free energy is closely correlated to probe-target identity (data not shown). Free energy can be considered as one of the primary oligonucleotide design criteria for the selection of gene-specific or group-specific probes. Our results indicated that −30 kcal/mol can be set as the threshold for the selection of 50-mer gene-specific probes and −60 kcal/mol for 50-mer group-specific probes. The cutoff values of free energy should be −40 and −90 kcal/mol for the selection of gene-specific and group-specific 70-mer probes, respectively.
A combination of multiple criteria may exclude fewer qualified probe candidates without a significant effect on specificity. The exclusion of all nonspecific probes with a single criterion (identity, stretch, free energy, and MMPD) would be difficult, and a possible solution would be to increase the criterion stringency until all experimental verified nonspecific probes were excluded. However, this would also exclude a large portion of specific probes and lead to a low gene coverage. A polyphasic approach would improve and standardize probe design, but the appropriate combination of multiple criteria for the design of specific probes and the exclusion of nonspecific probes has not been previously tested. In this study, the comparison of theoretical predictions and experimental hybridization results indicated that a combination of identity (85%), stretch (15 bases), and free energy (−30 kcal/mol) was able to exclude all nonspecific probes for 50-mer probes and a combination of identity (85%), stretch (20 bases), and free energy (−40 kcal/mol) for 70-mer probes. The results also suggested that the relaxation of one or more criteria may exclude all truly nonspecific probes and keep more qualified probe candidates.
Essential criteria for oligonucleotide probe design.
Based on our results, a set of essential criteria are suggested for gene-specific and group-specific oligonucleotide probe design (Table 1). First, all criteria must be comprehensively considered, and a single parameter was not stringent enough for the exclusion of all nonspecific probes. For example, a probe-target identity of 75% was suggested as a potential cutoff value (11), but our data suggested a sequence identity of 85% under the tested conditions. This value is particularly important for the design of probes for groups of highly homologous sequences, such as the construction of functional gene arrays (22, 29). Based on our results, specific hybridization can be obtained for probes without long stretches or low free energy even if the probe-target identity is 85 to 90%, and these results agree with previous experiments (22, 29).
TABLE 1.
Summary of essential probe design criteria for 50-mer and 70-mer oligonucleotides
| Probe type and parameter | 50-mer value | 70-mer value |
|---|---|---|
| Gene-specific probe | ||
| Max. identity with nontargets | 85% | 85% |
| Max. stretch length with nontargets | 15 bases | 20 bases |
| Min. binding energy with nontargets | −30 kcal/mol | −40 kcal/mol |
| Max. no. of self-binding nucleotides | 8 | 8 |
| Group-specific probe | ||
| Min. identity within the group | 96% | 96% |
| Min. stretch length within the group | 35 bases | 50 bases |
| Max. binding energy within the group | −60 kcal/mol | −90 kcal/mol |
| Max. no. of self-binding nucleotides | 8 | 8 |
In many probe design programs, the sequence identity is the most influential criterion, and stretch or free energy is rarely considered. Therefore, potential cross-hybridization could occur due to long stretch and/or low free energy problems even though a probe meets the identity criterion. This problem can be avoided when the proposed strategies are used (see Table S3 in the supplemental material). The “good” probes that meet all criteria have low signals and low SNR (<3.0) values, and the sequence identities are as high as 86% for 50-mer probes (e.g., S2-07-p1) and 87% for 70-mer probes (e.g., S3-09-p2). Other probes with significant hybridization signals and SNR of >3.0 were excluded by a single criterion (i.e., stretch or free energy) or by both criteria, even when the identity was set to 90%. However, it should be noted that some probes (e.g., S2-07-p3 and S3-09-p3) with low hybridization signals and low SNR values were excluded by free energy or stretch, and these results suggested that the establishment of universal probe design criteria is still a challenge. Further investigations of the probe-target interactions on glass microarray slides are needed.
For group-specific probes, all sequences must have high identity and a common long stretch within a group. Our experimental results showed that approximately 50% of the PM signals could be obtained at probe-target identities of 96% for 50-mer and 70-mer oligonucleotides. Therefore, for the group-specific probe design, the probe-target identity should be as high (98% to 100%) as possible. A group-specific probe should also have a minimal number of continuous stretches (35 bases for 50-mer and 50 bases for 70-mer probes) and a maximal binding free energy (−60 kcal/mol for 50-mer and −90 for 70-mer probes) within the group. It should also be noted that the maximal nucleotide number for self-binding should be considered to avoid strong secondary structures of designed probes (set to 8, as previously described by Wang and Seed [31]). In addition, other criteria such as GC content, Tm, and sequence complexity also need to be considered. Since GC content varies among different organisms, an oligonucleotide design tool should evaluate all sequences in the data set and determine Tm values that fall into a narrow range to ensure quantitative comparison of gene expression. Those parameters should be used as filters for excluding probe candidates.
