Abstract
Two α-amylase genes from the thermophilic alkaliphile Anaerobranca gottschalkii were cloned, and the corresponding enzymes, AmyA and AmyB, were investigated after purification of the recombinant proteins. Based on their amino acid sequences, AmyA is proposed to be a lipoprotein with extracellular localization and thus is exposed to the alkaline milieu, while AmyB apparently represents a cytoplasmic enzyme. The amino acid sequences of both enzymes bear high similarity to those of GHF13 proteins. The different cellular localizations of AmyA and AmyB are reflected in their physicochemical properties. The alkaline pH optimum (pH 8), as well as the broad pH range, of AmyA activity (more than 50% activity between pH 6 and pH 9.5) mirrors the conditions that are encountered by an extracellular enzyme exposed to the medium of A. gottschalkii, which grows between pH 6 and pH 10.5. AmyB, on the other hand, has a narrow pH range with a slightly acidic pH optimum at 6 to 6.5, which is presumably close to the pH in the cytoplasm. Also, the intracellular AmyB is less tolerant of high temperatures than the extracellular AmyA. While AmyA has a half-life of 48 h at 70°C, AmyB has a half-life of only about 10 min at that temperature, perhaps due to the lack of stabilizing constituents of the cytoplasm. AmyA and AmyB were very similar with respect to their substrate specificity profiles, clearly preferring amylose over amylopectin, pullulan, and glycogen. Both enzymes also hydrolyzed α-, β-, and γ-cyclodextrin. Very interestingly, AmyA, but not AmyB, displayed high transglycosylation activity on maltooligosaccharides and also had significant β-cyclodextrin glycosyltransferase (CGTase) activity. CGTase activity has not been reported for typical α-amylases before. The mechanism of cyclodextrin formation by AmyA is unknown.
Few anaerobic bacteria are able to grow at a combination of elevated temperature and alkaline pH, two factors which put considerable stress on the cells. Species known to grow optimally at pH 8.5 and 55°C or higher belong to the low-G+C gram-positive bacteria (4, 29), with the exception of “Thermopallium natronophilum” (proposed name). To our knowledge, they are Anaerobranca horikoshii, Anaerobranca gottschalkii, Anaerobranca californiensis, Clostridium paradoxum, Thermosyntropha lipolytica, and Desulfotomaculum alkaliphilum (5, 6, 13, 20, 21, 22).
Surprisingly, a number of enzymes isolated from mesophilic organisms are known to exhibit a combination of high thermostability and alkali tolerance. Several α-amylases, mostly from Bacillus species, have been described as displaying these rare properties (3, 14, 17). Such α-amylases and other saccharolytic enzymes in general are of considerable commercial interest for industrial processes, e.g., in the paper industry and in starch processing. Also, enzymes with these properties are of importance as additives to laundry and dishwashing detergents. Despite the potential of thermoalkaliphilic microorganisms as sources of biotechnologically interesting biocatalysts, very few of their enzymes have been characterized to date, among them an alkalitolerant and thermostable type I pullulanase and a β-cyclodextrin glycosyltransferase (CGTase) from A. gottschalkii (2, 23).
Gram-positve A. gottschalkii was isolated from humid soil at a hot inlet of an alkaline soda lake, Lake Bogoria, Kenya. The anaerobic organism grows in a range of pH 6.0 to 10.5 and 30 to 65°C with the optimum at pH 9.5 and 50 to 55°C and a supplementation of 230 mM Na+ ions. It is reported to grow heterotrophically on various mono- and polysaccharides and polypetides (21).
Microorganisms that live in an alkaline environment have to cope with a substantial pH gradient across the cytoplasm membrane. The pH conditions for extra- and intracellular enzymes differ greatly. In consequence, in an alkaline environment, the differences between the biochemical optimum conditions of extracellular and intracellular enzymes can be expected to be much greater than in neutrophiles, where the pH values outside and inside the cells are similar. The amylases of the neutrophile Thermoactinomyces vulgaris, for example, differ in their cellular localizations, but their amino acid sequences and physicochemical characteristics are similar (25).
In this communication, we report the heterologous expression, purification, and comparison of the biochemical characteristics of two α-amylases with different cellular localizations from the thermoalkaliphilic bacterium A. gottschalkii.
MATERIALS AND METHODS
Sequence analysis.
Protein sequence databases were searched with the BLAST algorithm available at the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov) (1). Lipoprotein signal sequence prediction was carried out using the LipoP 1.0 server of the Center for Biological Sequence Analysis, Technical University of Denmark (http://www.cbs.dtu.dk) (9).
Anaerobranca gottschalkii culture conditions.
Anaerobranca gottschalkii DSM 13577 was grown anaerobically as previously described (21) at 50°C with 0.5% starch as a carbon source. The supernatant of a 100-liter fermentor culture was concentrated to 57 ml by cross-flow filtration using a 30-kDa membrane (Filtron, Karlstein, Germany).
Construction of AmyA and AmyB expression clones.
The genes coding for AmyA and AmyB were amplified from a genomic library of A. gottschalkii (Epidauros, Bernried, Germany) (11). While for amyB the full-length sequence of the open reading frame (ORF) was obtained, the amyA gene was amplified without its putative signal peptide-encoding sequence. A new start codon was introduced. The primer sequences for amyA were 5′-GAA TTC ATG ACC AGT GAT AAG CAA GGT CCC CAA GAG ACC-3′ (fwd; the EcoRI site is underlined) and 5′-AAG AGC TCA ATT TTT AAG ACA AAT AGA AAA GGA AGA AGA ATT-3′ (rev), and those for amyB were 5′-ATT AGG ATT CAT ATG CAA CAA GAG ATT TTA TAT TTT-3′ (fwd; the NdeI site is underlined) and 5′-C AGA AAA AAG CTT TTT ATT TTT CAG GGG C-3′ (rev; the HindIII site is underlined). The amplified products were initially cloned into pTOPO (Invitrogen, Karlsruhe, Germany) and pBluescript (Stratagene, Heidelberg, Germany) vectors, respectively, before being recloned into pET21c overexpression vectors (Novagen, Schwalbach, Germany). The correct cloning of both amylase ORFs was confirmed by sequencing and restriction enzyme analyses. All recombinant techniques were performed in Escherichia coli XL1-blue, E. coli DH5α, and E. coli BL21.
