Abstract
A new real-time PCR assay was developed and validated in combination with an immunomagnetic separation system for the quantitative determination of Legionella pneumophila in water samples. Primers that amplify simultaneously an 80-bp fragment of the dotA gene from L. pneumophila and a recombinant fragment including a specific sequence of the gyrB gene from Aeromonas hydrophila, added as an internal positive control, were used. The specificity, limit of detection, limit of quantification, repetitivity, reproducibility, and accuracy of the method were calculated, and the values obtained confirmed the applicability of the method for the quantitative detection of L. pneumophila. Moreover, the efficiency of immunomagnetic separation in the recovery of L. pneumophila from different kinds of water was evaluated. The recovery rates decreased as the water contamination increased (ranging from 59.9% for distilled water to 36% for cooling tower water), and the reproducibility also decreased in parallel to water complexity. The feasibility of the method was evaluated by cell culture and real-time PCR analysis of 60 samples in parallel. All the samples found to be positive by cell culture were also positive by real-time PCR, while only eight samples were found to be positive only by PCR. Finally, the correlation of both methods showed that the number of cells calculated by PCR was 20-fold higher than the culture values. In conclusion, the real-time PCR method combined with immunomagnetic separation provides a sensitive, specific, and accurate method for the rapid quantification of L. pneumophila in water samples. However, the recovery efficiency of immunomagnetic separation should be considered in complex samples.
Legionella is one of the main causative agents of severe atypical pneumonias, particularly among people with impaired immune systems. Although the genus Legionella comprises more than 40 species with 64 serogroups (6), L. pneumophila is the most common pathogenic species, accounting for more than 90% of legionellosis cases. Present in soil and natural aquatic environments (12), legionellae sometimes survives as an intracellular parasite of amoebae and ciliates (7). Legionellae have also found a niche in several man-made aquatic environments such as potable water systems, cooling towers, and wastewater systems (9, 10). Consequently, the possible presence of legionellae in bioaerosols generated from soil or aquatic environments poses a significant hazard for human health (34).
Outbreaks of L. pneumophila occur throughout the world (39), impacting public health as well as various industrial, tourist, and social activities. For these reasons some countries specifically regulate the surveillance and control of L. pneumophila in water regularly and assess its presence by culture on a selective medium (25). However, this monitoring technique is time-consuming due to the slow growth rate of the bacterium, the inability to detect viable noncultivable bacteria, and the difficulty in isolating legionellae in samples contaminated with high levels of other microbiota. To avoid these problems, nucleic acid amplification techniques, mainly PCR, have been described as useful tools for the detection of L. pneumophila in clinical and environmental samples. Several PCR-based methods for the detection of L. pneumophila DNA have been described, but most of them are based on the amplification of the macrophage infectivity potentiator (mip) gene (4, 9) or the 16S or 5S rRNA gene (17, 36, 45).
In this work we proposed amplification of an alternative L. pneumophila gene, the defective organelle trafficking (dotA) gene. This gene is involved in L. pneumophila virulence and is regarded as a pathogenicity island, such as cagA in Helicobacter pylori, hly in uropathogenic Escherichia coli, or the vir complex in Agrobacterium tumefaciens (3). In this way, dotA and mip are part of the mechanism that mediates the initial invasion of eukaryotic cells and the subsequent intracellular survival and multiplication (8). The dotA gene product also regulates trafficking of the L. pneumophila phagosome, playing a fundamental role in regulating initial phagosome trafficking decisions either during or immediately after macrophage uptake (38). L. pneumophila strains that possess a mutation in dotA cannot replicate intracellularly because they are unable to alter the endocytic pathway of macrophages (21).
Despite the advantages of conventional PCR, two main obstacles remain. One is the presence of PCR inhibitors, such as humic and fulvic acids and metals, in environmental samples that can produce false-negative results. The second is that conventional PCR is a qualitative assay, informing only of the presence or absence of the microorganism. Various methods have been described that permit procurement of pure DNA lacking PCR inhibitors. These methods include rapid gel filtration to remove humic substances (1), filtration through chelating ion exchange resins to eliminate metal ions (19, 43), addition of polyvinylpyrrolidone to remove polyphenols (23), and cesium chloride density centrifugation to improve general DNA purity (23). The advantages and drawbacks of all of these methods have been reported (1).
By contrast, purification of the intact cell rather than purification of the DNA provides another strategy for eliminating PCR inhibitors. Thus, the use of immunomagnetic separation methodologies, which permit DNA isolation with a minimum of inhibitors (32), has been developed. Immunomagnetic separation relies on the interaction between antibodies attached to paramagnetic beads and cell surface antigens, permitting separation of specific cells by placing a bead-cell suspension in a strong magnetic field. In this way, immunomagnetic separation provides a simple but powerful method for extracting the desired microorganism from heterogeneous bacterial suspensions, such as those encountered in food, clinical, and environmental samples (11).
The recent advent of fluorescent probe-based PCR technology (real-time PCR) has led to the development of a quantitative assay, which was lacking in conventional PCR. With real-time quantitative PCR, signal fluorescence that is released during amplification is proportional to the amount of product generated, and the initial copy number can be estimated from the exponential phase of product accumulation by comparison to a standard curve.
With the above knowledge, the aims of this work were to evaluate an immunomagnetic separation method for the purification of L. pneumophila from water samples and to develop and validate a quantitative L. pneumophila PCR method based on amplification of the dotA gene.
MATERIALS AND METHODS
Bacterial strains and cultivation.
The Legionella strains and the other bacterial species used in this study are described in Table 1. Legionella strains were grown on buffered charcoal-yeast extract containing 0.1% α-ketoglutarate, adjusted to pH 6.9 with KOH, and supplemented (per liter) with 0.4 g l-cysteine and 0.25 g ferric pyrophosphate (BCYE). For the isolation of legionellae from environmental samples, GVPC medium was used. This medium is identical to BCYE except 3 g glycine, 1 mg vancomycin, 50,000 IU polymyxin B, and 80 mg cycloheximide are added to 1 liter of BCYE medium. Inoculated plates were incubated at 37°C in a humidified atmosphere containing 5% carbon dioxide.
TABLE 1.