Finally, the criteria set in Table 1 are generally conservative. The parameters may be cautiously relaxed when more probes or a high gene coverage rate is needed. For example, based on our experimental results, identity may be increased to 90% for both 50-mer and 70-mer probes, and free energy may be set up to −40 kcal/mol for 50-mer or −50 kcal/mol for 70-mer oligonucleotides if approximately 10% of the PM signal intensity is allowed and the SNR is generally below 3.0, but further relaxation of the criteria would need to be experimentally reevaluated. In addition, for fellow researchers to access the detailed data for various analyses, theoretically calculated values of parameters, such as sequence identity, maximal stretch length, minimal free energy, and MMPD, and experimentally determined hybridization results, such as relative signal intensity (percentage of the perfect match probe signals) and signal-to-noise ratios are summarized in Table S4 in the supplemental material.
Selection of optimal oligonucleotides.
Two approaches were considered for the selection of the best 50-mer or 70-mer oligonucleotide probes. One is to rank three essential criteria (identity, continuous stretch, and free energy) in different orders. For example, identity is considered first, then stretch, and then free energy. In this situation, probes with lowest identity to other sequences were usually obtained. Stretch and free energy were then considered in order if some probes had the same identity. The other approach was to assign each criterion (maybe other parameters as well) a weight, and all probe candidates were ranked by a final score for each gene or group of genes. In addition, the optimal probes could be selected based on the MMPD value. In this case, the best gene-specific probe should have the smallest MMPD value for each gene, and the best group-specific probe should have the largest MMPD value for a group of genes.
In summary, we investigated the effects of probe-target identity, continuous stretch, mismatch position, and free energy on the design of 50-mer and 70-mer probes and then experimentally compared the designed probes. The results produced weighted parameters for a polyphasic approach for oligonucleotide probe design and will facilitate the establishment of the criteria for gene-specific and group-specific oligonucleotide probes. Experimentally tested criteria are needed for the development of software and algorithms for oligonucleotide probe design. We are currently developing new algorithms and software to select optimal oligonucleotide probes for functional gene arrays as well as whole-genome microarrays.
Supplementary Material
Acknowledgments
We thank Xiufeng Wan for assistance with the design of mismatch oligonucleotide probes.
This research was supported by the U.S. Department of Energy under the Genomics: GTL, Microbial Genome Programs of the Office of Biological and Environmental Research and the Natural and Accelerated Bioremediation program, Office of Science. Oak Ridge National Laboratory is managed by University of Tennessee-Battelle LLC for the Department of Energy under contract DE-AC05-00OR22725.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Allawi, H. T., and J. SantaLucia, Jr. 1997. Thermodynamics and NMR of internal G-T mismatches in DNA. Biochemistry 36:10581-10594. [DOI] [PubMed] [Google Scholar]
- 2.Allawi, H. T., and J. SantaLucia, Jr. 1998. Nearest neighbor thermodynamic parameters for internal G-A mismatches in DNA. Biochemistry 37:2170-2179. [DOI] [PubMed] [Google Scholar]
- 3.Allawi, H. T., and J. SantaLucia, Jr. 1998. Nearest-neighbor thermodynamics of internal A-C mismatches in DNA: sequence dependence and pH effects. Biochemistry 37:9435-9444. [DOI] [PubMed] [Google Scholar]
- 4.Allawi, H. T., and J. SantaLucia, Jr. 1998. Thermodynamics of internal C-T mismatches in DNA. Nucleic Acids Res. 26:2694-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul, S., T. Madden, A. Schäffer, J. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 7.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 8.Hughes, T. R., M. Mao, A. R. Jones, J. Burchard, M. J. Marton, K. W. Shannon, S. M. Lefkowitz, M. Ziman, J. M. Schelter, M. R. Meyer, S. Kobayashi, C. Davis, H. Dai, Y. D. He, S. B. Stephaniants, G. Cavet, W. L. Walker, A. West, E. Coffey, D. D. Showmarker, R. Stoughton, A. P. Blanchard, S. H. Friend, and P. S. Linsley. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19:342-347. [DOI] [PubMed] [Google Scholar]
- 9.Ivanov, I., C. Schaab, S. Planitzer, U. Teichmann, A. Machl, S. Theml, S. Meier-Ewert, B. Seizinger, and H. Loferer. 2000. DNA microarray technology and antimicrobial drug discovery. Pharmacogenomics 1:169-178. [DOI] [PubMed] [Google Scholar]