Expression and crude-extract preparation of the recombinant α-amylases.
Liquid cultures (two, 1 liter each) of the recombinant E. coli clones carrying the pET21c-amyA construct and the pET21c-amyB construct were grown in 5-liter baffle flasks overnight at 37°C in Luria-Bertani medium (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl) supplemented with 50 μg ml−1 ampicillin. When the optical densities (600 nm) of the cultures reached approximately 0.6, the T7 promoter of the pET vectors was induced with 1 mM isopropyl-1-thio-β-d-galactoside (IPTG), and incubation was continued overnight. Cells were harvested by centrifugation (10,400 × g; 15 min; 4°C), washed, and resuspended in 30 ml phosphate-buffered saline (pH 8.5) in the case of the AmyA-expressing culture or 20 ml 20 mM Tris-HCl (pH 8.0) in the case of the AmyB-expressing culture. The cells were disrupted by twofold passage through a French pressure cell (American Instruments, Silver Springs, MD). After separation from the cell debris by centrifugation (20,000 × g; 30 min; 4°C), the supernatants were incubated at 65°C for 20 min in order to denature the thermolabile host proteins, which were sedimented at 20,000 × g for 15 min at 4°C.
Purification of AmyA.
The cleared supernatant from the heat-treated crude extract of E. coli BL21(pET21c-amyA) containing ∼95 mg protein was dialyzed against 20 mM Tris-HCl (pH 8.0) buffer and subjected to anion-exchange chromatography on a Source 15Q XK 26/10 column (Amersham Biosciences, Freiburg, Germany) equilibrated with the same buffer. Elution was done with a two-step gradient (0 to 0.4 M NaCl in 10 column volumes [CV]; 0.4 to 1 M NaCl in 1 CV) in the same buffer at a flow rate of 10 ml min−1. Amylase activity-containing fractions, which eluted at about 0.3 M NaCl, were pooled and dialyzed against 20 mM Tris-HCl (pH 8.0) buffer. In order to further purify the protein by hydrophobic interaction chromatography, the pool (∼32 mg protein) was adjusted to 5 M NaCl and applied to a phenyl-Sepharose HP XK 16/10 column (Amersham Biosciences) equilibrated with 20 mM Tris-HCl (pH 8.0) buffer containing 5 M NaCl. Elution was performed with a two-step gradient (5 to 3 M NaCl in 1 CV; 3 to 0 M NaCl in 9 CV) in 20 mM Tris-HCl (pH 8.0) buffer at a flow rate of 2 ml min−1. AmyA-containing fractions, which eluted at ∼0.5 M NaCl, were pooled and dialyzed against 20 mM Tris-HCl (pH 8.0). The purity of the resulting enzyme preparation was >95%, as confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Purification of AmyB.
The cleared supernatant from the heat-treated crude extract of E. coli BL21(pET21c-amyB), about 165 mg protein, was dialyzed against 20 mM Tris-HCl (pH 8.0) buffer and applied to a Source 15Q XK 26/10 column (Amersham Biosciences) equilibrated with the same buffer. Elution was done with a two-step gradient (0 to 0.4 M NaCl in 10 CV; 0.4 to 1 M NaCl in 2 CV) in the same buffer at a flow rate of 5 ml min−1. AmyB-containing fractions were pooled and dialyzed against 20 mM Tris-HCl (pH 8.0) buffer. The purity of the resulting enzyme preparation was >95%, as confirmed by SDS-PAGE.
Analytical size exclusion chromatography.
In order to determine the native molecular masses of the purified proteins, analytical size exclusion chromatography was carried out using a Superdex 200 prep grade HiLoad 16/60 column (Amersham Biosciences). An isocratic gradient of 20 mM Tris-HCl (pH 8.0) with the addition of 150 mM NaCl was applied.
Gel electrophoresis.
SDS-PAGE according to the method of Laemmli (12) was routinely performed to test the purity of the samples during purification and to estimate the molecular masses of proteins under denaturing conditions. SDS-PAGE was carried out with gels containing 12% (vol/vol) acrylamidebisacrylamide in Mini-Protean II electrophoresis units (Bio-Rad, Munich, Germany), and the gels were stained with Coomassie brilliant blue. Sizes of the analyzed proteins were estimated by comparison with the standard protein marker mixture SDS-6H (Sigma-Aldrich, Taufkirchen, Germany) separated on the same gel.
For activity staining, the SDS-PAGE gel was equilibrated in 0.2 M phosphate buffer (pH 7.0) for 45 min, followed by an overnight incubation in 0.2 M phosphate buffer containing 1% soluble starch. Amylase activity was determined by incubating the gel in this buffer at 75°C for 15 min. The remaining starch was stained with Lugol solution (0.3% [wt/vol] I2, 0.6% [wt/vol] KI), a process that left clear halos around sites of amylolytic activity.
Activity assay.
