Abstract
Perchlorate contamination is a concern because of the increasing frequency of its detection in soils and groundwater and its presumed inhibitory effect on human thyroid hormone production. Although significant perchlorate contamination occurs in the vadose (unsaturated) zone, little is known about perchlorate biodegradation potential by indigenous microorganisms in these soils. We measured the effects of electron donor (acetate and hydrogen) and nitrate addition on perchlorate reduction rates and microbial community composition in microcosm incubations of vadose soil. Acetate and hydrogen addition enhanced perchlorate reduction, and a longer lag period was observed for hydrogen (41 days) than for acetate (14 days). Initially, nitrate suppressed perchlorate reduction, but once perchlorate started to be degraded, the process was stimulated by nitrate. Changes in the bacterial community composition were observed in microcosms enriched with perchlorate and either acetate or hydrogen. Denaturing gradient gel electrophoresis analysis and partial sequencing of 16S rRNA genes recovered from these microcosms indicated that formerly reported perchlorate-reducing bacteria were present in the soil and that microbial community compositions were different between acetate- and hydrogen-amended microcosms. These results indicate that there is potential for perchlorate bioremediation by native microbial communities in vadose soil.
For over 50 years, perchlorate (ClO4−) salts have been manufactured and used as ingredients in solid rocket fuels, highway safety flares, air bag inflators, fireworks, and matches (20, 23, 35). Due to past improper disposal of these chemicals, perchlorate has become a widespread environmental contaminant in the United States (35). Perchlorate has been detected in soil, groundwater, drinking water supplies (9, 34, 35), and milk (19). Perchlorate ion is reported to interrupt human thyroid hormone production (33), and the U.S. Environmental Protection Agency's 2002 draft toxicological report recommended a drinking water limit of 1 ppb (4).
Although perchlorate is persistent in the environment, studies have proven microorganisms can reduce perchlorate, an electron acceptor, to innocuous chloride and oxygen under anaerobic conditions (5, 8, 12, 16, 18, 21, 22, 26, 27, 36, 38, 40). A widely accepted perchlorate-reducing pathway (26) is the following:
ClO4− (perchlorate) → ClO3− (chlorate) → ClO2− (chlorite) → Cl− (chloride) + O2
All previously isolated perchlorate-reducing bacteria belong to Proteobacteria (8, 12, 18, 21, 26, 27, 36, 37, 40). In earlier studies, most isolates are members of Dechloromonas and Dechlorosoma in the β-Proteobacteria (8, 12, 21, 22, 40). Many perchlorate-reducing Azospirillum spp., a group of α-Proteobacteria, have also been isolated recently (37). Acetate is an electron donor commonly used to isolate perchlorate-reducing bacteria. Strains of Dechloromonas spp. and Dechlorosoma spp. can use lactate, and strains of Dechlorosoma spp. can use ethanol as an electron donor (12, 21). On the other hand, autotrophic Dechloromonas sp. strain HZ uses hydrogen as an electron donor (40). All perchlorate-reducing isolates are facultative anaerobic or microaerobic, and most, with the exception of three Dechloromonas strains, can use nitrate as an electron acceptor (8, 12, 18, 21, 26, 36).
Perchlorate has been deposited in vadose (unsaturated) zone soils via disposal and spills, and it can be a substantial source for groundwater contamination through infiltration. Agricultural soil also may be impacted by irrigating with perchlorate-contaminated water. Nevertheless, relatively little is known about perchlorate degradation in vadose soil. The potential for perchlorate reduction has been shown in soils with or without prior perchlorate exposure by saturated soil microcosm studies and most probable number enumeration (12, 29, 32, 38). Microorganisms involved in perchlorate reduction in vadose soil, however, have not been identified.
In this study, we examined the effect of electron donor (acetate and hydrogen) and nitrate additions on perchlorate reduction by native microbial communities in vadose soil by conducting microcosm studies. Nitrate, usually present in soil, is likely to compete with perchlorate because of the capability of many organisms to use both as electron acceptors. Microorganisms presumably involved in perchlorate reduction were detected by observing changes in microbial community composition after perchlorate exposure.
MATERIALS AND METHODS
Soil sampling.
Yolo silt loam soil was collected from the top 0 to 15 cm in an agricultural field in Yolo County, California, in November 2001. The soil had no known history of exposure to anthropogenic sources of perchlorate and contained approximately 13 mmol · kg dry soil−1 of nitrate (NO3−) and 3.3 mmol · kg dry soil−1 of chloride (Cl−) endogenously. The pH was 7.2 ± 0.4. The soil was air dried, passed through a 2-mm sieve, and stored at 4°C until experiments were performed.
Effect of acetate and hydrogen amendments.
The effect of electron donor addition on perchlorate reduction was examined in a soil microcosm experiment. Soil microcosms were prepared in 30-ml serum bottles with butyl-rubber septa and aluminum caps. About 30 bottles of microcosms with each condition were set up so that triplicates were sampled at each sampling time. Five different combinations of carbon compounds and headspace gases were used in the microcosms, and two conditions were used for sterile controls (Table 1). Acetate (10 mmol · kg dry soil−1; CH3COONa solution) and hydrogen gas (H2) were added as additional electron donors to support perchlorate reduction. Bicarbonate (20 mmol · kg dry soil−1; NaHCO3 solution) also was used as an inorganic carbon source when hydrogen was an electron donor. A mixture of 10 g dry equivalent soil, carbon and/or electron donor additives, and perchlorate (1.0 mmol · kg dry soil−1; NH4ClO4 solution) was added in a serum bottle. The measured perchlorate concentration was 1.09 ± 0.07 mmol · kg dry soil−1. Gravimetric moisture was adjusted to 20%. Sterile soils for abiotic controls were also prepared by repeating 1-h autoclaving three times with 23-h waiting intervals. Air in the headspace of bottles was purged with either nitrogen or hydrogen gas through a 0.2-μm sterile filter. Microcosms were incubated at room temperature (23 ± 1°C). The soil pH was not adjusted and ranged from 6.8 to 8.5 during the experiment.