Bacterial strains used in this study
| Species | Strain identificationa | Serogroup | Source |
|---|---|---|---|
| L. pneumophila | PHLS. (A) April 2001 | External quality assesment | |
| PHLS. (B) April 2001 | External quality assesment | ||
| PHLS. (C) April 2001 | External quality assesment | ||
| L401 | 1 | Environment | |
| L403 | Environment | ||
| L410 | 1 | Environment | |
| L437 | 1 | Environment | |
| L446 | Environment | ||
| L500 | 1 | Environment | |
| L510 | 2-14 | Environment | |
| L511 | 1 | Environment | |
| L512 | 2-14 | Environment | |
| L514 | 1 | Environment | |
| L533 | 1 | Environment | |
| L534 | 2-14 | Environment | |
| L558 | 2-14 | Environment | |
| L560 | 2-14 | Environment | |
| L561 | 2-14 | Environment | |
| L565 | 2-14 | Environment | |
| L568 | 2-14 | Environment | |
| L594 | 2-14 | Environment | |
| L674 | 1 | Environment | |
| L922 | 1 or 9 | Environment | |
| L923 | 1 or 9 | Environment | |
| L924 | 1 or 9 | Environment | |
| L926 | 1 or 9 | Environment | |
| L1587 | 1 | Environment | |
| L1588 | 2-14 | Environment | |
| L1598 | 2-14 | Environment | |
| L1599 | 2-14 | Environment | |
| L1635 | 1 | Environment | |
| L1636 | 1 | Environment | |
| L1638 | 2-14 | Environment | |
| L1642 | 1 | Environment | |
| L1657 | 1 | Environment | |
| L131 | 1 | Environment | |
| L1130 | 4 | Environment | |
| L1298 | 6 | Environment | |
| L1253 | 2,3 | Environment | |
| L226 | 10 | Environment | |
| L53 | 12 | Environment | |
| L63 | 14 | Environment | |
| L1809 | 8 | Environment | |
| L1572 | 6 | Environment | |
| L1528 | 3 | Environment | |
| L1527 | 3 | Environment | |
| L1413 | 9 | Environment | |
| NCTC 11406 | 6 | Collection | |
| NCTC 11230 | 2 | Collection | |
| NCTC 11192 | 1 | Collection | |
| Legionella sp. | L782 | Environment | |
| Legionella sp. | L593 | Environment | |
| Legionella sp. | L1568 | Environment | |
| Legionella sp. | L1592 | Environment | |
| Legionella sp. | L1586 | Environment | |
| L. bozemanii | NCTC 11368 | Collection | |
| L. bozemanii | NCTC 11360 | Collection | |
| L. gravella feeli | NCTC 12022 | Collection | |
| L. jordanis | NCTC 11533 | Collection | |
| L. micdadei | NCTC 11371 | Collection | |
| L. gormanii | NCTC 11401 | Collection | |
| L. dumoffii | NCTC 11370 | Collection | |
| Escherichia coli | CECT 434 | Collection | |
| Vibrio cholerae | CECT 557 | Collection | |
| Enterococcus faecium | CECT 410 | Collection | |
| Salmonella enterica | NCTC 12848 | Collection | |
| Mycobacterium tuberculosis | NCTC 7417 | Collection | |
| Clostridium perfringens | CECT 376 | Collection | |
| Aeromonas hydrophila | CECT 839 | Collection |
CECT, Spanish Type Culture Collection, Valencia, Spain; NCTC, National Collection of Type Cultures, Central Public Health Laboratory, London, United Kingdom; PHLS, Public Health Laboratory Service, London, United Kingdom.
Escherichia coli was cultivated on Tergitol-7 agar (Oxoid), Salmonella enterica serovar Typhimurium on Rambach agar (Merck), Mycobacterium tuberculosis on Seven H11 agar (Becton Dickinson), Clostridium perfringens on thioglycolate broth, and Enterococcus faecium on Slanetz and Bartley medium (Oxoid). All inoculated plates were incubated at 37°C for the period of time appropriate for each species.
Samples.
To prepare spiked samples, legionellae were grown for 3 days on BCYE medium, cells were harvested, and a peptone-water suspension was prepared. This suspension was serially diluted 10-fold in peptone water. Next, 2 ml of dilutions from 10−5 to 10−9 were added to 1 liter of each water matrix to obtain the contaminated samples.
In addition, we also analyzed 25 samples from potable hot water systems belonging to different hotels in Alicante, Spain, and 35 water samples from different cooling tower systems.
Immunomagnetic bead preparation.
An anti-Legionella pneumophila polyclonal antibody (OBT0943; Oxford Biotechnology) was coupled to superparamagnetic beads (Dynabeads M-280 Tosylactivated; Dynal Biotech) by incubating 3 μg of antibody with 107 Dynabeads for 24 h at 37°C with slow tilt rotation. After washing with phosphate-buffered saline, pH 7.4, containing 0.1% bovine serum albumin, the suspension reached a final concentration of 4 × 108 particles per ml of wash buffer.
Sample processing and immunomagnetic purification.
Water samples were concentrated by filtration through 0.4-μm-pore-size, 47-mm-diameter polycarbonate membranes, which were placed in 10 ml of sterile deionized water in a screw-cap tube. To release the cells from the membrane, the tube was vortexed for 3 min, and the 10 ml of solution was further concentrated to 1 ml using Amicon Ultra-15 filters (Millipore, Molsheim, France). Each concentrated sample was added to a microcentrifuge tube containing 50 μl of bacterium-binding beads to reach a final concentration of 2 × 107 bacterium-binding beads per ml of sample. Samples were then incubated for 60 min at room temperature with gentle agitation. Bacteria bound to magnetic beads were drawn to the wall of the microcentrifuge tube by a magnetic particle concentrator (Dynal MPC-M; Dynal). Finally, the supernatant was carefully removed, with a micropipette, without disrupting the bacterium-bead complexes.
DNA isolation from environmental samples and pure cultures.
The processed water samples and bacterial colonies from pure cultures were resuspended in 200 μl of 20% Chelex 100 resin (Bio-Rad Laboratories, Richmond, CA). DNA was then extracted by three freeze-thaw cycles (−75°C for 10 min and 94°C for 10 min), and cellular debris was removed by pelleting at 10,000 × g for 1 min. The quantity of genomic DNA was measured by spectrophotometry at 260 nm in triplicate, and DNA purity was checked using the A260/A280 ratio (29).
The number of copies of the dotA gene in purified DNA samples was calculated by assuming an average molecular mass of 660 Da for 1 bp of double-stranded DNA (PCR Applications Manual, 2nd ed., Roche Diagnostics GmbH, Mannheim, Germany, 1999). The calculation was performed with the following equation: number of copies = quantity of DNA (fg)/mean mass of the L. pneumophila genome. The mean mass of the L. pneumophila genome was calculated from the mean size of the genome, which is assumed to be 3.9 Mb (5).