A 3,5-dinitrosalicylic acid-based method for measuring the enzymatic release of reducing groups from starch was used as a standard test (19). The reaction mixture contained 250 μl substrate (0.5% [wt/vol] soluble starch [Sigma-Aldrich] or other polysaccharides as specified in the text); 100 μl 200 mM McIlvaine buffer adjusted to pH 8.0 for AmyA and pH 6.0 for AmyB, respectively; 150-x μl distilled H2O; and x μl appropriately diluted enzyme (where x represents the volume of enzyme preparation). After incubation for 20 min at 60°C for AmyA and 15 min at 50°C for AmyB, 750 μl of 3,5-dinitrosalicylic acid solution (1% [wt/vol] 3,5-dinitrosalicylic acid, 0.2% [vol/vol] phenol, 0.05% [wt/vol] Na2SO4, 20% KNa-tartrate, 1% [wt/vol] NaOH in H2Obidest) was added, and the mixture was incubated for 15 min at 98°C. Absorption was measured at 575 nm, and the specific activity was calculated using maltose as a standard. One unit of α-amylase is defined as the amount of enzyme that liberates 1 μmol of reducing groups per min.
Influences of pH and temperature on activity.
The influence of pH on AmyA was measured at 60°C and on AmyB at 50°C with McIlvaine buffer adjusted to various pH values, using the standard assay described above. The influence of temperature on the enzyme activity was determined using standard assay reactions incubated at temperatures ranging from 35 to 80°C.
Thermoinactivation.
The enzymes were diluted to a protein concentration of about 50 μg ml−1 in McIlvaine buffer adjusted to pH 8.0 (AmyA) or pH 6.0 (AmyB) at selected temperatures. The kinetics of thermoinactivation of the recombinant enzymes was determined through preincubation of the diluted enzyme at various temperatures for different intervals of time, followed by determination of residual activity with the standard enzyme assay.
Influence of metal ions.
The effect of metal ions was analyzed at 1 mM final concentrations of a variety of mono- and divalent cations in a HEPES (pH 8.0) and a 2-morpholinoethanesulfonic acid (MES) (pH 6.0) buffer system for AmyA and AmyB, respectively, in order to avoid precipitation of some of the ions with the McIlvaine buffer components.
The effect of EDTA on AmyA activity was analyzed in HEPES, McIlvaine, and Tris buffers, all at 50 mM at pH 8.0. The enzyme was diluted to 50 μg ml−1 with buffer and incubated with 5 mM EDTA for 1 h prior to the determination of activity in the same buffer system.
Analysis of sugars by TLC.
Thin-layer chromatography (TLC) of mono- and oligosaccharides was done on 0.2 mm silica gel-coated aluminum sheets (Type 60; Merck, Darmstadt, Germany) with a solvent system containing 1-propanol-ethyl acetate-H2O at a volume ratio of 6:1:3 or 1-propanol-nitromethane-H2O at a volume ratio of 5:3:2. The chromatograms were sprayed with anilinediphenylamine reagent (1% [wt/vol] diphenylamine, 1% [vol/vol] aniline in acetone mixed with 0.1 volume 87% [vol/vol] phosphoric acid prior to use) and baked for 10 min at 160°C to visualize the carbohydrate spots.
β-Cyclodextrin assay.
The presence of cylodextrins was determined using a phenolphthalein colorimetric assay (27). After incubation for 1 h at 60°C, a standard reaction mixture aliquot of 100 μl was added to 2 ml phenolphthalein test solution ready mixed from 10 ml Na2CO3 (0.2 M) and 17 μl of phenolphthalein stock solution (1% [wt/vol] phenolphthalein in 50% [vol/vol] ethanol). Absorption was measured at 537 nm against a blank of 100 μl maltooligosaccharide mixture (0.1% [wt/vol]) and 2 ml phenolphthalein test solution. The concentration of β-cyclodextrin was calculated by comparison with assay results obtained with defined amounts of β-cyclodextrin standard.
RESULTS
Sequence analysis and enzyme localization.
The amyA gene (gb:AY842299) from A. gottschalkii has a size of 1,599 bp and codes for a 532-amino-acid polypeptide. A lipoprotein signal peptide of 21 residues was found by using the LipoP 1.0 prediction program for lipoproteins, which uses a hidden Markov model to distinguish between lipoproteins (SPase II-cleaved proteins), SPase I-cleaved proteins, cytoplasmic proteins, and transmembrane proteins. The potential cleavage site in the sequence MTSKILRVLLVFLLIFAIVG-CTS(…) is predicted between glycine 20 and cysteine 21. In order to avoid expression problems, amyA was cloned without the signal peptide-encoding sequence, and cysteine 21 was mutated to a methionine. The resulting ORF on the pET21-amyA vector had a length of 1,539 bp and coded for a protein of 512 amino acids.
The amino acid sequence of AmyA shows between 42 and 45% identity with a putatively exported α-amylase from Bacillus megaterium (gb:P20845) (18), a neopullulanase from Paenibacillus sp. strain KCTC8848P (gb:AAL07400) (10), and α-amylase 3 from Dictyoglomus thermophilum, another putative extracellular protein (gb:P14899) (8), which constitute the three best-scored hits with the BLAST algorithm.
No indications of a signal peptide could be found in the 443-residue polypeptide that is encoded by the 1,331-bp amyB ORF (gb:AY842298), indicating that AmyB presumably is a cytoplasmic protein. The AmyB amino acid sequence shows 43% identity with that of the α-amylase from Thermoactinomyces vulgaris (gb:CAA49465) (7), 39% identity with a β-/α-amylase precursor from Bacillus polymyxa (gb:A29130) (26), and 34% identity with a putative glycoside hydrolase from Deinococcus radiodurans (gb:AAF11034) (28). Altogether, the amino acid sequences of both AmyA and AmyB bear high similarity to glycoside hydrolase family 13 (GHF13) proteins; the four highly conserved motifs typical of GHF13 enzymes were found in both A. gottschalkii α-amylases (Table 1). Beyond the conserved regions, the two studied proteins had an overall sequence identity of 33%.