TABLE 1.
Summary of the conditions in microcosm test with acetate and hydrogen
| Assigned set no. | Soil | Carbon addition | Headspace gas |
|---|---|---|---|
| 1 | Fresh | None | Nitrogen |
| 2 | None | Hydrogena | |
| 3 | Acetatea | Nitrogen | |
| 4 | Bicarbonate | Hydrogena | |
| 5 | Acetatea | Hydrogena | |
| 6 | Sterile | Acetatea | Nitrogen |
| 7 | Bicarbonate | Hydrogena |
Acetate and hydrogen were used as electron donors to reduce perchlorate.
Effect of nitrate.
Another microcosm series was prepared to examine the effect of nitrate on perchlorate reduction. To eliminate the high concentration of naturally occurring nitrate, the soil was washed with 3 pore volumes of autoclaved purified water by vacuum extraction. The soil nitrate concentration decreased by 1.7 mmol · kg dry soil−1 after washing. This method also leached other soluble substances in soil; for instance, chloride concentration decreased by 0.27 mmol · kg dry soil−1. The soil pH was raised to 8.1 ± 0.2 by leaching, though it was lowered to 7.7 ± 0.1 by adding solutions of perchlorate, acetate, and nitrate. Microcosms were set up in the same manner as previously, but nitrate (NaNO3 solution) was added to achieve three different concentrations (approximately 1.7, 2.9, and 5.0 mmol · kg dry soil−1), and only acetate was used as an electron donor. The moisture content was adjusted to 23% this time, since the washing step raised the soil moisture content (∼21%). Due to possible negative effects by soil washing, 0.5 mmol · kg dry soil−1 of perchlorate, half of the load of the first experiment, was added to soil. The measured perchlorate concentration was 0.461 ± 0.022 mmol · kg dry soil−1. Five mmol · kg dry soil−1 of acetate was added to keep the same ClO4−/CH3COO− ratio as in the first experiment. NO3−/ClO4− molar ratios were approximately 3.7, 6.6, and 11, respectively, in each set of microcosms. A sterile control microcosm was set up with 5.0 mmol · kg dry soil−1 of nitrate. The serum bottle headspace was filled with 0.2 μm filtered nitrogen gas. The pH was not adjusted and ranged from 7.2 to 8.1 during the experiment.
Chemical analyses in microcosms.
Perchlorate degradation, chloride formation, and nitrate degradation were monitored by destructively sampling three microcosms at each sampling time. Perchlorate and chloride were extracted from 10 g dry equivalent soil by shaking with 20 ml of deionized water for 6 h. A preliminary experiment demonstrated that more than 90% of perchlorate was recovered by this extraction method. Nitrate was extracted by shaking with 20 ml of 0.5 N potassium sulfate for 0.5 h (3). The extracts were centrifuged and filtered to remove soil particles. Perchlorate concentrations in the extracts were measured by a perchlorate ion-selective electrode (Thermo Orion, Beverly, Mass.). Chloride concentrations were measured by a digital chloridometer. Nitrate concentrations in the extracts were analyzed colorimetrically (3). Ion chromatography (Dionex, Sunnyvale, Calif.) also was used to measure lower concentrations of perchlorate with an IonPac AS16 column, and chloride, chlorite, chlorate, nitrate, and nitrite were measured with an IonPac AS14 column.
Kinetic analysis of perchlorate degradation.
Perchlorate biodegradation kinetic parameters were determined by assuming no-growth Monod-type biodegradation (the same form as that of the Michaelis-Menten equation) limited only by perchlorate concentration with the equation dC/dt = −νm C/(Ks + C), where t is the time (days), νm is the maximum perchlorate degradation rate (mmol · kg dry soil−1 · day−1), C is the perchlorate concentration (mmol · kg dry soil−1), and Ks is the saturation constant (mmol · kg dry soil−1) (6). Lag periods were determined by the comparison of mean degradation rates between adjacent time points. The period of active degradation was assumed to begin when the difference in degradation between two time points was at a maximum. The reaction times were recalculated by subtracting the lag periods, and the kinetic parameters of perchlorate degradation (νm and Ks) were calculated with an integral form of the Monod equation, t = [Ks log (C0/C) + C0 − C]/νm, where C0 is the perchlorate concentration at t = 0 (the end of lag period), by using the nonlinear curve-fitting function in KaleidaGraph 3.6 (Synergy Software, Reading, Pa.). Pseudo-first-order rate constants (νm/Ks, where C ≪ Ks) also were calculated.
Enrichment culture and growth medium.
