Abstract
The methyl parathion hydrolase (MPH)-encoding gene mpd was placed under the control of the P43 promoter and Bacillus subtilis nprB signal peptide-encoding sequence. High-level expression and secretion of mature, authentic, and stable MPH were achieved using the protease-deficient strain B. subtilis WB800 as the host.
Organophosphate compounds are used extensively in agricultural and domestic pest control and as chemical warfare agents; these compounds inhibit the acetylcholinesterase of animals (8). Mass application of these compounds in the environment causes serious problems. Organophosphate hydrolase (OPH) isolated from soil microorganisms detoxified organophosphates effectively (14, 18), and bioremediation of organophosphate compounds in the environment by using bacterial enzymes may provide an efficient, convenient, and economical method for detoxification. We previously isolated Plesiomonas sp. strain M6, which is capable of hydrolyzing methyl parathion at high efficiency (5), and the gene (mpd) encoding a novel methyl parathion hydrolase (MPH) was cloned (5) and expressed in Escherichia coli (9). However, the previous work showed that E. coli could process only a small proportion of the inactive precursor polypeptide, comprising the signal peptide and the mature enzyme, to produce the active MPH (9).
Most microorganisms that produce OPH are gram-negative bacteria, and the OPH is located intracellularly or secreted into the periplasm. The outer membrane acts as a permeability barrier and limits interaction between the pesticides and OPH residing within the cells (12). This bottleneck reduces the application efficacy. Bacillus subtilis can serve as an efficient and safe host for recombinant protein secretion (3, 4, 7, 16, 17). Secreted proteins usually remain in biologically active forms (13, 15), and downstream purification is greatly simplified. In this work, the mpd gene was fused with the nprB signal peptide-encoding sequence, and the organophosphate hydrolase was expressed and secreted in B. subtilis under the control of the P43 promoter (20).
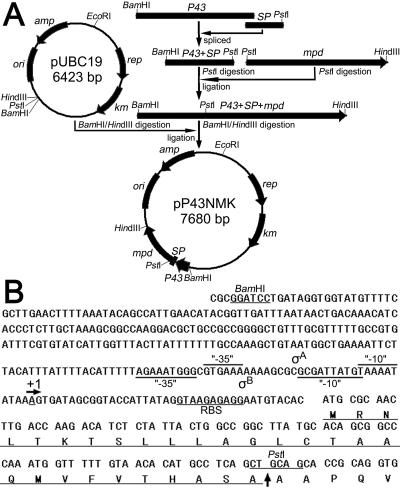
The methyl parathion hydrolase-encoding gene mpd (GenBank accession no. AF338729) cannot be expressed in B. subtilis by using its own promoter. To achieve expression and secretion of MPH in B. subtilis, the secretion signal peptide-encoding sequence of the nprB gene was fused with the mature mpd gene. A typical cleavage site (ASA-A) (19) for signal peptidase I was designed for the release from B. subtilis of MPH with an authentic N terminus (Fig. 1B). The mature mpd gene sequence (9) was cloned using the primer pair P1 (5′-GCGCTGCAGCACCGCAGGTG-3′) and P2 (5′-CGCAAGCTTTCATCATCACTTGGGGTTGACGACCGA-3′) from plasmid pMT1 (5). To introduce the PstI site (underlined) upstream, the codon GCC encoding the first amino acid of mature MPH was modified to GCA (Fig. 1B). Two additional stop codons (TGATGA) and the HindIII site (underlined) were introduced with primer P2. Primers P3 (5′-CGCGGATCCTGATAGGTGGTATGTTTTCGC-3′) and P4 (5′-CTTGGTCAAGTTGCGCATGTGTACATTCCTCTCTT-3′) were used to clone the P43 promoter sequence from B. subtilis 168 (2) chromosomal DNA, and the BamHI site (underlined) was introduced in the forward primer P3. Primers P5 (5′-AAGAGAGGAATGTACACATGCGCAACTTGACCAAG-3′) and P6 (5′-GCGCTGCAGCTGAGGCATG-3′) were used to clone the signal peptide-encoding sequence of the nprB gene from B. subtilis 168 chromosomal DNA, and the PstI site (underlined) was introduced in the reverse primer P6. The initial codon (TTG) of the nprB signal peptide-encoding sequence was modified to ATG in primer P5. The two PCR fragments of the P43 promoter and the nprB signal peptide-encoding sequence were spliced by overlap extension (11). The spliced fragment and the mature mpd gene PCR fragment were then digested with PstI and ligated together. The ligation product was purified and digested with BamHI/HindIII. The mpd gene with the P43 promoter and nprB signal peptide-encoding sequence was cloned into pUBC19, and the recombinant plasmid was designated pP43NMK (Fig. 1A).