TABLE 1.
Comparison of the four highly conserved α-amylase regions of AmyA and AmyB from A. gottschalkii with TAKA-amylase from Aspergillus oryzae (gb:0901305A) (24)a
| Enzyme | Region I | Region II | Region III | Region IV |
|---|---|---|---|---|
| TAKA-amylase | 117 DVVANH | 202 GLRIDTVKH | 229 GEVLD | 292 FVENHD |
| AmyA | 150 DLVVNH | 238 GFRLDAAKH | 276 GEIWD | 339 FLSNHD |
| AmyB | 108 DIVVNH | 180 GFRIDTVKH | 212 GEVWH | 274 FIDNHD |
The three invariable catalytic residues are in boldface. Numbering of the amino acid sequences starts at the amino-terminal residue of each mature enzyme.
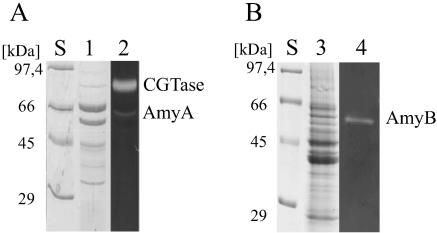
The presumed localizations of AmyA and AmyB in the culture supernatant and the cytosolic fraction of an A. gottschalkii culture were validated by SDS-PAGE and subsequent activity staining. The mobilities of the bands with amylolytic activity corresponded well with the expected sizes of the enzymes. Obviously, at least part of putative lipoprotein AmyA is released into the culture supernatant (Fig. 1).
FIG. 1.
SDS-PAGE analysis and activity staining of A. gottschalkii culture supernatant (A) and cytosolic crude extract (B). Lanes S, size standard; lane 1, Coomassie-stained culture supernatant; lane 2, activity-stained culture supernatant; lane 3, Coomassie-stained cytosolic crude extract; lane 4, activity-stained cytosolic crude extract.
Purification.
After induction with 1 mM IPTG, the overnight flask cultures of E. coli BL21 cells transformed with pET21c-amyA yielded approximately 1.6 g cells liter−1. Following the disruption of the E. coli cells, recombinant AmyA was initially concentrated by heat treatment at 65°C for 20 min, which resulted in denaturation of the majority of the host proteins. After two subsequent chromatographic separation steps, i.e., anion-exchange followed by a hydrophobic interaction chromatography, an enzyme preparation appearing as a single band in SDS-PAGE was obtained. AmyB was purified through heat treatment of the crude extract, followed by anion-exchange chromatography, leading to a homogenous enzyme preparation as judged by SDS-PAGE (data not shown). With starch as the substrate, the specific activities were 52 U/mg for purified AmyA and 220 U/mg for purified AmyB.
The apparent sizes of the proteins according to SDS-PAGE are in agreement with the molecular masses derived from the amino acid sequences. The conceptual translation masses of the full-length open reading frames of the amyA (as cloned without the signal peptide-encoding sequence) and amyB genes are 59,146 Da and 51,702 Da, respectively. Recombinant AmyB had a molecular mass of 108 kDa as determined by its elution behavior during gel filtration chromatography, which suggests the active molecule exists as a dimer. For AmyA, a molecular mass of 58 kDa was determined, which points to a monomer as the active quaternary form.
Biochemical characterization and substrate specificity.
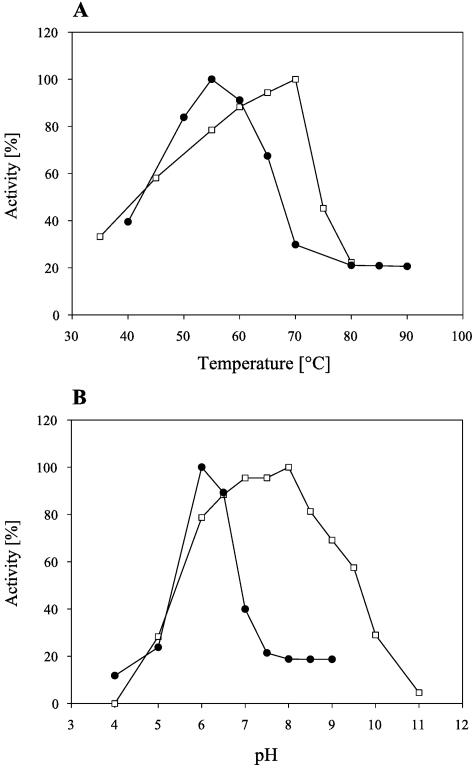
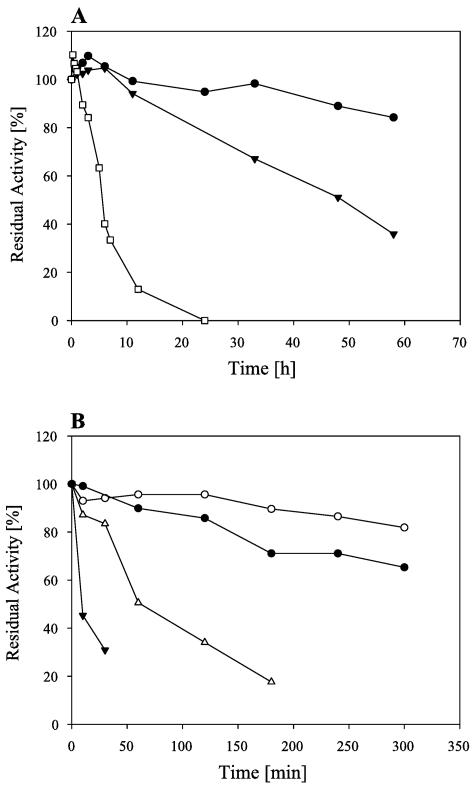
With a 20-min assay using starch as the substrate, the maximum enzymatic activity of AmyA was measured at 70°C and pH 8.0. For AmyB, maximum activity was found at 55°C and pH 6.0 (Fig. 2). The half-life of AmyA at 70°C was 48 h. At 60°C, no significant loss of activity was found even after 60 h of preincubation. AmyB retained more than 80% activity after 5 h of preincubation at 50°C. At 65°C, the maximum growth temperature of A. gottschalkii (21), AmyB had a half-life of 60 min (Fig. 3).