To further examine changes in microbial community compositions following exposure to perchlorate, an enrichment culture experiment was conducted. The soil was inoculated with a mineral liquid medium (0.08 g of KH2PO4, 0.11 g of K2HPO4, 0.42 g of NaHCO3, 0.36 g of NH4Cl, 0.15 g of MgSO4 · 7H2O, 0.1g of CaCl2 · 2H2O, 0.003 g of FeCl3 · 6H2O, 2.86 mg of H3BO3, 1.81 mg of MnCl2, 0.22 mg of ZnSO4, 0.08 mg of CuSO4, 0.06 mg of CoCl2, and 25 μg of Na2MoO4 in 1 liter with pH adjusted to 7.2 to 7.4 with phosphoric acid) containing either acetate or bicarbonate (10 mM each) and four different perchlorate concentrations (0, 0.25, 1.0, and 2.5 mM). The ratio of soil to media was 1:4 by weight. Two replicates per perchlorate concentration were set up in 50-ml serum bottles. Acetate-amended cultures were filled without leaving headspace. For hydrogen enrichment, the bottles were filled to half capacity with bicarbonate-amended cultures and the headspaces were filled with 0.2-μm-filtered hydrogen gas. The cultures were incubated at room temperature (23 ± 1°C) for 4 to 5 months. Perchlorate concentration was monitored, and the enrichments were subcultured by transferring a one-fifth volume of the culture to a fresh medium when perchlorate was degraded by more than 90%.
Soil DNA extraction.
Unexposed soils and microcosm soils incubated with or without perchlorate exposure were collected for DNA extraction and stored at −20°C. Soil enriched with perchlorate-containing media was centrifuged and collected for DNA extraction. Soil DNA was extracted from 0.5-g samples using a FastDNA SPIN kit for soil (Qbiogene, Carlsbad, Calif.) as described by the manufacturer. Extracted DNA was dissolved in 75 μl of nuclease-free water, and the quality of the extracted DNA was analyzed by electrophoresis on a 1.5% agarose gel.
DGGE analysis.
Bacterial community DNA extracted from soil was amplified by PCR with the primer pair 357F with a G-C clamp and 534R (24). Denaturing gel gradient electrophoresis (DGGE) of bacterial partial 16S rRNA gene PCR products was performed by using 8% acrylamide gel with a 30-to-60% denaturing gradient, where 100% denaturant was defined as 7 M urea and 40% formamide. After PCR products were loaded, the gel was run in 0.5× Tris-acetate-EDTA (TAE) buffer in a D-Code system (Bio-Rad, Hercules, Calif.) for 3.25 h at 150V and 0.75 h at 200V. The gel was stained with SYBR green solution for 30 min and observed on a UV transilluminator. The cluster analysis of microcosm DGGE profiles was performed by GelcomparII (Applied Maths, Kortrijk, Belgium) using the Pearson product-moment correlation and the unweighted pair-group method using arithmetic averages.
Cloning and sequence analysis.
Excised DGGE bands were crushed and soaked in 450 μl of Tris-EDTA overnight to elute PCR products. Diluted solutions were used to reamplify bacterial partial 16S rRNA gene with the same primer set as described above without the G-C clamp. The PCR products were cloned into pCR 2.1-TOPO vector and transformed to Escherichia coli by using a TOPO TA cloning kit (Invitrogen, Carlsbad, Calif.). Positive clones were selected randomly and screened by the lengths of the PCR products with M13 primers located in the vector. Plasmids were extracted from clones by using a Plasmid Mini Kit (QIAGEN, Valencia, Calif.), and the insertion was sequenced with an ABI Prism automatic sequencer (Applied Biosystems, Foster City, Calif.) at Davis Sequencing Inc., Davis, Calif. Sequence identification was performed by using the BLAST program (2), and the partial 16S rRNA gene sequences (168 to 194 bp) of the DGGE band clones, the closest relatives, and perchlorate-reducing bacteria were aligned by CLUSTAL W (30). A phylogenetic tree was constructed by using the neighbor-joining method (28) and visualized with NJplot (25).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the GenBank database under accession numbers AY905570 to AY905580.
RESULTS
Perchlorate reduction in unsaturated soil microcosms.
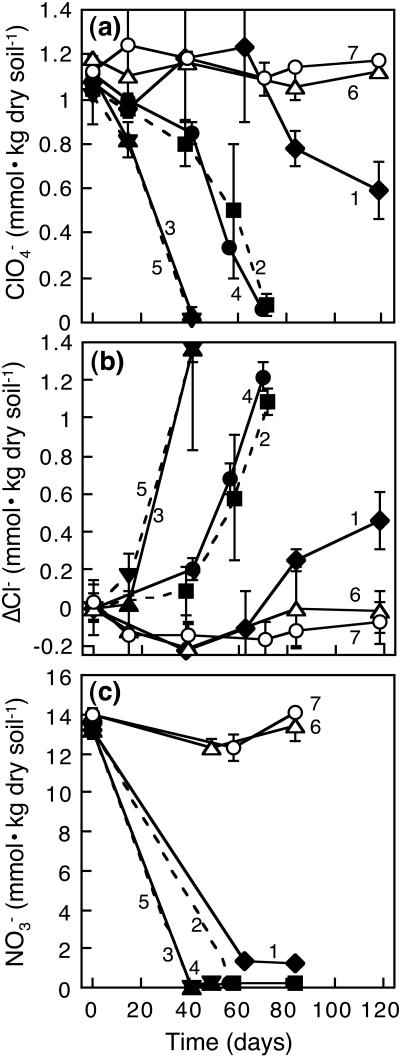
Perchlorate was degraded by native microbial communities in microcosms containing soil not known to be previously exposed to anthropogenic perchlorate, whereas no degradation was observed in sterile controls. Addition of electron donors such as acetate and hydrogen enhanced perchlorate degradation (Fig. 1a). A considerably long lag period (63 days) was observed in microcosms without adding any electron donor or carbon source; only about 44% of perchlorate was degraded after 120 days in these microcosms. Addition of acetate enhanced perchlorate degradation the most, regardless of the presence or absence of hydrogen. When hydrogen was added as an electron donor, longer lag periods (41 days) were observed without organic carbon addition than when acetate was supplied (14 days). Addition of inorganic carbon did not make a substantial difference under a hydrogen headspace, although greater variability of replicates was observed for perchlorate degradation among microcosms without bicarbonate. Increases in the concentration of chloride during microcosm incubation indicated perchlorate was reduced to chloride by soil microorganisms (Fig. 1b). Mean conversion ratios to chloride were in the range of 95 to 135% at the end of the experiment. Chlorate (ClO3−) was only transitionally detected and at very low concentrations (<0.5 mmol · kg dry soil−1); chlorite (ClO2−) was not detected (data not shown). Nitrate, naturally occurring in the soil, was mostly transformed by or prior to the end of perchlorate reduction in active microcosms (Fig. 1c). Nitrite (NO2−) remained at low concentrations throughout the course of the experiment (<0.1 mmol · kg dry soil−1; data not shown).