- 10.Jaeger, J., D. H. Turner, and M. Zuker. 1989. Improved predictions of secondary structures for RNA. Proc. Natl. Acad. Sci. USA 86:7706-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kane, M. D., T. A. Jatkoe, C. R. Stumpf, J. Lu, J. D. Thomas, and J. M. Madore. 2000. Assessment of the specificity and sensitivity of oligonucleotide (50mer) microarrays. Nucleic Acid Res. 28:4552-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, Y., J. Zhou, M. Omelchenko, A. Beliaev, A. Venkateswaran, J. Stair, L. Wu, D. K. Thompson D. Xu, I. B. Rogozin, E. K. Gaidamakova, M. Zhai, K. S. Makarova, E. V. Koonin, and M. J. Daly. 2003. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl. Acad. Sci. USA 100:4191-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 14.Lyngso, R. B., M. Zuker, and C. N. Pedersen. 1999. Fast evaluation of internal loops in RNA secondary structure prediction. Bioinformatics 15:440-445. [DOI] [PubMed] [Google Scholar]
- 15.Marshall, E. 2004. Getting the noise out of gene arrays. Science 306:630-631. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen, H. B., R. Wernersson, and S. Knudsen. 2003. Design of oligonucleotides for microarrays and perspectives for design of multi-transcriptome arrays. Nucleic Acids Res. 31:3491-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ochs, M. F., and A. K. Godwin. 2003. Microarray in cancer: research and application. BioTechniques 34:S4-S15. [Google Scholar]
- 18.Peritz, A. E., R. Kierzek, N. Sugimoto, and D. H. Turner. 1991. Thermodynamic study of internal loops in oligoribonucleotides: symmetric loops are more stable than asymmetric loops. Biochemistry 30:6428-6436. [DOI] [PubMed] [Google Scholar]
- 19.Petricoin, E. F., J. L. Hackett, L. J. Lesko, R. K. Puri, S. I. Gutman, K. Chumakov, J. Woodcock, D. W. Feigal, Jr., K. C. Zoon, and F. D. Sistare. 2002. Medical application of microarray technologies: a regulatory science perspective. Nat. Genet. 32:474-479. [DOI] [PubMed] [Google Scholar]
- 20.Peyret, N., P. A. Seneviratne, H. T. Allawi, and J. SantaLucia, Jr. 1999. Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A-A, C-C, G-G, and T-T mismatches. Biochemistry 38:3468-3477. [DOI] [PubMed] [Google Scholar]
- 21.Relógio, A., C. Schwager, A. Richter, W. Ansorge, and A. Valcarcel. 2002. Optimization of oligonucleotide-based DNA microarrays. Nucleic Acid Res. 30:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhee, S. K., X. Liu, L. Wu, S. C. Chong, X. Wan, and J. Zhou. 2004. Detection of biodegradation and biotransformation genes in microbial communities using 50-mer oligonucleotide microarrays. Appl. Environ. Microbiol. 70:4303-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rouillard, J. M., C. Herbert, and M. Zuker. 2002. OligoArray: genome-scale oligonucleotide design for microarrays. Bioinformatics 18:486-487. [DOI] [PubMed] [Google Scholar]
- 25.Rouillard, J. M., M. Zuker, and E. Gulari. 2003. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using thermodynamic approach. Nucleic Acids Res. 31:3057-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shoemaker, D. D., E. E. Schadt, C. D. Armour, Y. D. He, P. Garrett-Engele, P. D. McDonagh, P. M. Loerch, A. Leonardson, P. Y. Lum, G. Cavet, L. F. Wu, S. J. Altschuler, S. Edwards, J. King, J. S. Tsang, G. Schimmack, J. M. Scheliter, J. Koch, M. Ziman, M. J. Marton, B. Li, P. Cundiff, T. Ward, J. Castle, M. Krolewski, M. R. Meyer, M. Mao, J. Burchard, M. J. Kidd, H. Dai, J. W. Phillips, P. S. Linsley, R. Stoughton, S. Scherler, and M. S. Boguski. 2001. Experimental annotation of the human genome using microarray technology. Nature 409:922-927. [DOI] [PubMed] [Google Scholar]
- 27.Sugimoto, N., S. Nakano, M. Yoneyama, and K. Honda. 1996. Improved thermodynamic parameters and helix initiation factor to predict stability of DNA duplexes. Nucleic Acids Res. 24:4501-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taroncher-Oldedburg, G., E. M. Griner, C. A. Francis, and B. B. Ward. 2003. Oligonucleotide microarray for the study of functional gene diversity in the nitrogen cycle in the environment. Appl. Environ. Microbiol. 69:1159-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiquia, S. M., L. Wu, S. C. Chong, S. Passovets, D. Xu, Y. Xu, and J. Zhou. 2004. Evaluation of 50-mer oligonucleotide arrays for detecting microbial populations in environmental samples. BioTechniques 36:664-675. [DOI] [PubMed] [Google Scholar]
- 30.Verdick, D., S. Handran, and S. Pickett. 2002. Key considerations for accurate microarray scanning and image analysis, p. 83-98. In G. Kamberova (ed.), DNA image analysis: nuts and bolts. DNA Press LLC, Salem, Mass.
- 31.Wang, X., and B. Seed. 2003. Selection of oligonucleotide probes for protein coding sequences. Bioinformatics 19:796-802. [DOI] [PubMed] [Google Scholar]
- 32.Wodicka, L., H. Dong, M. Mittmann, M. H. Ho, and D. J. Lockhart. 1997. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 15:1359-1367. [DOI] [PubMed] [Google Scholar]
- 33.Xu, D., G. Li, L. Wu, J. Zhou, and Y. Xu. 2002. PRIMEGENS: robust and efficient design of gene-specific probes for microarray analysis. Bioinformatics 18:1432-1437. [DOI] [PubMed] [Google Scholar]
- 34.Zhou, J. 2003. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]
- 35.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.