FIG. 2.
Effects of temperature (A) and pH (B) on the activities of AmyA (white squares) and AmyB (black squares). All measurements were carried out in McIlvaine buffer calibrated at the respective pH and temperature.
FIG. 3.
Effects of long-term incubation at elevated temperatures on AmyA (A) and AmyB (B). AmyA was incubated at 60°C (black circles), 70°C (black inverted triangles), and 75°C (white squares). AmyB was incubated at 50°C (white circles), 60°C (black circles), 65°C (white triangles), and 70°C (black inverted triangles).
AmyA and AmyB were hydrolytically active on a variety of α-1,4-linked carbohydrates, with amylose being the best substrate in both cases (Table 2). The main products of AmyA hydrolysis were maltose and glucose, except with pullulan as a substrate. Substrates exclusively containing non-α-1,4-glycosidic bonds, such as mannan, dextran, and cellulose, were not degraded. AmyB action resulted in glucose, maltose, and maltotriose as final products, with maltose being the major product. Both enzymes hydrolyzed amylose with an endomechanism, which was determined by thin-layer chromatography with samples drawn at different time points during amylose degradation. First, maltooligosaccharides of various lengths were generated, and later, glucose and maltose or glucose, maltose, and maltotriose were accumulated as final products (data not shown). On the basis of their substrate specificities and characteristics of product formation by cleavage of α-1,4-glycosidic bonds in polysaccharides, the A. gottschalkii enzymes AmyA and AmyB were classified as α-amylases (EC 3.2.1.1).
TABLE 2.
Substrate specificities of the recombinant α-amylases
| Substrate | Relative sp act (%)
|
|
|---|---|---|
| AmyA | AmyB | |
| Starch | 100 | 100 |
| Amylose | 157 | 202 |
| Amylopectin | 64 | 61 |
| Glycogen | 16 | 14 |
| Pullulan | 17 | 44 |
| α-Cyclodextrin | 13 | 11 |
| β-Cyclodextrin | 59 | 52 |
| γ-Cyclodextrin | 58 | 47 |
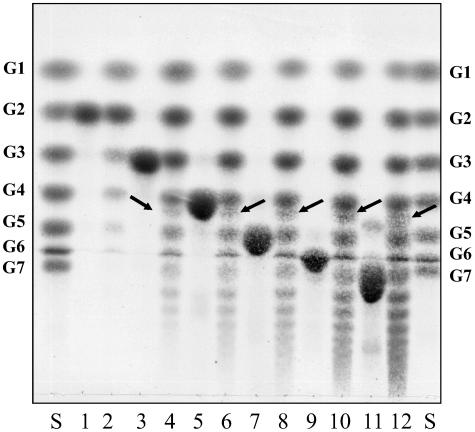
With short maltodextrins as substrates, AmyA displayed significant transferase activity, as well as hydrolysis (Fig. 4). Strikingly, in addition to linear maltodextrins, a small amount of cyclodextrin was formed, which was identified as β-cyclodextrin by its mobility in TLC analysis and its complexation property in a colorimetric assay with phenolphthalein. In a standard assay with 23.5 μg ml−1 AmyA, about 1.6% of the initial 25 mM maltotriose was converted to β-cyclodextrin with a specific activity of 0.3 U/mg. Over time, the maltodextrins formed, as well as the β-cyclodextrin, were degraded again by the enzyme. For AmyB, no significant transferase activity or cyclodextrin formation was observed.
FIG. 4.
TLC analysis of product formation from various maltodextrins by AmyA. Maltodextrins (25 mM) were digested with recombinant AmyA at 24 μg/ml in McIlvaine buffer for 1 h at 60°C. The products were separated on a silica gel TLC plate in 1-propanol-ethyl acetate-H2O (6:1:3) (vol/vol/vol). Lane S, G1-G7 oligosaccharide standard; lane 1, maltose; lane 2, maltose after incubation with AmyA; lane 3, maltotriose; lane 4, maltotriose after incubation with AmyA; lane 5, maltotetraose; lane 6, maltotetraose after incubation with AmyA; lane 7, maltopentaose; lane 8, maltopentaose after incubation with AmyA; lane 9, maltohexaose; lane 10, maltohexaose after incubation with AmyA; lane 11, maltoheptaose; lane 12, maltoheptaose after incubation with AmyA. The arrows indicate the formed β-cyclodextrin.
Effects of metal ions and other reagents.