FIG. 1.
Effect of acetate and hydrogen amendments on perchlorate reduction in unsaturated soil microcosms. (a) Perchlorate degradation, (b) chloride formation, and (c) nitrate transformation. Numbers in the figure refer to the assigned set numbers in Table 1. Data are means ± standard deviations (n = 3).
Effect of nitrate on perchlorate reduction.
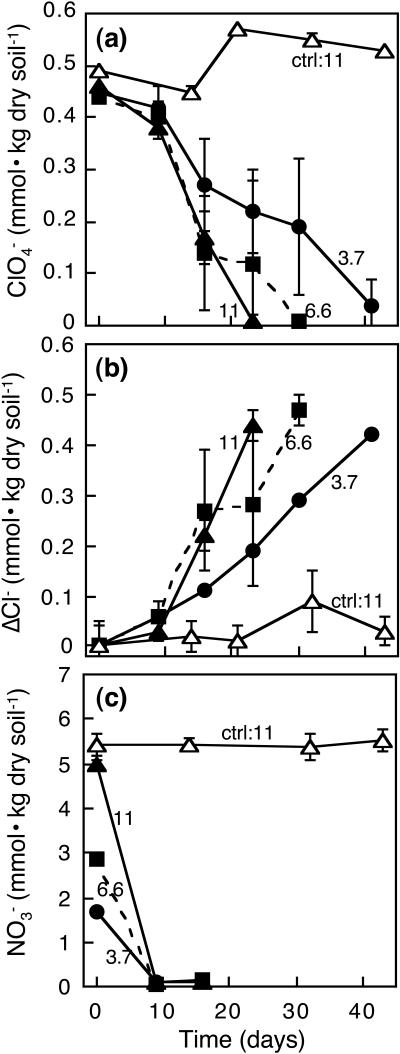
Based on the results above and a previous microcosm study using the same soil (32), we hypothesized that the lag periods observed for perchlorate reduction were due to the presence of nitrate and that the elimination of nitrate would shorten the lag periods and enhance perchlorate reduction rates. Therefore, a separate microcosm experiment with perchlorate and three different nitrate concentrations was conducted to test this hypothesis, with acetate as an additional electron donor. Nitrate was depleted in 9 days in all active microcosms with different nitrate concentrations, whereas little perchlorate degradation was observed during this period (Fig. 2a and c). Differences in perchlorate degradation lag periods among different nitrate amendments were not observed in this experiment, although the perchlorate degradation rates were slightly higher with higher nitrate concentrations. After the initial 9-day lag period, substantial perchlorate degradation occurred in all active microcosms. However, degradation occurred most rapidly in the high-nitrate-concentration treatment (NO3−/ClO4− ratio of 11), with over 95% reduced in 23 days. In contrast, 30 days and more than 41 days were needed for over 95% degradation in the microcosms with lower nitrate concentrations (NO3−/ClO4− ratios of 6.6 and 3.7, respectively), and high variability was observed among triplicates during perchlorate degradation. Chloride was produced as perchlorate was degraded, and the mean conversion ratios to chloride were 98 to 110% at the end points (Fig. 2b).
FIG. 2.
Effect of nitrate on perchlorate reduction in acetate-amended soil microcosms. (a) Perchlorate degradation, (b) chloride formation, and (c) nitrate transformation. Numbers in the figure refer to molar ratios of NO3− to ClO4−. “ctrl” refers to sterile control. Data are means ± standard deviations (n = 3).
Kinetic parameters of perchlorate reduction.
Lag periods and kinetic parameters of perchlorate degradation were determined from the data obtained from the microcosms with hydrogen and bicarbonate and from the microcosms with acetate and three different nitrate concentrations (Table 2). The higher νm and νm/Ks values were obtained as initial nitrate concentrations increased in the acetate-amended microcosms.
TABLE 2.