Amplification conditions and calculation of efficiencies.
To design oligonucleotides specific for L. pneumophila, a multiple alignment of dotA sequences deposited in the GenBank database (accession numbers AY36018 to AY36035 [21], AF095231 to AF095235, AF440205 to AF440215, and AY280173 to AY280338) was performed using CLUSTAL X software (42). The sequences of the output regions were searched against GenBank sequences with the BLAST family program package (28) to ensure the specificity of primers and probes. Finally, primers and probes were analyzed for the requirements imposed by real-time quantitative PCR using Primer Express (version 2.0) of Applied Biosystems. When an optimal design was found (Table 1), primers were synthesized commercially (Applied Biosystems).
The amplification reactions were performed in optical microplates using a total volume of 25 μl. The reaction mixtures contained 1× TaqMan universal PCR master mix (PCR buffer, deoxynucleoside triphosphates, AmpliTaq Gold polymerase, internal reference signal 6-carboxy-x-rhodamine [ROX], Amp Erase uracil N-glycosylase [UNG], MgCl2; Applied Biosystems, Foster City, CA), 300 nM of each L. pneumophila-specific oligonucleotide primer, and 250 nM TaqMan Minor Grove Binding (MGB) L. pneumophila-specific probe labeled with 6-carboxy fluorescein (FAM). An MGB probe is a modification of a TaqMan probe that forms hyperstabilized duplexes with cDNA (2). It is specifically recommended when there are few regions available for probe selection and it is impossible to get a high enough melting temperature (Tm) to match the selected primers. For instance, a 12-mer probe with an MGB group has a melting temperature identical to that of a 27-mer DNA probe lacking an MGB group (22).
To detect PCR inhibitors, a hybrid internal control was constructed that could be amplified simultaneously with the target DNA by using the same primers set. The control sequence contained a fragment from gyrB of Aeromonas hydrophila (Spanish Type Culture Collection, CECT 839) that was linked at both ends with the same sequences as the dotAF and dotAR primers. Two hybrid primers, dotAFgyrB (dotAF sequence CAAGGCGTTCGTCGAATACC) (positions 562 to 583) and dotARgyrB (dotAR sequence GCTGCGGAATGTTGTTGGT) (positions 776 to 757) were synthesized, and DNA from A. hydrophila was amplified as described previously (35). The resultant amplicon was a fragment containing 161 bp of the A. hydrophila gyrB gene, flanked at both ends by two sequences of the L. pneumophila dotA gene (primers dotAF and dotAR). To use this hybrid DNA in real-time PCR, a TaqMan probe was designed in the gyrB region and labeled with VIC (PE Biosystems) (Table 3).
TABLE 3.
Oligonucleotides used for real-time amplification of L. pneumophila and the internal positive control
| Name | Positions on genea | Sequence (5′ → 3′) | Accession no. |
|---|---|---|---|
| dotAF | 986-1004 | ATTGTCTCGCGCGATTGC | AY720956 |
| dotAR | 1066-1043 | CCGGATCATTATTAACCATCACC | AY720956 |
| dotA probe | 1006-1027 | ATACAGCAAATGTATGTGACTT | AY720956 |
| gyrB probe | 590-614 | AACAAGACCCCGATCCACCCGAAG | AY101778 |
Primer and probe nucleotide positions are given according to the complete sequence of the dotA gene of L. pneumophila ATCC 33152 (AF095231). In the case of the internal control (gyrB gene of A. hydrophila CECT 839), the probe positions are given using the Escherichia coli numbering.
Amplification reactions were performed using an ABI Prism 7000 sequence detector (Applied Biosystems). The thermal profile was 2 min at 50°C (activation of the UNG) and 10 min at 95°C (activation of the AmpliTaq Gold DNA polymerase), followed by 15 s at 95°C and 1 min at 60°C for 42 cycles.
The ABI Prism 7000 detection software permits quantification of PCR products in real time, as revealed by the increase of fluorescence signal by 5′-nuclease activity during the amplification process. The threshold cycle (Ct), the cycle at which the fluorescence in the sample increases above a defined threshold, is inversely proportional to the starting amount of nucleic acid. The threshold (Ct) for each standard was plotted against the log10 of the starting DNA quantity to generate a standard curve.
The amplification efficiency (E) was estimated by using the slope of the standard curve and the formula E = (10−1/slope). A reaction with 100% efficiency will generate a slope of −3.32 and has an efficiency of 2.
Statistical analysis.
Applied Biosystems real-time quantitative PCR data were analyzed by the Applied SDS software version 1.1 (Applied Biosystems). Statistical analysis was performed using Statgraphics Plus version 5 (Manugistics).
RESULTS
Purification of L. pneumophila by immunomagnetic separation.
To test the ability of immunomagnetic separation to recover different strains of legionellae from water, different distilled water samples were spiked with 50 different strains of L. pneumophila belonging to the serogroups described in Table 1. L. pneumophila was then concentrated by filtration using polycarbonate membranes, further concentrated with Amicon Ultra-15 filters, and isolated by immunomagnetic separation as indicated above. All the strains tested were retained by the immunomagnetic beads, demonstrating the ability of the polyclonal antibody used to isolate all the serogroups studied.
Since the method of purification involved the isolation of intact cells by an immunomagnetic separation procedure, the percentage of L. pneumophila cells recovered from clean water, such as distilled and potable water, was determined by culture isolation. Known concentrations of L. pneumophila (NCTC 11192) were used to spike distilled water and potable water, and legionellae were then concentrated and purified by immunomagnetic separation as described above. We counted the number of colonies isolated from the supernatant and from the bacterium-binding beads during the immunomagnetic purification. Because the abundant growth of microbiota found in cooling tower samples hampers the isolation of legionellae by culture methods, the efficiency of the purification method could not be tested in the same way. Water samples containing high levels of microbiota can be treated with acid or heat to reduce microbiota, but Legionella concentrations are also reduced. To overcome this hurdle, we used real-time PCR to determine the efficiency of Legionella purification from cooling tower samples.