The effects of various metal ions added to the reaction mixture were tested. In order to avoid precipitation, McIlvaine buffer was replaced with HEPES (AmyA) and MES (AmyB), although AmyA displayed significantly lower activity in HEPES buffer than in McIlvaine buffer. Ca2+, Sr2+, and Ba2+ at a concentration of 1 mM had a positive effect on AmyA. With Ca2+ supplementation, its activity was increased twofold, while Sr2+ and Ba2+ had a slightly weaker effect on the enzyme. Mn2+ and Co2+ reduced AmyA activity, while Ni2+, Cu2+, Zn2+, and Cd2+ completely inhibited the enzyme. NaCl in concentrations above 50 mM slightly enhanced the activity of AmyA. The most prominent effects on AmyB were generated by the addition of 1 mM Mn2+, Co2+, Cu2+, and Zn2+, with all of these ions strongly inhibiting the recombinant enzyme (data not shown).
The addition of ethanol (5% [vol/vol]) and 2-mercaptoethanol (2 mM) had no effect on AmyA activity, whereas glycerol (5% [vol/vol]), dithiothreitol (10 mM), and NAD (6 mM) had minor inhibitory effects on the recombinant enzyme (data not shown).
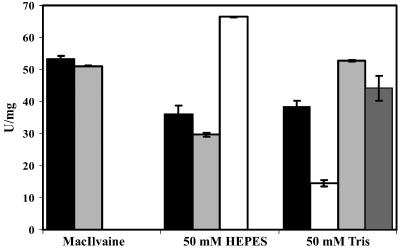
EDTA was not found to be inhibitory for AmyA in HEPES and McIlvaine buffer systems. Even after preincubation for 1 h at 60°C with 5 mM EDTA, the enzyme activity was not significantly reduced, indicating that Ca2+, although stimulating, was not required for activity of the recombinant enzyme in these buffers. Curiously, when Tris buffer was used, EDTA had an inhibitory effect, which could be completely reversed by the addition of 10 mM Ca2+ (Fig. 5).
FIG. 5.
Effects of EDTA on AmyA under different buffer conditions. The enzyme was diluted to 50 μg ml−1 with the corresponding buffer and preincubated with CaCl2 or EDTA at 60°C for 1 h. Black bars, AmyA without additive; light-gray bars, AmyA with 5 mM EDTA; white bars, AmyA with 5 mM CaCl2; dark-gray bar, AmyA with 5 mM EDTA plus 10 mM CaCl2 added to the activity test. The effect of CaCl2 in MacIlvaine buffer was not determined due to precipitation.
DISCUSSION
The α-amylases AmyA and AmyB from the anaerobic thermoalkaliphilic bacterium A. gottschalkii were heterologously overexpressed in E. coli and biochemically characterized. AmyA, a putative lipoprotein, was expressed without its signal peptide, resulting in a monomer of 59 kDa. AmyB was found to be a homodimer with two 52-kDa subunits. Based on their modes of hydrolytic action (endomechanism) and their substrates (α-1,4-linked polysaccharides), both enzymes were classified as α-amylases. They both showed sequence similarity to members of glycoside hydrolase family 13. It has been argued that the last two amino acid residues in the conserved region II of GHF13 proteins are of critical importance for substrate specificity (15). Both α-amylases examined have a lysine and a histidine in those positions, which supports the theory that these two residues appear in enzymes with specificity for α-1,4-glycosidic bonds.
Apparently, AmyA features, in addition to the typical α-amylase hydrolytic activity, the ability to act as a 4-α-glucanotransferase, a combination of activities not unusual for α-amylases. However, it is noteworthy that AmyA has significant β-cyclodextrin-forming activity, which is unprecedented for typical α-amylases. On the other hand, cyclodextrin glycosyltransferases are known to have various degrees of transferase activity on linear maltodextrins (“disproportionation activity”), as well as hydrolytic activity, in addition to their main activity, i.e., cyclodextrin formation. Some CGTases have quite high α-amylase activity, e.g., the CGTase from Thermoanaerobacterium thermosulfurigenes EM1 (30), whose hydrolytic activity (23 U/mg) is only about fourfold smaller than its CGTase activity. AmyA from A. gottschalkii has broadened the spectrum of cyclodextrin-forming enzymes, being an enzyme with low cyclodextrin glycosyltransferase activity (0.3 U/mg) compared to its hydrolytic activity (52 U/mg). Apart from belonging to the same glycoside hydrolase family (GHF13), the primary structure of AmyA is not similar to those of typical CGTases. It will be interesting to compare the crystal structure of AmyA, once available, with the high-resolution structures of CGTases in order to understand how cyclodextrin formation is accomplished by AmyA. The physiological relevance of the cyclodextrin-forming activity of AmyA, however, is arguable, since A. gottschalkii has an extracellular, recently characterized CGTase with a specific activity of 210 U mg−1 (23).
The different cellular localizations of AmyA and AmyB are reflected in their physicochemical characteristics; in particular, the pH optima, as well as the range of pH values, of the A. gottschalkii amylases mirror the pH conditions at the cellular locations where the enzymes are predicted to be found. AmyA shows activity over a broad pH range from 6.0 to 9.5, with an optimum at pH 8.5. As a probable lipoprotein located outside the cell, the enzyme is exposed to the alkaline milieu of the habitat. A. gottschalkii grows between pH 6.0 and pH 10.5, a range corresponding to the pH range of the protein. Another extracellular enzyme of A. gottschalkii, a type I pullulanase (PulAg), which is also predicted to be a lipoprotein, is active from pH 7.0 to 9.5, with an optimum at pH 8.0 (2). This activity range coincides very well with that of the recombinant AmyA. The same is true for the temperature-activity profiles of both amylolytic enzymes. Using a 20-min and a 10-min assay, respectively, at their optimal pHs, both AmyA and PulAg displayed maximum rates of substrate hydrolysis at 70°C. The physicochemical characteristics of the extracellular CGTase from A. gottschalkii differ only slightly from this. The enzyme shows maximum activity at pH 6 to 9 and 65°C in a 45-min assay (23).