Experimental conditions and kinetic constants of perchlorate reduction in laboratory microcosm studies
| Soil | Microcosm experimental condition
|
Kinetic parameter for perchlorate degradationb
|
Source or reference | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Water | Additive | Initial ClO4− (mmol · kg dry soil−1) | Initial NO3− (mmol · kg dry soil−1) | Initial NO3−/ ClO4− ratio | Lag period (days) | νm (mmol · kg dry soil−1 · day−1) | Ks (mmol · kg dry soil−1) | νm/Ks or first-order rate constants (day −1) | ||
| Yolo silt loam | Unsaturated | H2/HCO3− | 1.11 ± 0.02 | 13.2 ± 0.5 | 12 | 41 | 4.8 | 24 | 0.20 | This study |
| (moisture, | Acetate | 0.453 ± 0.009 | 1.69 ± 0.09 | 3.7 | 9 | 0.016 | 0.066 | 0.25 | ||
| 20-23%) | 0.434 ± 0.011 | 2.86 ± 0.07 | 6.6 | 9 | 0.033 | 0.073 | 0.46 | |||
| 0.463 ± 0.017 | 4.98 ± 0.09 | 11 | 9 | 0.035 | 0.035 | 1.0 | ||||
| Longhorn Army | Saturated | None | 0.050 | 0.45 | 9 | 35.7 ± 4.0a | n/a | n/a | 0.16 ± 0.08 | 29 |
| Ammunition | (soil:water, | 0.56 | 11 | 35.7 ± 4.0a | n/a | n/a | 0.14 ± 0.02 | |||
| Plant site | 1:3) | 1.22 | 24 | >60.0a | n/a | n/a | n/a | |||
Lag periods were determined as those where ≥5% of perchlorate was lost.
n/a, not available.
DGGE profiles of soil microcosms with or without perchlorate.
Changes in bacterial community composition were analyzed by comparing DGGE profiles among the original soil and the microcosm soils at the end points of perchlorate degradation (data not shown). DGGE banding indicated a high level of diversity in bacterial communities before and after incubation. Shifts in communities were observed regardless of perchlorate exposure, and few unique bands were observed in the presence of perchlorate. Cluster analysis showed that the perchlorate-exposed community with acetate added as a sole electron donor was the most dissimilar to the community in the original soil; nevertheless, overall, there was little variation in community composition following treatments (>80% similarity).
Identification of bacteria involved in perchlorate reduction from enriched soil cultures.
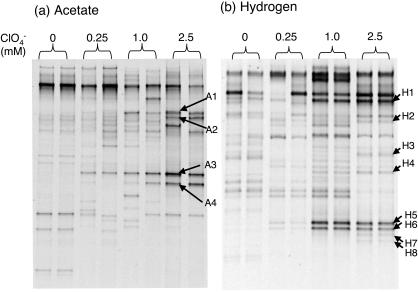
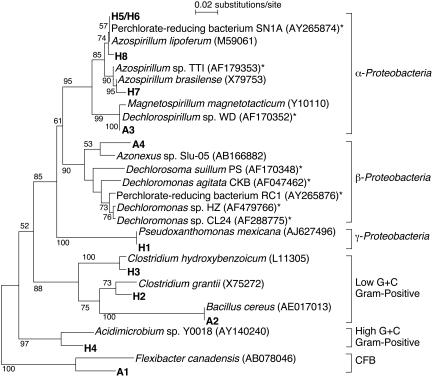
Since few differences were observed in microcosm community compositions after perchlorate exposure, further enrichment of perchlorate degraders in the soil was conducted. DGGE patterns were compared among soils enriched with different concentrations of perchlorate and acetate (Fig. 3a). Four bands with stronger intensities in soil enriched with 2.5 mM perchlorate were selected for sequencing: some of the bands were also observed in soil enriched with 0.25 and 1.0 mM perchlorate and none of the four bands was observed in soil enriched with 0 mM perchlorate (Fig. 3a). Similarly, eight bands observed in communities after enrichment with hydrogen and higher perchlorate concentrations were selected for sequencing (Fig. 3b). DGGE patterns of enrichment cultures with 2.5 mM perchlorate and hydrogen were similar to DGGE patterns of hydrogen-amended soil microcosms. In contrast, the DGGE patterns of the acetate enrichments showed few similarities to the microcosm DGGE patterns (data not shown). The phylogenetic analysis indicated that several sequences from the enrichments were identical to 16S rRNA gene sequences of previously reported perchlorate-reducing bacteria: Dechlorospirillum sp. with acetate and Azospirillum spp. with hydrogen (Fig. 4). There were no sequences in common between the enrichments with acetate or hydrogen.
FIG. 3.
Comparison of DGGE patterns of PCR-amplified bacterial 16S rRNA gene fragments in soil enriched with perchlorate and either (a) acetate or (b) hydrogen. The bands A1 to A4 and H1 to H8 were excised and cloned for sequencing.
FIG. 4.
Phylogenetic relationships of bacterial 16S rRNA gene fragments obtained from soil enriched with perchlorate and either acetate or hydrogen. The GenBank accession numbers of reference sequences are shown in parentheses. Asterisks indicate known perchlorate-reducing bacteria. Bootstrap values greater than 50 based on a total of 100 replicate resamplings are shown at the nodes.
DISCUSSION
Potential for perchlorate biodegradation and the effect of electron donor addition.