FIG. 1.
(A) Construction of pP43NMK, a B. subtilis expression vector for the production and secretion of MPH. See the text for a detailed description of the construction of pP43NMK. P43 and SP represent the P43 promoter and the nprB signal peptide-encoding sequence from B. subtilis, respectively. ori, amp, rep, and km represent the sequences coding for the ColE1 replication origin, ampicillin resistance marker, replicase, and kanamycin resistance marker, respectively. The arrows show the transcription directions for these genes. (B) Nucleotide sequences of the P43 promoter fragment, nprB signal peptide-encoding sequence, and part of the mpd gene. The deduced amino acid sequence of the neutral protease B signal peptide is underlined. The “−10” and “−35” regions of the promoters (recognized by σA and σB factors), the transcription start site (“+1”), and the ribosome binding site (RBS) are indicated. The vertical arrow indicates the predicted cleavage site for the signal peptidase I.
Competent cells of B. subtilis WB800 (21) were transformed with pP43NMK. Transformants able to hydrolyze methyl parathion were screened on super-rich medium (10) agar plates containing 10 μg/ml kanamycin and 100 μg/ml methyl parathion, where they form yellow halos of p-nitrophenol around the colonies. A preculture of WB800(pP43NMK) was grown overnight at 37°C in super-rich medium with kanamycin (10 μg/ml), and 1% (vol/vol) of this preculture was used to inoculate 500 ml of the same culture medium in a 1-liter flask. The culture was maintained at 37°C, and 20-μl aliquots were taken over time and assayed for MPH according to the method of Cui et al. (6).
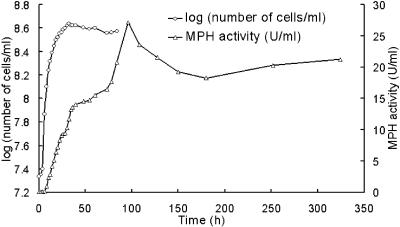
In previous studies, MPH was expressed and secreted in B. subtilis 168, but because the host produced numerous proteases, MPH was expressed only at low levels (0.2565 U/ml after 12 h, decreasing to 0.0967 U/ml after 24 h). B. subtilis WB800 is deficient in eight proteases and therefore can serve as an excellent host for the expression of foreign proteins. In this work, recombinant MPH was secreted into the culture medium via the nprB signal peptide with some apparent improvement in enzyme stability. Time courses for bacterial growth and secreted MPH activity are shown in Fig. 2. The P43 promoter is a well-characterized overlapping promoter (Fig. 1B) that is functional during both the exponential and stationary growth phases (20). The results show that, under the control of the P43 promoter, the mpd gene was continuously expressed throughout the exponential growth phase and into the late stationary phase. MPH activity in the culture medium accumulated to a maximum level after 96 h of cultivation, declined to approximately 20 U/ml at 180 h, and then remained at a relatively stable level for a further 150 h. A genome-based survey of the secretome of B. subtilis has shown that a total of at least 27 proteases are present in the membrane, cell wall, and culture medium (19). The expression host used in this study is deficient in eight proteases. However, a certain degree of MPH degradation was observed, indicating that the enzyme may be sensitive to an as-yet-uncharacterized protease produced by the host during the late stationary phase or to intracellular proteases released during cell lysis.