Few α-amylases are active at both temperatures above 65°C and alkaline pH values of 7.5 or higher. All of them are proteins from Bacillus species, such as the α-amylases from Bacillus sp. strain TS-23, with an activity optimum at pH 9.0 and 70°C; from B. megaterium, with an activity optimum at pH 8.9 and 70°C; and from Bacillus licheniformis CUMC 305, with an activity optimum at pH 9.0 and 90°C. AmyA from A. gottschalkii, a non-Bacillus organism, is a notable exception (3, 14, 17).
AmyB, the intracellular α-amylase, has an optimum at pH 6.0 and a much narrower pH range than AmyA. Since the intracellular pH of alkaliphiles is more acidic than the medium, intracellular enzymes can be expected to be somewhat less alkaliphilic than their extracellular counterparts. Due to cell homeostasis, changes of the internal pH are small, which allows enzymes to be less tolerant of pH changes. In addition, the intracellular AmyB is less tolerant of high temperatures than the extracellular AmyA. While AmyA has a half-life of 48 h at 70°C, AmyB has a half-life of only about 10 min at that temperature. However, in vivo chaperones, low-molecular-mass stabilizers, or other cytoplasmic compounds may have a stabilizing effect on AmyB. The precise physiological role of AmyB is not known, but the enzyme may be involved in the breakdown of maltooligosaccharides transported into the cell or in the turnover of storage polysaccharides.
Many α-amylases are known to be Ca2+ dependent. In the case of AmyA, the addition of Ca2+ had a positive effect on activity. On the other hand, EDTA reduced the enzyme activity only under certain conditions, namely, in Tris buffer. In HEPES and McIlvaine buffers, EDTA was unable to decrease the activity significantly even after incubation with the enzyme at elevated temperatures. The reason for the observed buffer-dependent effect of EDTA is unclear, but it may point to tightly bound Ca2+ that is released only under certain circumstances.
Similarly, resistance to chelating agents was found for the A. gottschalkii type I pullulanase (2). While Ca2+ enhances the stability of the CGTase from A. gottschalkii, effects on activity were not reported (23). The soda lake from which A. gottschalkii was originally isolated (Lake Bogoria, Kenya) is severely depleted of Ca2+ (4), which makes a dependence of extracellular enzymes on loosely bound Ca2+ seem inappropriate. It has even been reported for two Bacillus-related strains isolated from soda lakes in Ethiopia that amylase activity in the culture supernatant was stimulated by EDTA and decreased by Ca2+ ions, possibly due to adaptation to the Ca2+-deficient environment (16).
Thermoalkaliphilic organisms are thus a promising source for chelating-reagent-resistant glycoside hydrolases, which are active at alkaline pH values and tolerant of elevated temperatures.
Acknowledgments
R. Sterner and R. Freudl are thanked for helpful discussions. We are grateful to H.-P. Klenk for his aid in sequence analysis.
Financial support from the Deutsche Bundesstiftung Umwelt (DBU) and the Deutsche Forschungsgemeinschaft (DFG; Li398/6-3) is greatly appreciated.
Footnotes
Dedicated to H. G. Schlegel on the occasion of his 80th birthday.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bertoldo, C., M. Armbrecht, F. Becker, T. Schafer, G. Antranikian, and W. Liebl. 2004. Cloning, sequencing, and characterization of a heat- and alkali-stable type I pullulanase from Anaerobranca gottschalkii. Appl. Environ. Microbiol. 70:3407-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brumm, P. J., R. E. Hebeda, and W. M. Teague. 1996. Purification and characterisation of the commercialized, cloned Bacillus megaterium α-amylase. Part II: transferase properties. Starch/Staerke 43:319-323. [Google Scholar]
- 4.Duckworth, A. W., W. D. Grant, B. E. Jones, and R. P. Van Steenbergen. 1996. Phylogenetic diversity of soda lake alkaliphiles. FEMS Microbiol. Ecol. 19:181-191. [Google Scholar]
- 5.Engle, M., Y. Li, C. Woese, and J. Wiegel. 1995. Isolation and characterization of a novel alkalitolerant thermophile, Anaerobranca horikoshii gen. nov., sp. nov. Int. J. Syst. Bacteriol. 45:454-461. [DOI] [PubMed] [Google Scholar]
- 6.Gorlenko, V., A. Tsapin, Z. Namsaraev, T. Teal, T. Tourova, D. Engler, R. Mielke, and K. Nealson. 2004. Anaerobranca californiensis sp. nov., an anaerobic, alkalithermophilic, fermentative bacterium isolated from a hot spring on Mono Lake. Int. J. Syst. Evol. Microbiol. 54:739-743. [DOI] [PubMed] [Google Scholar]
- 7.Hofemeister, B., S. Konig, V. Hoang, J. Engel, G. Mayer, G. Hansen, and J. Hofemeister. 1994. The gene amyE(TV1) codes for a nonglucogenic alpha-amylase from Thermoactinomyces vulgaris 94-2A in Bacillus subtilis. Appl. Environ. Microbiol. 60:3381-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horinouchi, S., S. Fukusumi, T. Ohshima, and T. Beppu. 1988. Cloning and expression in Escherichia coli of two additional amylase genes of a strictly anaerobic thermophile, Dictyoglomus thermophilum, and their nucleotide sequences with extremely low guanine-plus-cytosine contents. Eur. J. Biochem. 176:243-253. [DOI] [PubMed] [Google Scholar]
- 9.Juncker, A. S., H. Willenbrock, G. von Heijne, H. Nielsen, S. Brunak, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, H. J., J. N. Park, H. O. Kim, H. B. Lee, S. B. Chun, and S. Bai. Unpublished data.