Bioremediation of perchlorate by native microbial communities may be feasible when enhanced by adding electron donors in the vadose and especially surface soils, where large and active populations of microorganisms can be maintained. Enhancement of perchlorate reduction by adding acetate or lactate was also observed in Yolo loam vadose soil as well as in Columbia loam surface soil (32) and in surface soil taken from a public park in Pennsylvania with no known prior perchlorate exposure (38). Shorter lag periods for perchlorate degradation with acetate than with hydrogen/bicarbonate imply more heterotrophic than autotrophic perchlorate-reducing populations in the soil.
Effect of nitrate on perchlorate biodegradation.
Although perchlorate reduction was originally thought to be catalyzed by nitrate reductase (17), recent studies using perchlorate-reducing bacteria Dechloromonas sp. strain KJ (39) and Dechlorosoma suillum strains PS (11) and perc1ace (15) have suggested the presence of two distinct inducible enzymatic systems for perchlorate and nitrate reduction. Nitrate-grown strain KJ showed much lower levels of perchlorate than of nitrate reductase activity, and perchlorate reduction was delayed after exposure to nitrate (39). In strain PS, the expression of chlorite dismutase, one of the enzymes for perchlorate reduction, was suppressed by nitrate (11). Unlike strains KJ and PS, nitrate-grown strain perc1ace showed both nitrate and perchlorate reductase activities, although the level of perchlorate reductase activity was slightly lower on nitrate than on perchlorate (15). At the community level, Tipton et al. (32) reported a preferential reduction of nitrate over perchlorate in soil without prior exposure to perchlorate, similar to what was observed in our study. The preferential reduction of nitrate over perchlorate in their experiments and ours was probably due to the prior induction of nitrate-reducing enzymes: induction of perchlorate-reduction enzymes may have occurred after adding perchlorate. Tipton et al. (32) also demonstrated simultaneous degradations of perchlorate and nitrate by spiking them subsequently, suggesting that the soil microbial community can reduce both electron acceptors simultaneously once the appropriate enzymatic systems are induced.
The removal of nitrate from soil did not shorten the perchlorate lag period or enhance the rate of perchlorate removal as we expected. The observed enhancement of perchlorate reduction by nitrate may be explained by increases in the microbial populations capable of using both nitrate and perchlorate as electron acceptors during nitrate reduction. Many perchlorate-reducing bacteria are also capable of growing using nitrate as an electron acceptor (12, 18, 21, 26, 36, 40). Therefore, native perchlorate-reducing populations may have been able to grow on the added nitrate, resulting in higher rates of perchlorate reduction. Stoichiometrically, 5/3 mol of acetate is required to reduce 1 mol of nitrate completely to N2 (denitrification), whereas 1 mol of acetate is needed for the reduction of 1 mol of perchlorate (26). Assuming the same growth yields as those of strain GR-1 (11.9 ± 0.4 and 14.4 ± 2.2 g dry weight · mol acetate−1 with nitrate and perchlorate, respectively) (26), increases in microbial biomass were calculated in the range of 34 to 99 mg dry weight · kg dry soil−1 by nitrate reduction, in contrast to approximately 6.5 mg dry weight · kg dry soil−1 by perchlorate reduction in the second experiment. Thus, it is possible that the density of perchlorate-reducing populations is higher in the high-nitrate-concentration soils than what could be supported in the low-nitrate-concentration soils.
Kinetic parameters of perchlorate reduction.
A Monod-type biodegradation model, used in this study, can be applied to a wider range of the concentrations than the commonly used first-order model, which is applicable when the initial concentration of the limiting substrate is much lower than the Ks (6). The initial perchlorate concentrations in this study were much higher than the calculated Ks, confirming that a first-order approximation was not suitable. We used a no-growth, rather than a growth-based, form of the equation because there was not sufficient data to describe the initial growth-limited portion as well as no direct measurement of the parameters required to describe growth, i.e., population density and growth yield. Though this decision prevented investigation of how microbial population growth interacted with perchlorate degradation kinetics, our approach did permit comparisons of perchlorate reduction rates both within our study and with results from other published studies.
Comparison of kinetic parameters among microcosms with different initial nitrate concentrations indicated that the elimination of nitrate negatively affected rates of, but not lag periods for, perchlorate reduction. In contrast, Tan et al. (29) observed a longer lag period in soil samples from an ammunition plant, where solid-propellant rocket motors were manufactured, after adding 0.81 mM NO3− (the cumulative concentration was 1.22 mM) (Table 2). Although differences between experimental designs may explain the observed differences, another possible explanation is that the ammunition soil contained a large perchlorate-reducing population enriched by prior perchlorate exposure (concentration not shown) (29) and that nitrate addition did not further increase the population density.
Changes in microbial communities and detection of bacteria involved in perchlorate reduction.
Perchlorate enrichment cultures revealed sequences of organisms known to reduce perchlorate. In acetate enrichments, band A3 was 100% identical to the sequence of Dechlorospirillum sp. strain WD, isolated from a swine lagoon with acetate (12). Band A4 was closely related to the sequences of Dechloromonas/Dechlorosoma, two β-Proteobacteria, the group to which many perchlorate-reducing bacteria that have been isolated from wastewater treatment systems and environmental samples belong (1, 8, 12, 21, 37, 40), although its closest relative was the sequence of an Azonexus sp. (95%). The organism whose sequence was most closely related to the sequence of band A2, Bacillus cereus, has also been reported to be a perchlorate reducer (17).