FIG. 2.
MPH activity in B. subtilis WB800(pP43NMK) cultures during the exponential and stationary growth phases.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and zymogram analysis of the MPH were performed as described previously (9). Recombinant MPH secreted by B. subtilis WB800(pP43NMK) was evident as a distinct band at a position equivalent to a molecular mass of approximately 34 kDa (Fig. 3), confirming that the recombinant MPH was identical in size to the MPH from Plesiomonas strain M6. The biological activity of the recombinant MPH band was confirmed by zymogram analysis as a single band with a yellow halo (Fig. 3, lane 7). Using bovine serum albumin as the standard, the amount of MPH in the SDS-polyacrylamide gel was estimated with the densitometry analysis software BandScan (Glyko). This indicated, on extrapolation, that 53 mg/liter of enzyme was present in the culture medium after 96 h of cultivation, which was equivalent to an activity of 27.1 U/ml. MPH activities in cell culture lysates (treated with sonication) and in the culture supernatant of the original Plesiomonas strain M6 were only 0.96 U/ml and 0.029 U/ml, respectively. Therefore, a 28.3-fold increase in recombinant enzyme was achieved, and the activity of extracellular MPH produced by the engineered B. subtilis strain was 945.4-fold higher than that found in the original producer bacterium.
FIG. 3.
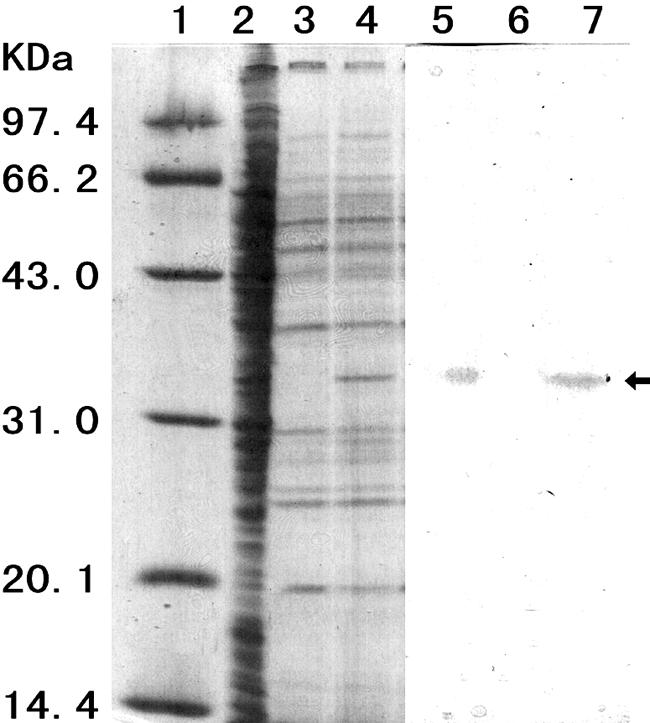
SDS-PAGE and zymogram analysis of MPH production by M6 and WB800 carrying pUBC19 and pP43NMK. The supernatant of the M6 cell fraction and the culture supernatant of WB800 harboring different vectors were analyzed on a 12% SDS-polyacrylamide gel. The partial gel containing lanes 5 to 7 was excised for the zymogram analysis, and the remaining gel was stained with Coomassie brilliant blue R250. Lane 1, molecular mass markers; lanes 2 and 5, M6; lanes 3 and 6, WB800(pUBC19) (negative control); lanes 4 and 7, WB800(pP43NMK).