- 11.Klenk, H. P., A. Ruepp, M. Stark, A. Zibat, and I. Becker. 2002. Screening for enzyme encoding genes in genomes of extremophilic prokaryotes, p. 23. In R. Grote and G. Antranikian (ed.), Book of abstracts, 1st International Congress of Biocatalysis, Hamburg, 2002. TUHH-Technology GmbH, Hamburg, Germany.
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Li, Y., L. Mandelco, and J. Wiegel. 1993. Isolation and characterization of a moderately thermophilic anaerobic alkaliphile, Clostridium paradoxum sp. nov. Int. J. Syst. Bacteriol. 43:450-460. [Google Scholar]
- 14.Lin, L. L., C. C. Chyau, and W. H. Hsu. 1998. Production and properties of a raw-starch-degrading amylase from the thermophilic and alkaliphilic Bacillus sp. TS-23. Biotechnol. Appl. Biochem. 28:61-68. [PubMed] [Google Scholar]
- 15.MacGregor, A., S. Janecek, and B. Svensson. 2001. Relationship of sequence and structure specificity in the α-amylase family of enzymes. Biochem. Biophys. Acta 1545:1-20. [DOI] [PubMed] [Google Scholar]
- 16.Martins, R. F., W. Davids, W. Abu Al-Soud, F. Lavender, P. Radstrom, and R. Hatti-Kaul. 2001. Starch-hydrolyzing bacteria from Ethiopian soda lakes. Extremophiles 5:135-144. [DOI] [PubMed] [Google Scholar]
- 17.Medda, S., and A. K. Chandra. 1980. New strains of Bacillus licheniformis and Bacillus coagulans producing thermostable α-amylase active at alkaline pH. J. Appl. Bacteriol. 48:47-58. [DOI] [PubMed] [Google Scholar]
- 18.Metz, R. J., L. N. Allen, T. M. Cao, and N. W. Zeman. 1988. Nucleotide sequence of an amylase gene from Bacillus megaterium. Nucleic Acids Res. 16:5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426. [Google Scholar]
- 20.Pikuta, E., A. Lysenko, N. Suzina, G. Osipov, B. Kuznetsov, T. Tourova, V. Akimenko, and K. Laurinavichius. 2000. Desulfotomaculum alkaliphilum sp. nov., a new alkaliphilic, moderately thermophilic, sulfate-reducing bacterium. Int. J. Syst. Evol. Microbiol. 50:25-33. [DOI] [PubMed] [Google Scholar]
- 21.Prowe, S., and G. Antranikian. 2001. Anaerobranca gottschalkii sp. nov., a novel thermoalkaliphilic bacterium that grows anaerobically at high pH and temperature. Int. J. Syst. Evol. Microbiol. 51:457-465. [DOI] [PubMed] [Google Scholar]
- 22.Svetlitshnyi, V., F. Rainey, and J. Wiegel. 1996. Thermosyntropha lipolytica gen. nov., sp. nov., a lipolytic, anaerobic, alkalitolerant, thermophilic bacterium utilizing short- and long-chain fatty acids in syntrophic coculture with a methanogenic archaeum. Int. J. Syst. Bacteriol. 46:1131-1137. [DOI] [PubMed] [Google Scholar]
- 23.Thiemann, V., C. Dönges, S. G. Prowe, R. Sterner, and G. Antranikian. 2004. Characterisation of a thermoalkalistable cyclodextrin glycosyltransferase from the anaerobic thermoalkaliphilic bacterium Anaerobranca gottschalkii. Arch. Microbiol. 182:226-235. [DOI] [PubMed] [Google Scholar]
- 24.Toda, H., K. Kondo, and K. Narita. 1982. The complete amino acid sequence of Taka-amylase A. Proc. Jpn. Acad. 58B:208-212. [Google Scholar]
- 25.Tonozuka, T., S. Mogi, Y. Shimura, A. Ibuka, H. Sakai, H. Matsuzawa, Y. Sakano, and T. Ohta. 1995. Comparison of primary structures and substrate specificities of two pullulan-hydrolyzing alpha-amylases, TVA I and TVA II, from Thermoactinomyces vulgaris R-47. Biochim. Biophys. Acta 1252:35-42. [DOI] [PubMed] [Google Scholar]
- 26.Uozumi, N., K. Sakurai, T. Sasaki, S. Takekawa, H. Yamagata, N. Tsukagoshi, and S. Udaka. 1989. A single gene directs synthesis of a precursor protein with beta- and alpha-amylase activities in Bacillus polymyxa. J. Bacteriol. 171:375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vikmon, M. 1981. Rapid and simple spectrophotometric method for determination of micro-amounts of cyclodextrins, p. 69-74. In J. Szejtli (ed.), Advances in inclusion science, vol. 1. Proceedings of the First International Symposium on Cyclodextrins, Budapest, Hungary, 30 September-2 October 1981. D. Reidel, Dordrecht, The Netherlands. [Google Scholar]
- 28.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, et al. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiegel, J., and V. Kevbrin. 2004. Alkalithermophiles. Biochem. Soc. Trans. 32:93-98. [DOI] [PubMed] [Google Scholar]
- 30.Wind, R. D., W. Liebl, R. M. Buitelaar, D. Penninga, A. Spreinat, L. Dijkhuizen, and H. Bahl. 1995. Cyclodextrin formation by the thermostable α-amylase of Thermoanaerobacterium thermosulfurigenes EM1 and reclassification of the enzyme as a cyclodextrin glycosyltransferase. Appl. Environ. Microbiol. 61:1257-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]