In enrichments with hydrogen, bands H5 and H6 could not be separated in the process of cloning. The PCR product of each band clone resulted in two bands in the same position as H5 and H6 on DGGE gel. However, sequencing of the clones indicated the presence of just one sequence. All previously isolated perchlorate-reducing Azospirillum sp. strains use acetate as an electron donor (12, 37); the use of hydrogen for perchlorate reduction has not been tested. Bacteria belonging to the genus Azospirillum are capable of oxidizing hydrogen microaerobically or anaerobically under denitrifying conditions (7, 31). The presence of the sequences closely related to that of an Azospirillum sp. (bands H5/H6, H7, and H8) in the perchlorate-reducing hydrogen enrichment may imply that members of this genus can directly couple perchlorate reduction with hydrogen oxidation; however, this requires further study to confirm. So far, the only previously isolated and investigated bacterium capable of reducing perchlorate using H2/CO2 is a Dechloromonas sp. (40). We did not obtain any band sequence similar to those of members of β-Proteobacteria, including Dechloromonas, from the hydrogen enrichment.
The DGGE analysis of hydrogen enrichments also indicated the presence of Clostridium spp. (band H2 and H3), bacteria known to produce acetate from hydrogen and CO2 (13). Acetate produced by Clostridium spp. could support acetate-utilizing perchlorate-reducing organisms in the hydrogen enrichment community. On the other hand, bands indicative of the presence of Clostridium spp. were not observed in the enrichment without perchlorate despite the presence of hydrogen and bicarbonate. Furthermore, members of the genus Clostridium are capable of reducing nitrate (10), and their acetogenic ability is strongly inhibited by the presence of nitrate (14). Therefore, it is possible that Clostridium spp. were directly involved in perchlorate reduction.
The majority of previously isolated perchlorate-reducing bacteria belong to β-Proteobacteria, but Waller et al. (37) have recently found nearly half of isolates from 12 perchlorate-contaminated aquifers were closely related to Azospirillum spp. (α-Proteobacteria). In both our acetate and hydrogen enrichments, we similarly detected sequences identical to those of known perchlorate reducers in α-Proteobacteria.
Acknowledgments
The project described was supported by grant number 5 P42 ES04699-16 from the National Institute of Environmental Health Sciences (NIEHS), NIH, with funding provided by EPA, and by the Showa Shell Sekiyu Foundation for the Promotion of Environmental Research.
Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS, NIH, or EPA.
We thank K. R. Hristova for helpful suggestions on the DGGE analysis and R. E. Drenovsky and K. P. Feris for critical reading of the manuscript. The comments of three anonymous reviewers significantly improved the manuscript.
REFERENCES
- 1.Achenbach, L. A., U. Michaelidou, R. A. Bruce, J. Fryman, and J. D. Coates. 2001. Dechloromonas agitata gen. nov., sp. nov. and Dechlorosoma suillum gen. nov., sp. nov., two novel environmentally dominant (per)chlorate-reducing bacteria and their phylogenetic position. Int. J. Syst. Evol. Microbiol. 51:527-533. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, J. M., and J. S. I. Ingram (ed.). 1993. Tropical soil biology and fertility: a handbook of methods, 2nd ed., p. 74-75. Oxford University Press, Oxford, United Kingdom.
- 4.Anonymous. 2002. Low-level perchlorate exposures. Environ. Sci. Technol. 36:125A-126A.11831214 [Google Scholar]
- 5.Attaway, H., and M. Smith. 1993. Reduction of perchlorate by an anaerobic enrichment culture. J. Ind. Microbiol. 12:408-412. [Google Scholar]
- 6.Bekins, B. A., E. Warren, and E. M. Godsy. 1998. A comparison of zero-order, first-order, and Monod biotransformation models. Ground Water 36:261-268. [Google Scholar]
- 7.Bowien, B., and H. G. Schlegel. 1981. Physiology and biochemistry of aerobic hydrogen-oxidizing bacteria. Annu. Rev. Microbiol. 35:405-452. [DOI] [PubMed] [Google Scholar]
- 8.Bruce, R. A., L. A. Achenbach, and J. D. Coates. 1999. Reduction of (per)chlorate by a novel organism isolated from paper mill waste. Environ. Microbiol. 1:319-329. [DOI] [PubMed] [Google Scholar]
- 9.California Department of Health Services. 2004. Perchlorate in California drinking water: monitoring update. [Online.] http://www.dhs.ca.gov/ps/ddwem/chemicals/perchl/monitoringupdate.htm.
- 10.Caskey, W. H., and J. M. Tiedje. 1979. Evidence for clostridia as agents of dissimilatory reduction of nitrate to ammonium in soils. Soil Sci. Soc. Am. J. 43:931-936. [Google Scholar]
- 11.Chaudhuri, S. K., S. M. O'Connor, R. L. Gustavson, L. A. Achenbach, and J. D. Coates. 2002. Environmental factors that control microbial perchlorate reduction. Appl. Environ. Microbiol. 68:4425-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coates, J. D., U. Michaelidou, R. A. Bruce, S. M. O'Connor, J. N. Crespi, and L. A. Achenbach. 1999. Ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 65:5234-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drake, H. L. 1994. Acetogenesis, acetogenic bacteria, and the acetyl-Co A “Wood/Ljungdahl” pathway: past and current perspectives, p. 3-60. In H. L. Drake (ed.), Acetogenesis. Chapman & Hall, New York, N.Y.