The efficiency of immunomagnetic bead isolation of L. pneumophila from spiked distilled water was higher than that of spiked potable water, which was higher than that of cooling tower water (Table 2). The average recovery rate for distilled water was 59.9%, for potable water 42.0%, and for cooling tower water 36.0%. The recovery rates decreased as the complexity, i.e., the number of components, of the water increased. In parallel, the reproducibility of the recovery rates also decreased. In the case of distilled water, the values of recovery were very similar for all samples tested (standard deviation, 8%). By contrast, the recovery rates for potable water ranged from 74.1% to 16% (standard deviation, 17.7%), and for cooling tower water from 89% to 7.7% (standard deviation, 32.8%).
TABLE 2.
Recovery of L. pneumophila by immunomagnetic purification from distilled and potable water (CFU) and cooling tower water (number of copies)
| Sample | CFU or no. of copies
|
Recovery (%) | |
|---|---|---|---|
| Supernatant | Bacterium-beads | ||
| Distilled water | 80 × 106 | 12 × 107 | 60.0 |
| 70 × 106 | 12 × 107 | 63.0 | |
| 40 × 106 | 40 × 106 | 71.0 | |
| 840 | 1,800 | 68.2 | |
| 105 | 120 | 53.3 | |
| 870 | 1,200 | 58.0 | |
| 80 | 70 | 46.7 | |
| 8 | 12 | 58.8 | |
| Potable water | 294 | 230 | 43.9 |
| 1,260 | 1,600 | 55.9 | |
| 1,260 | 1,600 | 55.9 | |
| 126 | 160 | 55.9 | |
| 126 | 160 | 55.9 | |
| 4 | 12 | 74.1 | |
| 2 | 2 | 48.8 | |
| 105 | 120 | 53.3 | |
| 8 | 12 | 58.8 | |
| 80 | 70 | 46.7 | |
| 46 | 28 | 37.7 | |
| 40 × 107 | 40 × 107 | 50.0 | |
| 40 × 106 | 40 × 106 | 50.0 | |
| 1,050 | 280 | 21.0 | |
| 105 | 28 | 21.0 | |
| 11 | 2 | 16.1 | |
| Cooling tower water | 462.2 | 3,778.8 | 89.1 |
| 22,501.6 | 6,734.7 | 23.0 | |
| 951.7 | 452.9 | 32.2 | |
| 184.2 | 15.6 | 7.8 | |
| 17,3975.9 | 14,626.3 | 7.7 | |
| 418.7 | 1,254.7 | 75.0 | |
| 5,069.8 | 1,053.4 | 17.2 | |
Design, optimization, and specificity of primers and probes.
The primers and probes were designed based on a conserved region of the L. pneumophila dotA gene. The sequences were compared to all the nucleotide databases of GenBank (http://www.ncbi.nlm.nih.gov) to ensure their specificity, and the sequences of the L. pneumophila dotA gene were unique. The selected regions were analyzed for the requirements imposed by real-time quantitative PCR using Primer Express software (version 2.0), with the resulting primer set (Table 3) producing a product of 80 bp. Nevertheless, a conventional TaqMan probe did not fulfill the requirements imposed by the software, so an MGB probe was designed to maintain the same nucleotide sequence but have a higher melting temperature. In our case, the TaqMan probe had a Tm of 46°C, whereas the MGB probe had a Tm of 66.4°C.
The specificity of the primers and the MGB probe was verified experimentally by using all the species listed in Table 1. The primers set and the probe amplified L. pneumophila strains but not any other bacteria or Legionella spp. tested in this work.
The concentration of primers was optimized by using 6 pg of DNA from L. pneumophila (NCTC 11192) as a template and performing 5′-nuclease assay reactions with different concentrations of forward and reverse primers. This allowed determination of the primer concentrations that gave the lowest Ct values and the highest fluorescence intensity for a normalized reported value. The primer concentrations tested ranged from 300 to 900 nM, while the rest of the parameters were kept invariant, including the annealing temperature and the MGB probe concentration (250 nM, excess following the manufacturer's recommendations). The Ct values we obtained were approximately the same with all combinations, so we used the lowest concentration (300 nM) of each primer.
The optimal concentration of the internal positive control was determined to minimize its competition with the L. pneumophila target. Serial 10-fold dilutions of the internal positive control amplicon were combined with different concentrations of L. pneumophila DNA and amplified with the primers dotAF and dotAR. The concentration containing 39 copies of the internal positive control was selected to be used in each run since no competition was observed with the Legionella target (Table 6).
TABLE 6.
Optimization of internal positive control (IPC) concentration
| No. of copies of L. pneumophila dotA |
Ct with IPC and dotA probes at the indicated no. of IPC copies:
|
|||||
|---|---|---|---|---|---|---|
| 390
|
39
|
0
|
||||
| IPC | dotA | IPC | dotA | IPC | dotA | |
| 1,000 | 35.41 | 30.83 | 37.55 | 30.87 | 30.68 | |
| 100 | 36.22 | 34.22 | 36.17 | 32.79 | 32.67 | |
| 10 | 34.05 | 36.69 | 35.59 | 36.38 | 35.11 | |
| 0 | 34.24 | 34.48 | ||||
Determination of the standard curve.
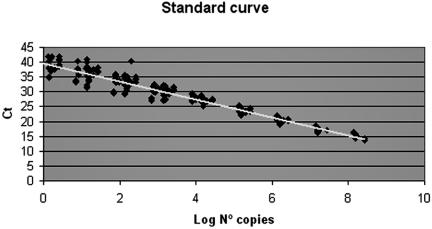
To develop an external standard curve for quantitative PCR, 10-fold serial dilutions of known L. pneumophila DNA concentrations were prepared. Three different analysts performed 21 different amplification experiments in triplicate, for a total of 507 samples. Linear regression analysis, plotting the obtained Ct values versus the logarithm of the dotA gene copy number, gave a straight-line plot and a correlation coefficient (r) of −0. 973 (R2 = 0.947). The equation resulting from the regression curve obtained was Ct = −3.03 log10 [copy number] + 39.61, and it was linear over at least a 9-log10 range of DNA concentrations, and the reaction had an efficiency of 2.13 (Fig. 1).
FIG. 1.
External standard curve of L. pneumophila dotA gene.
The results obtained from the evaluation of the within-run (intra-assay) and between-run (interassay) variations in the multiple assays are shown in Table 4. As determined from the values, the coefficient of variation for the Ct was 1.85% in the intra-assay study (repetitivity) and 3.09% in the interassay study (reproducibility). In contrast, when the intra- and interassay variations were calculated for the number of copies of target DNA, the values were 39.61% and 65.06%, respectively, which are higher than the Ct values. Moreover, differences were found in the coefficient of variation values for both intra- and interassay between samples containing high and low copy numbers: as the number of copies decreased, the coefficient of variation values increased (Table 4 and Fig. 1).