For purification of the recombinant MPH from B. subtilis WB800(pP43NMK), 50 ml super-rich medium in a 150-ml flask was inoculated with 0.5 ml preculture and incubated at 37°C for 96 h. The culture supernatant was concentrated 10-fold by freeze-drying and dialyzed overnight against 20 mM HEPES buffer (pH 7.5) with two changes of buffer. The supernatant was then applied to a DEAE-Sephadex A-50 column (2.5 by 10 cm), which had been preequilibrated with 20 mM HEPES buffer (pH 7.5) at 4°C. By adjusting the pH of the loading and elution buffers to pH 7.5, most of the contaminating proteins were bound to the column. MPH was eluted with the same buffer at a flow rate of 0.5 ml/min, and only four distinct bands could be detected by SDS-PAGE after this purification step. Total protein in the fractions was detected by the Bradford method (1). The yield of the partially purified MPH was 86.3% (the deviation between different batches was less than 10%), and samples were purified fivefold to a specific activity of 155.6 U/mg. Active fractions were concentrated and further purified by preparative SDS-PAGE and electroelution. The electroeluted enzyme produced a single band following SDS-PAGE.
The calculated molecular mass of MPH is 31,397 Da, but because of the relatively low migration rate, MPH had an apparent molecular mass of 34 kDa as determined by SDS-PAGE. The exact molecular mass of recombinant MPH was determined by matrix-assisted laser desorption ionization-time of flight mass spectrometry to be 31,384 Da. The N-terminal sequence of purified MPH was determined by high-performance liquid chromatography-electrospray ionization mass spectrometry as AAPQVR, demonstrating that the B. subtilis nprB signal peptide sequence had been processed correctly.
For the first time, we have achieved a high level of expression and secretion of active MPH in B. subtilis WB800. The expression level of MPH was markedly higher than the 20.5 U/ml obtained using an E. coli expression system (9) that produced no detectable extracellular activity. This will greatly improve the application efficiency of the engineered strain to degrade organophosphorus pesticides in farm products and in contaminated soil and water. The new expression/secretion vector, containing the nprB typical signal peptide I cleavage site, that we have constructed should also facilitate the expression and secretion of other useful proteins in forms that retain their authentic N-terminal sequences. However, enzyme production efficiency is not yet high enough for practical industrial applications, e.g., for enzyme-based products, and research to further increase MPH yields is in progress.
Acknowledgments
We thank Daniel Zeigler from the Bacillus Genetic Stock Center and Sui-Lam Wong from the University of Calgary for providing bacterial strains and plasmid vectors. We are also grateful to John Buswell from the Edible Fungi Institute, Shanghai Academy of Agricultural Sciences, for the helpful comments and linguistic revision of the manuscript.
This work was supported by grants from the Jiangsu Province Natural Science Foundation of the People's Republic of China (no. BK200171) and the National Natural Science Foundation (no. 30300005) and by the 863 Hi-Tech Research and Development Program of the People's Republic of China (project no. 2004AA214102 and 2004AA246070).
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kinston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Short protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Burkholder, P. R., and N. H. Giles. 1947. Induced biochemical mutations in Bacillus subtilis. Am. J. Bot. 33:345-348. [PubMed] [Google Scholar]
- 3.Chang, S. 1987. Engineering for protein secretion in gram-positive bacteria. Methods Enzymol. 153:507-516. [DOI] [PubMed] [Google Scholar]
- 4.Chang, S., O. Gary, D. Ho, J. Kroyer, S.-Y. Chany, J. McLaughlin, and D. Mark. 1982. Expression of eukaryotic genes in Bacillus subtilis using signals of penP, p. 159-169. In A. T. Ganesan, S. Chang, and J. A. Hoch (ed.), Molecular cloning and gene regulation in bacilli. Academic Press, Inc., San Diego, Calif.