- 14.Frostl, J. M., C. Seifritz, and H. L. Drake. 1996. Effect of nitrate on the autotrophic metabolism of the acetogens Clostridium thermoautotrophicum and Clostridium thermoaceticum. J. Bacteriol. 178:4597-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giblin, T., and W. T. Frankenberger. 2001. Perchlorate and nitrate reductase activity in the perchlorate-respiring bacterium perclace. Microbiol. Res. 156:311-315. [DOI] [PubMed] [Google Scholar]
- 16.Giblin, T. L., D. C. Herman, and W. T. Frankenberger. 2000. Removal of perchlorate from ground water by hydrogen-utilizing bacteria. J. Environ. Qual. 29:1057-1062. [Google Scholar]
- 17.Hackenthal, E., W. Mannheim, R. Hackenthal, and R. Becher. 1964. Die Reduktion von Perchlorat durch Bakterien. I. Untersuchungen an intakten Zellen. Biochem. Pharmacol. 13:195-206. [DOI] [PubMed] [Google Scholar]
- 18.Herman, D. C., and W. T. Frankenberger. 1999. Bacterial reduction of perchlorate and nitrate in water. J. Environ. Qual. 28:1018-1024. [Google Scholar]
- 19.Kirk, A. B., E. E. Smith, K. Tian, T. A. Anderson, and P. K. Dasgupta. 2003. Perchlorate in milk. Environ. Sci. Technol. 37:4979-4981. [DOI] [PubMed] [Google Scholar]
- 20.Logan, B. E. 2001. Assessing the outlook for perchlorate remediation. Environ. Sci. Technol. 35:482A-487A. [DOI] [PubMed] [Google Scholar]
- 21.Logan, B. E., H. S. Zhang, P. Mulvaney, M. G. Milner, I. M. Head, and R. F. Unz. 2001. Kinetics of perchlorate- and chlorate-respiring bacteria. Appl. Environ. Microbiol. 67:2499-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, J. P., and B. E. Logan. 2000. Sustained perchlorate degradation in an autotrophic, gas-phase, packed-bed bioreactor. Environ. Sci. Technol. 34:3018-3022. [Google Scholar]
- 23.Motzer, W. E. 2001. Perchlorate: problems, detection, and solutions. Environ. Forensics 2:301-311. [Google Scholar]
- 24.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 25.Perrière, G., and M. Gouy. 1996. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 26.Rikken, G. B., A. G. M. Kroon, and C. G. Van Ginkel. 1996. Transformation of (per)chlorate into chloride by a newly isolated bacterium: reduction and dismutation. Appl. Microbiol. Biotechnol. 45:420-426. [Google Scholar]
- 27.Romanenko, V. I., V. N. Korenkov, and S. I. Kuznetsov. 1976. Bacterial decomposition of ammonium perchlorate. Mikrobiologiya 45:204-209. [PubMed] [Google Scholar]
- 28.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 29.Tan, K., T. A. Anderson, and W. A. Jackson. 2004. Degradation kinetics of perchlorate in sediments and soils. Water Air Soil Pollut. 151:245-259. [Google Scholar]
- 30.Thompson, J., D. Higgins, and T. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tibelius, K. H., and R. Knowles. 1984. Uptake hydrogenase activity in denitrifying Azospirillum brasilense grown anaerobically with nitrous oxide or nitrate. J. Bacteriol. 157:84-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tipton, D. K., D. E. Rolston, and K. M. Scow. 2003. Transport and biodegradation of perchlorate in soils. J. Environ. Qual. 32:40-46. [DOI] [PubMed] [Google Scholar]
- 33.Urbansky, E. T. 1998. Perchlorate chemistry: implications for analysis and remediation. Bioremediat. J. 2:81-95. [Google Scholar]
- 34.U.S. Environmental Protection Agency. 2004. Known perchlorate releases in the U.S.—March 23, 2004. [Online.] http://www.epa.gov/swerffrr/pdf/perchlorate_releases.pdf.
- 35.U.S. Environmental Protection Agency. 1999. Region 9 perchlorate update. [Online.] http://yosemite.epa.gov/r9/sfund/fsheet.nsf/0/aa09570d64878/ba882568e300594caf/$FILE/Perch-99.pdf.
- 36.Wallace, W., T. Ward, A. Breen, and H. Attaway. 1996. Identification of an anaerobic bacterium which reduces perchlorate and chlorate as Wolinella succinogenes. J. Ind. Microbiol. 16:68-72. [Google Scholar]
- 37.Waller, A. S., E. E. Cox, and E. A. Edwards. 2004. Perchlorate-reducing microorganisms isolated from contaminated sites. Environ. Microbiol. 6:517-527. [DOI] [PubMed] [Google Scholar]
- 38.Wu, J., R. F. Unz, H. Zhang, and B. E. Logan. 2001. Persistence of perchlorate and the relative numbers of perchlorate- and chlorate-respiring microorganisms in natural waters, soils, and wastewater. Bioremediat. J. 5:119-130. [Google Scholar]
- 39.Xu, J. L., J. J. Trimble, L. Steinberg, and B. E. Logan. 2004. Chlorate and nitrate reduction pathways are separately induced in the perchlorate-respiring bacterium Dechlorosoma sp. KJ and the chlorate-respiring bacterium Pseudomonas sp. PDA. Water Res. 38:673-680. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, H. S., M. A. Bruns, and B. E. Logan. 2002. Perchlorate reduction by a novel chemolithoautotrophic, hydrogen-oxidizing bacterium. Environ. Microbiol. 4:570-576. [DOI] [PubMed] [Google Scholar]