TABLE 4.
Reproducibility of dotA real-time PCR
| No. of copies | Coefficient of variation (%)
|
|||
|---|---|---|---|---|
| Intra-assaya
|
Interassayb
|
|||
| Ct | No. of copies | Ct | No. of copies | |
| 1,894,405.6 | 0.47 | 5.97 | 0.61 | 8.0 |
| 189,440.6 | 1.14 | 17.85 | 2.96 | 45.58 |
| 18,944.1 | 0.75 | 13.12 | 3.65 | 60.84 |
| 1,894.4 | 1.59 | 32.65 | 3.24 | 60.31 |
| 189.4 | 2.91 | 67.37 | 3.57 | 91.29 |
| 18.9 | 1.93 | 50.87 | 3.42 | 87.06 |
Dilution series of DNA of L. pneumophila were analyzed in triplicate by two analysts in four different runs. The coefficients of variation (CVs) represent the quadratic mean CVs of the personal means. The personal mean CV was calculated from the 12 triplicates analyzed by one analyst.
The CVs represent the mean of the CVs obtained in samples with the same number of copies by two analysts in four separate runs (four triplicates).
Concerning the limit of detection of the PCR method, L. pneumophila was detected in 100% of samples containing as few as 6.9 copies of target DNA, in 85% of samples containing 2.6 copies, in 71% of samples containing 1.6 copies, and in 60% of samples containing 1.4 copies. Due to the low reproducibility at these extremely low concentrations, the limit of quantification was calculated from the standard deviation of Ct values at the limit of detection. For this reason, when the average Ct value at this concentration was increased by three times the standard deviation, the new Ct value was 36.16. Thus, in accordance with the standard curve, the number of copies calculated was 13.8, indicating that reliable quantification was possible above this limit.
Relative accuracy of quantification.
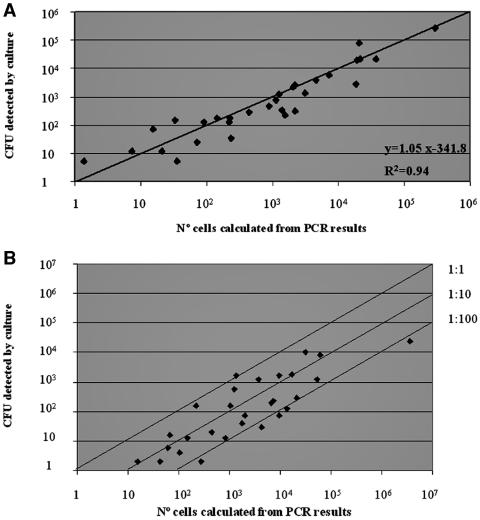
A series of experiments were conducted with L. pneumophila cultures to determine the correlation between the results obtained by real-time quantitative PCR assays and those obtained by the culture isolation technique. The Ct values obtained from the analysis of 10-fold serial dilutions of L. pneumophila cultures were extrapolated to the corresponding external standard curve, previously calculated experimentally. The resulting theoretical number of cells was compared to the CFU obtained from culture isolation (Table 5). A strong positive correlation was found for both methods for the total sample group (R2 = 0.94; r = 0.97) (Fig. 2). The slope of the corresponding curve was 1.05.
TABLE 5.
Accuracy of real-time PCR assay for L. pneumophila in dilutions of L. pneumophila cultures and water samples
| Sample | Theoretical no. of cells | CFU |
|---|---|---|
| Dilutionsa | 290,009.33 | 260,000 |
| 20,571.56 | 80,000 | |
| 890.19 | 460 | |
| 144.63 | 170 | |
| 7.41 | 12 | |
| 7,149.40 | 5,700 | |
| 1,126.81 | 730 | |
| 91.65 | 125 | |
| 19,065.39 | 20,100 | |
| 2,070.24 | 2,100 | |
| 223.10 | 170 | |
| 15.35 | 70 | |
| 37,224.95 | 21,200 | |
| 1,253.37 | 1,200 | |
| 33.34 | 150 | |
| 4,634.98 | 3,800 | |
| 1,404.79 | 340 | |
| 71.86 | 25 | |
| 1.35 | 5 | |
| 21,046.20 | 22,300 | |
| 3,169.13 | 1,340 | |
| 216.41 | 120 | |
| 20.65 | 12 | |
| 2,233.79 | 2,500 | |
| 442.27 | 290 | |
| 18,802.82 | 2,670 | |
| 1,549.30 | 220 | |
| 2,183.10 | 310 | |
| 232.39 | 33 | |
| 35.21 | 5 | |
| Water samplesb | 17,029 | 1,800 |
| 13,555 | 120 | |
| 54,088 | 1,200 | |
| 2,057 | 70 | |
| 826 | 12 | |
| 6,735 | 200 | |
| 453 | 20 | |
| 16 | 2 | |
| 13,555 | 120 | |
| 826 | 12 | |
| 2,057 | 70 | |
| 62 | 6 | |
| 274 | 2 | |
| 4,268 | 28 | |
| 21,230 | 280 | |
| 4,268 | 28 | |
| 274 | 2 | |
| 7,267 | 230 | |
| 62 | 6 | |
| 9,775 | 1,600 | |
| 1,364 | 1,600 | |
| 1,061 | 160 | |
| 227 | 160 | |
| 146 | 12 | |
| 43 | 2 | |
| 1,761 | 40 | |
| 106 | 4 | |
| 9,731 | 70 | |
| 67 | 16 | |
| 11,568,079 | 236,000 | |
| 60,898 | 8,148 | |
| 1,256 | 580 | |
| 3,620,056 | 24,638 | |
| 31,442 | 10,670 | |
| 3,819 | 1,261 |
Ct values determined from the analysis of different dilutions of L. pneumophila cultures were extrapolated to the external standard regression curve to calculate the theoretical number of cells per reaction.
Ct values determined from the analysis of different water samples were extrapolated to the external standard regression curve to calculate the theoretical number of cells per reaction.
FIG. 2.
Correlation between real-time PCR and culture for L. pneumophila. (A) Dilutions of L. pneumophila cultures. (B) Water samples.
Quantitative detection of L. pneumophila in water samples.