- 5.Cui, Z., S. Li, and G. Fu. 2001. Isolation of methyl parathion-degrading strain M6 and cloning of the methyl parathion hydrolase gene. Appl. Environ. Microbiol. 67:4922-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui, Z.-L., X.-Z. Zhang, Z.-H. Zhang, and S.-P. Li. 2004. Construction and application of a promoter-trapping vector with methyl parathion hydrolase gene mpd as the reporter. Biotechnol. Lett. 26:1115-1118. [DOI] [PubMed] [Google Scholar]
- 7.Doi, R. H., S.-L. Wong, and F. Kawamura. 1986. Potential use of Bacillus subtilis for secretion and production of foreign proteins. Trends Biotechnol. 4:232-235. [Google Scholar]
- 8.Donarski, W. J., D. P. Dumas, D. P. Heitmeyer, V. E. Lewis, and F. M. Raushel. 1989. Structure-activity relationship in the hydrolysis of substrates by the phosphotriesterase from Pseudomonas diminuta. Biochemistry 28:4650-4655. [DOI] [PubMed] [Google Scholar]
- 9.Fu, G., Z. Cui, T. Huang, and S. Li. 2004. Expression, purification, and characterization of a novel methyl parathion hydrolase. Protein Expr. Purif. 36:170-176. [DOI] [PubMed] [Google Scholar]
- 10.Halling, S. M., F. J. Sanchez-Anzaldo, R. Fukuda, R. H. Doi, and C. F. Meares. 1977. Zinc is associated with the beta subunit of DNA dependent RNA polymerase of Bacillus subtilis. Biochemistry 16:2880-2884. [DOI] [PubMed] [Google Scholar]
- 11.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 12.Hung, S.-C., and J. C. Liao. 1996. Effects of ultraviolet light irradiation in biotreatment of organophosphates. Appl. Biochem. Biotechnol. 56:37-47. [DOI] [PubMed] [Google Scholar]
- 13.Motley, S. T., and S. Graham. 1988. Expression and secretion of human interleukin-1 in Bacillus subtilis, p. 371-375. In A. T. Ganesan and J. A. Hoch (ed.), Genetics and biotechnology of bacilli, vol. 2. Academic Press, Inc., San Diego, Calif. [Google Scholar]
- 14.Mulbry, W. W., and J. S. Karns. 1989. Parathion hydrolase specified by the Flavobacterium opd gene: relationship between the gene and protein. J. Bacteriol. 171:6740-6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakayama, A., K. Ando, K. Kawamura, I. Mita, K. Fukazawa, M. Hori, M. Honjo, and Y. Furutani. 1988. Efficient secretion of the authentic mature human growth hormone by Bacillus subtilis. J. Biotechnol. 8:123-134. [Google Scholar]
- 16.Palva, I., M. Sarvas, P. Lehtovaara, M. Sibakov, and L. Kaarianinen. 1982. Secretion of Escherichia coli beta-lactamase from Bacillus subtilis. Proc. Natl. Acad. Sci. USA 79:5582-5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Priest, F. G. 1977. Extracellular enzyme synthesis in the genus Bacillus. Bacteriol. Rev. 41:711-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serdar, C. M., and D. T. Gibson. 1985. Enzymatic hydrolysis of organophosphates: cloning and expression of a parathion hydrolase gene from Pseudomonas diminuta. Bio/Technology. 3:567-571. [Google Scholar]
- 19.Tjalsma, H., H. Antelmann, J. D. Jongbloed, P. G. Braun, E. Darmon, R. Dorenbos, J. Y. Dubois, H. Westers, G. Zanen, W. J. Quax, O. P. Kuipers, S. Bron, M. Hecker, and J. M. Van Dijl. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68:207-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, P. Z., and R. H. Doi. 1984. Overlapping promoters transcribed by Bacillus subtilis σ55 and σ37 RNA polymerase holoenzymes during growth and stationary phase. J. Biol. Chem. 259:8619-8625. [PubMed] [Google Scholar]
- 21.Wu, S.-C., J. C. Yeung, Y. Duan, R. Ye, S. J. Szarka, H. R. Habibi, and S.-L. Wong. 2002. Functional production and characterization of a fibrin-specific single-chain antibody fragment from Bacillus subtilis: effects of molecular chaperones and a wall-bound protease on antibody fragment production. Appl. Environ. Microbiol. 68:3261-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]