The purification of L. pneumophila by immunomagnetic separation coupled with the detection by real-time PCR was used to analyze 25 potable water samples and 35 cooling tower water samples. To determine the number of L. pneumophila cells presents in each sample, the MGB probe fluorescence was processed, and the Ct values were compared to the external standard curve. For the routine quantification of unknown samples, two known concentrations of L. pneumophila, containing 16 and 160 copies of target DNA, were included in every run to control the robustness of the external standard curve (Table 6). The same samples were analyzed in parallel by culture isolation. For potable water, 13 samples yielded positive results by both methods, whereas two were positive only by PCR. In the case of cooling tower water, 22 samples were positive by both methods and 6 samples only by PCR.
In all samples found positive by both culture isolation and real-time PCR, the number of cells obtained by real-time PCR was consistently higher than the CFU obtained by culture counts (Table 5). The regression analysis showed that the average number of cells calculated from the PCR analysis was 20-fold higher than the culture value.
DISCUSSION
Nucleic acid amplification by PCR and more recently real-time PCR can successfully detect legionellae in environmental samples, avoiding the inherent problems of conventional culture isolation (4, 15, 36, 45). PCR techniques have targeted the mip gene (4, 15), 5S ribosomal DNA (15), and 16S ribosomal DNA (17, 36, 45). More recently, methods were developed for the simultaneous detection of L. pneumophila and Legionella spp. based on the 23S-5S internal spacer (17). In this work we have described a new assay based on the specific amplification by real-time PCR of the dotA gene, chosen due to its relationship to the virulence of L. pneumophila. The specificity of the primers and the MGB probe was confirmed by performing multiple searches of nucleotide databases and by screening 50 different strains of L. pneumophila, 12 other Legionella species, and six different bacteria common in water.
In this work we have proposed an external standard curve for the quantification of L. pneumophila DNA in different real-time PCR runs. By using two control DNA sequences in each run, we can verify the robustness of the standard curve. This external standard curve provides laboratories an easier, faster, and cheaper method for the precise and reproducible quantification of DNA by real-time PCR.
The regression coefficient of the standard curve indicated a good correlation between the number of copies of target DNA and the amount of amplified product (represented by the Ct value). The obtained value for the slope of the standard curve (−3.03) indicated that the amplification efficiency was very near the optimal slope value of −3.32. Moreover, the R2 value was 0.947, indicating that the PCR system was highly linear. Therefore, the high correlation and linearity of the standard curve indicated that the assay was suitable for quantitative measurements.
The detection limit of our assay was approximately 6.85 copies of target DNA (29.9 fg), slightly more than the 4.3-fg L. pneumophila genome (5), and is comparable with data obtained by other authors (16, 31, 36, 37, 41, 46). Recently, it has been described that minor groove binding probes, such as we have designed, increase the sensitivity of bacterial detection 10- to 100-fold (33).
On the other hand, the standard curve is accurate enough to be employed for different runs since it is generated from a wide range of concentrations that were analyzed in triplicate and repeated several times. Moreover, the robustness of the standard curve is controlled in each run by the amplification of two different L. pneumophila DNA standards. The method for creating the calibration curve has recently been introduced into the Light Cycler software (24). Thus, the previous publication, combined with our current results, demonstrates that it is not necessary to prepare a standard curve with each run. This new methodology provides a much easier and faster method for precise and reproducible quantification of both DNA and target cell numbers present in a given sample.
The main problem for real-time PCR of samples containing low DNA concentrations is that the Ct values are greatly scattered, even for triplicates of the same concentration. The reproducibility of our PCR method was evaluated using the coefficient of variation values of several intra- and interassay studies. The coefficient of variation was calculated for Ct as well as for calculated copy numbers. However, because of the exponential nature of PCR amplification, the concentration and Ct have a log-linear relationship, so the expected coefficients of variation for Ct are lower than those for the concentration. Additionally, the imprecision of PCR assays is unavoidably larger than that observed in classical clinical chemistry or immunological assays. The results we obtained in this work are comparable to those obtained by other authors (16, 36, 41).
When Legionella cultures were used, we found that the correlation between our PCR method and the standard culture isolation method agreed with previous findings by Rodriguez-Lázaro et al. (37). By contrast, when unknown water samples were analyzed, we found that PCR results yielded higher values in all cases, in agreement with other reports (18, 45). All of these results appear plausible since amplification of exponential cultures of legionellae occurs with the majority of cells in a viable cultivable state. Thus, PCR and culture values should coincide. By contrast, when environmental samples are analyzed by PCR, all DNA is amplified, including DNA from dead bacteria and from viable but noncultivable bacteria. Moreover, in the case of legionellae, recovery rates of culture are usually less than 100% due to specific requirements of growth, overgrowth by other bacteria, and legionella loss and damage during sample preparation. For these reasons the number of cells calculated by PCR is always higher than the number of CFU.
The abundant presence of PCR inhibitors, such as heavy metals and organic matter, has been demonstrated in environmental samples. Different methods of DNA purification have been developed (1, 19, 23, 43), but despite their advantages, inhibition of PCRs is frequent in some problematic samples. For this reason the use of an internal positive control in PCRs is very important to monitor the efficiency of the reaction and to evaluate the possible presence of false-negative results. In this work, a real-time PCR method has been developed that uses the same primer set for simultaneous amplification of the L. pneumophila dotA gene and an internal positive control. This method has the advantage of rapid detection of both the target gene and possible PCR inhibitors while maintaining the sensitivity of the PCR assay. We have also reported the usefulness of an immunomagnetic separation method to isolate L. pneumophila from heterogeneous bacterial suspensions in water, avoiding PCR inhibitors. In this way the strategy was to purify intact cells rather than DNA. Such immunomagnetic separation techniques have been required to facilitate the rapid DNA detection of some bacteria, viruses, and parasites (26, 30, 32, 40).
In this work we used polyclonal antibodies specific for a number of L. pneumophila surface antigens to coat magnetic beads. This avoided the problem of the high level of specificity that can sometimes occur with monoclonal antibodies, and it also increased the likelihood of isolating the desired organism.
The efficiency of recovering L. pneumophila cells was similar to that obtained by various authors, ranging from 47% to 87% for microorganisms as different as Mycobacterium avium, Campylobacter jejuni, and Cryptococcus neoformans (20, 26, 44). The efficiency we obtained was for water samples spiked with known concentrations of L. pneumophila and without other enrichment techniques commonly performed for other pathogens (11). Some bacteria always remained in the supernatant because the immunobeads could not bind all cells even at the higher dilutions. Water from cooling towers and highly contaminated samples exhibited still smaller recoveries. The efficiency of the method was likely reduced by the presence of debris and other microorganisms that severely compromise the method by acting as nontarget objects (13, 27). In some samples the beads were hardly attracted to the magnetic particle concentrator because of physical impediments. Sample turbidity, for example, is known to reduce the sensitivity of the method (27). Also, subsequent PCRs were inhibited in some of these samples, as shown by the addition of internal control DNA to the PCR mixtures. Although immunomagnetic separation can separate microorganisms of interest from polymerase-inhibitory factors (26) present in environmentally contaminated samples, this did not occur in all cases (27), possibly due to the environmental origin of the samples.
In conclusion, the real-time PCR system that was evaluated and validated in this study, combined with immunomagnetic separation, provides many benefits (speed, specificity, accuracy, sensitivity, stability, and cost-effectiveness) for the quantitative detection of L. pneumophila in potable water and other relatively clean environmental water samples. The use of immunomagnetic purification may be limited in extremely contaminated samples, where the recovery efficiencies are highly variable due mainly to the principle of the method and the special characteristics of the sample. For these reasons, more efficient purification methods should be investigated and current purification methods should be improved. The integration of rapid and efficient sample preparation methods with rapid amplification and detection technologies, such as those described here, should improve the management and prevention of Legionella outbreaks.
Acknowledgments
This study was supported in part by grants from the Science and Technology Ministry (FIT-010000-2003-63) and from the Fundación Agbar (B185/1.999 and B221/1.998). V. Barberá and C. Carrasco-Serrano received a studentship from the Fundación Agbar.
REFERENCES
- 1.Abbaszadegan, M., M. S. Huber, C. P. Gerba, and I. L. Pepper. 1993. Detection of enteroviruses in groundwater with the polymerase chain reaction. Appl. Environ. Microbiol. 59:1318-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonina, I., M. Zivarts, L. Kutyavin, E. Lukhatanov, H. Gramper, and R. B. Meyer. 1997. Efficient priming of PCR with short oligonucleotides conjugated to a Minor Groove Binder. Nucleic Acids Res. 25:2657-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews, H. L., J. P. Vogel, and R. R. Isberg. 1988. Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infect. Immun. 66:950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballard, A. I., N. K., Fry, L. Chan, S. B. Surman, J. V. Lee, T. G. Harrison, and K. J. Towner. 2000. Detection of Legionella pneumophila using a real-time PCR hybridization assay. J. Clin. Microbiol. 38:4215-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender, L., M. Ott, R. Marre, and J. Hacker. 1990. Genome analysis of Legionella ssp. by orthogonal field alternation gel electrophoresis (OFAGE). FEMS Microbiol. Lett. 60:253-257. [DOI] [PubMed] [Google Scholar]
- 6.Benson, R. F., and B. S. Fields. 1998. Classification of the genus Legionella. Semin. Respir. Infect. 13:90-99. [PubMed] [Google Scholar]
- 7.Brand, B. C., and J. Haker. 1996. The biology of Legionella infection. p. 291-312. In S. H. E. Kaufmann (ed.), Host response to intracellular pathogens. R. G. Landes Company, Austin, Tex.
- 8.Bumbaugh, A. C., E. A. McGraw, K. L. Page, R. K. Selander, and T. S. Whittam. 2002. Sequence polymorphism of dotA and mip alleles mediating invasion and intracellular replication of Legionella pneumophila. Curr. Microbiol. 44:314-322. [DOI] [PubMed] [Google Scholar]
- 9.Catalán, V., F. García, C. Moreno, and D. Apraiz. 1997. Detection of Legionella pneumophila in water by Nested Polymerase Chain Reaction. Res. Microbiol. 148:71-78. [DOI] [PubMed] [Google Scholar]
- 10.Colbourne, J. S., P. J. Dennis, R. M. Trew, C. Berry, and G. Vesey. 1988. Legionella in public water supplies. Water Sci. Technol. 20:11-20. [Google Scholar]
- 11.Cudjoe, K. S., R. Krona, and E. Olsen. 1994. IMS: a new selective enrichment technique for detection of Salmonella in foods. Int. J. Food Microbiol. 23:159-165. [DOI] [PubMed] [Google Scholar]
- 12.Fliermans, C. B., W. B. Cherry, L. H. Orrison, S. J. Smith, D. L. Tison, and D. H. Pope. 1981. Ecological distribution of Legionella pneumophila. Appl. Environ. Microbiol. 41:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fricker, C., and H. Smith. 1997. Cryptosporidium and crytosporidiosis. SGM Q. 24:52-53. [Google Scholar]
- 14.Grimm, D. H., W. Merkert, K. Ludwig, J. Schleifer, J. Hacker, and B. C. Brand. 1998. Specific detection of Legionella pneumophila: Construction of a new 16S rRNA-targeted oligonucleotide probe. Appl. Environ. Microbiol. 64:2686-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayden, R. T., J. R. Uhl, X. Quian, M. K. Hopkins, M. C. Aubry, A. H. Limper, R. V. Lloyd, and F. R. Cockerill. 2001. Direct detection of Legionella species from bronchoalveolar lavage and open lung biopsy specimens: comparison of LightCycler PCR, in situ hybridization, direct fluorescence antigen detection, and culture. J. Clin. Microbiol. 37:2618-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, Q., J. P. Wang, M. Osato, and L. B. Lachman. 2002. Real-time quantitative PCR for the detection of Helicobacter pylori. J. Clin. Microbiol. 40:3720-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herpers, B. J., B. M. Jongh, K. Der Zwaluw, and E. J. Hannen. 2003. Real-Time PCR assay targets the 23S-5S spacer for direct detection and differentiation of Legionella spp. and Legionella pneumophila. J. Clin. Microbiol. 41:4815-4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huijsdens, X. W., R. K. Linskens, M. Mak, S. G. Meuwissen, C. M. Vandenbroucke-Grauls, and P. H. Savelkoul. 2002. Quantification of bacteria adherent to gastrointestinal mucosa by real-time PCR. J. Clin. Microbiol. 40:4423-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurt, R. A., X. Qiu, L. Wu, Y. Roh, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 67:4495-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katsu, M., A. Ando, R. Ikeda, Y. Mikami, and K. Nishimura. 2003. Immunomagnetic isolation of Cryptococcus neoformans by beads coated with anti-Cryptococcus serum. Jpn. J. Med. Mycol. 44:139-144. [DOI] [PubMed] [Google Scholar]
- 21.Ko, K. S., H. K. Lee, M. Y. Park, M. S. Park, K. H. Lee, S. Y. Woo, Y. J. Yu, and Y. H. Kook. 2002. Population genetic structure of Legionella pneumophila inferred from RNA polymerase (rpoB) and dotA gene sequences. J. Bacteriol. 184:2123-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kok, J. B., E. T. Wiegerinck, B. A. Giesendorf, and D. W. Swinkels. 2002. Rapid genotyping of single nucleotide polymorphisms using novel Minor Groove Binding DNA oligonucleotides (MGB probes). Hum. Mutat. 19:554-559. [DOI] [PubMed] [Google Scholar]
- 23.Koonjul, P. K., W.F. Brandt, J. M. Farrant, and G. G. Lindsey. 1999. Inclusion of polyvinylpyrrolidone in the polymerase chain reaction reverses the inhibitory effects of polyphenolic contamination of RNA. Nucleic Acids Res. 27:915-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kühne, B. 2003. Quantitative real-Time RT-PCR using imported plasmid standard curves. Biochimica 3:9-11. [Google Scholar]
- 25.Lee, T. C., R. M. Vickers, V. L. Yu, and M. M. Wagener. 1993. Growth of 28 Legionella species culture media: a comparative study. J. Clin. Microbiol. 31:2764-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Z., G. Han Bai, C. F. von Reyn, P. Marino, M. J. Brennan, N. Gine, and S. L. Morris. 1996. Rapid detection of Mycobaterium avium in stool samples from AIDS patients by immunomagnetic PCR. J. Clin. Microbiol. 34:1903-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowery, C. J., J. E. Moore, B. C. Millar, D. P. Burke, K. A. J. McCorry, E. Crothers, and J. S. G. Dooley. 2000. Detection and speciation of Cryptosporidium spp. in environmental water samples by immunomagnetic separation, PCR and endonuclease restriction. J. Med. Microbiol. 49:779-785. [DOI] [PubMed] [Google Scholar]
- 28.Madden, T. L., R. L. Tatusov, and J. Zhang. 1996. Applications of network BLAST server. Methods Enzymol. 266:131-141. [DOI] [PubMed] [Google Scholar]
- 29.Maniatis, T., E. F. Fritsch, and K. J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Muir, P., J. Nicholson, M. Jhetam, S. Neogi, and J. E. Banatvala. 1993. Rapid diagnosis of enterovirus infection by magnetic bead extraction and polymerase chain reaction detection of enterovirus RNA in clinical specimens. J. Clin. Microbiol. 31:13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicolas, L., G. Milon, and E. Prina. 2002. Rapid differentiation of Old World Leishmania species by LightCycler polymerase chain reaction and melting curve analysis. J. Microbiol. Methods 51:295-299. [DOI] [PubMed] [Google Scholar]
- 32.Olsvik, O., T. Popovic, E. Skjerve, S. Cudjoe, E. Hornes, J. Ugelstad, and M. Uhlen. 1994. Magnetic separation techniques in diagnostic microbiology. Clin. Microbiol. Rev. 7:43-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ott, S. J., M. Musfeldt, U. Ullmann, J. Hampe, and S. Schreiber. 2004. Quantification of intestinal bacterial populations by real-time PCR with a universal primer set and minor groove binder probes: a global approach to the enteric flora. J. Clin. Microbiol. 42:2566-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pascual, L., S. Pérez-Luz, A. Amo, C. Moreno, D. Apraiz, and V. Catalan. 2001. Detection of Legionella pneumophila in bioaerosols by polymerase chain reaction. Can. J. Microbiol. 47:341-347. [PubMed] [Google Scholar]
- 35.Pham, D. G., G. E. Madico, T. Quinn, M. J. Enzler, T. Smith, and C. Gaydos. 1998. Use of lambda phage DNA as a hybrid internal control in a PCR-enzyme immunoassay to detect Chlamydia pneumoniae. J. Clin. Microbiol. 36:1919-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rantakokko-Jalava, K., and J. Jalava. 2001. Development of conventional and real-time PCR assays for detection of Legionella DNA respiratory specimens. J. Clin. Microbiol. 39:2904-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodríguez-Lázaro, D., M. Hernández, M. Scortti, T. Esteve, J. A. Vázquez-Boland, and M. Pla. 2004. Quantitative detection of Listeria monocytogenes and Listeria innocua by real-time PCR: assessment of hly, iap, and lin02483 targets and AmpliFluor technology. Appl. Environ. Microbiol. 70:1366-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy, C. R. 2002. The Dot/Icm transporter of Legionella pneumophila: a bacteria conductor of vesicle trafficking that orchestrates the establishment of a replicative organelle in eukaryotic hosts. Int. J. Med. Microbiol. 291:463-467. [DOI] [PubMed] [Google Scholar]
- 39.Sabria, M., and M. Campins. 2003. Legionnaires′ disease: update on epidemiology and manage options. Am. J. Respir. Med. 2:235-243. [DOI] [PubMed] [Google Scholar]
- 40.Seesod, N., J. Lunderberg, A. Hedrum, L. Aslund, A. Holder, S. Thaithong, and M. Uhlem. 1993. Immunomagnetic purification to facilitate DNA diagnosis of Plasmodium falciparum. J. Clin. Microbiol. 31:2715-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Templeton, K. E., S. A. Scheltinga, P. Sillekens, J. W. Crielaard, A.P. van Dam, H. Goossens, and E. C. Claas. 2003. Development and clinical evaluation of an internally controlled single-tube multiplex real-time PCR assay for detection of Legionella pneumophila and other Legionella species. J. Clin. Microbiol. 41:4016-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. H. Higging. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai, Y. L., C. J. Palmer, and L. R. Sangermano. 1993. Detection of Escherichia coli in sewage and sludge by polymerase chain reaction. Appl. Environ. Microbiol. 59:353-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waller, D. F., and S. A. Ogata. 2000. Quantitative immunocapture PCR assay for detection of Campylobacter jejuni in foods. Appl. Environ. Microbiol. 66:4115-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wellinghausen, N., C. Frost, and R. Marre. 2001. Detection of Legionellae in hospital water samples by quantitative real-time LightCycler PCR. Appl. Environ. Microbiol. 67:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson, D. A., B. Yen-Lieberman, U. Reischl, S. M. Gordon, and G. Procop. 2003. Detection of Legionella pneumophila by real-time PCR for the mip gene. J. Clin. Microbiol. 41:3327-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]