Abstract
Population analyses in water samples obtained from the chemocline of crenogenic, meromictic Lake Cadagno, Switzerland, in October for the years 1994 to 2003 were studied using in situ hybridization with specific probes. During this 10-year period, large shifts in abundance between purple and green sulfur bacteria and among different populations were obtained. Purple sulfur bacteria were the numerically most prominent phototrophic sulfur bacteria in samples obtained from 1994 to 2001, when they represented between 70 and 95% of the phototrophic sulfur bacteria. All populations of purple sulfur bacteria showed large fluctuations in time with populations belonging to the genus Lamprocystis being numerically much more important than those of the genera Chromatium and Thiocystis. Green sulfur bacteria were initially represented by Chlorobium phaeobacteroides but were replaced by Chlorobium clathratiforme by the end of the study. C. clathratiforme was the only green sulfur bacterium detected during the last 2 years of the analysis, when a shift in dominance from purple sulfur bacteria to green sulfur bacteria was observed in the chemocline. At this time, numbers of purple sulfur bacteria had decreased and those of green sulfur bacteria increased by about 1 order of magnitude and C. clathratiforme represented about 95% of the phototrophic sulfur bacteria. This major change in community structure in the chemocline was accompanied by changes in profiles of turbidity and photosynthetically available radiation, as well as for sulfide concentrations and light intensity. Overall, these findings suggest that a disruption of the chemocline in 2000 may have altered environmental niches and populations in subsequent years.
Lake Cadagno is a crenogenic meromictic lake located in the catchment area of a dolomite vein rich in gypsum in the Piora valley in the southern Alps of Switzerland (46°33′N, 8°43′E). This lake is characterized by a compact chemocline with high concentrations of sulfate; steep gradients of oxygen, sulfide, and light; and a turbidity maximum that correlates to large numbers of bacteria (up to 107 cells ml−1) (5, 21, 31). Molecular methods such as 16S rRNA gene clone library analysis and subsequent in situ hybridization with specific fluorescent probes identified the most abundant bacteria in the chemocline as large- and small-celled purple sulfur bacteria. These accounted for up to 35% of all bacteria (34) and consisted of Chromatium okenii as the only representative of the large-celled purple sulfur bacteria and of four populations related to the genus Lamprocystis that represented the small-celled purple sulfur bacteria (31). Small-celled purple sulfur bacteria were most abundant with two populations accounting for 40 to 80% of the purple sulfur bacteria depending on the season (31). These populations, designated D and F, were not represented by a cultured relative, whereas the remaining populations were identified as Lamprocystis purpurea and Lamprocystis roseopersicina. L. purpurea and L. roseopersicina were found in numbers about 1 order of magnitude lower than populations D and F (34), similar to numbers for C. okenii and those for green sulfur bacteria that were entirely represented by Chlorobium phaeobacteroides (34).
The fluorescent probes designed in those studies (31, 34) were subsequently used to monitor enrichment cultures in an attempt to isolate representative strains of all populations (26). For this purpose, water samples were obtained from the chemocline in October 1999 and October 2003. These samples differed significantly in color with the typical purple-red color characteristic for purple sulfur bacteria in samples from 1999 and a much more brownish color typical for brown members of the green sulfur bacteria in samples from 2003, indicating large shifts in community structure in time between purple sulfur and green sulfur bacteria, and not only among populations of purple sulfur bacteria as previously described for an annual cycle (34). In addition, several enrichments of purple and green sulfur bacteria were obtained that did not hybridize with the established probes, indicating a larger diversity of the community structure of phototrophic sulfur bacteria in the chemocline than revealed in our previous analyses (7, 31, 32).
The aim of this study was to employ comparative 16S rRNA gene sequence analysis to characterize the potentially new populations of purple and green sulfur bacteria over an extended period of time. We determined their significance in water samples of the chemocline by in situ hybridization with specific probes and the extent of changes in community structure of phototrophic sulfur bacteria in time in water samples obtained from the chemocline of Lake Cadagno in October for the years 1994 to 2003.
MATERIALS AND METHODS
Sequence retrieval from enrichment cultures.
Enrichment cultures of phototrophic sulfur bacteria were obtained from water samples taken from the chemocline of Lake Cadagno with a “Friedinger” type bottle (Zuellig AG, Rheineck, Switzerland) at the maximum of turbidity corresponding to the highest bacterial density in October 1999 and 2003. As described in detail previously (26), phototrophic sulfur bacteria were initially enriched in liquid medium containing 0. 5 g KH2PO4 liter−1, 0.34 g NH4Cl liter−1, 0.5 g MgSO4 · 7H2O liter−1, 0.25 g CaCl2 · 2H2O liter−1, 0.34 g KCl liter−1, 1.5 g NaHCO3 liter−1, 0.5 ml trace element solution SL10, and 0.02 mg vitamin B12 (10). The medium was reduced with 0.3 g liter−1 Na2S · 9H2O (1.10 mM final concentration) and adjusted to a pH around 7.2 or 6.8 for Chromatiaceae (33) and Chlorobiaceae (23), respectively. Media were prepared in a 2-liter bottle with an N2/CO2 (80%/20%) gas phase according to Widdel and Bak (36). Enrichments in liquid medium were resuspended in 5 ml liquid medium before inoculation into agar shake dilution series (1%, vol/vol) prepared by the Hungate technique (27, 36). All cultures were incubated at room temperature (20 to 23°C) with a 6 h light/6 h dark photoperiod at light intensities of 4 to 8 and 5 to 20 μE m−2 s−1 for Chlorobiaceae and Chromatiaceae, respectively, generated with an incandescent 40-W bulb placed at a distance of 60 cm from the cultures (10). Enrichments and resuspended colonies of the agar shake dilution series were analyzed for target organisms by in situ hybridization with Cy3-labeled oligonucleotide probes listed in Table 1 (31, 32). Probes were synthesized at MWG Biotech, Reinach, Switzerland.
TABLE 1.
Oligonucleotide probes
| Probe | Targeta | Sequence (5′→3′) (% formamide in hybridization buffer) | Reference or source |
|---|---|---|---|
| Purple sulfur bacteria | |||
| Cmok453 | Chromatium okenii (DSM 169) 16S rRNA, pos. 453-479 | AGCCGATGGGTATTAACCACCAGGTT (35%) | 31 |
| Apur453 | Lamprocystis purpurea (DSM 4197) 16S rRNA, pos. 453-479 | TCGCCCAGGGTATTATCCCAAACGAC (40%) | 31 |
| Laro453 | Lamprocystis roseopersicina (DSM 229) 16S rRNA, pos. 453-479 | CATTCCAGGGTATTAACCCAAAATGC (30%) | 31 |
| S453D | Clone 261 from Lake Cadagno 16S rRNA, pos. 453-479 | CAGCCCAGGGTATTAACCCAAGCCGC (40%) | 31 |
| S453F | Clone 371 from Lake Cadagno 16S rRNA, pos. 453-479 | CCCTCATGGGTATTARCCACAAGGCG (40%) | 31 |
| S453H | Clone 222 from Lake Cadagno 16S rRNA, pos. 453-478 | GACGGAACGGTATTAACGCCCCGCTT (10%) | This study |
| S448 | Clone 249 from Lake Cadagno 16S rRNA, pos. 448-468 | TATTGACCCCGCGCTTTTCTT (25%) | This study |
| Green sulfur bacteria | |||
| Chlp441 | Chlorobium phaeobacteroides (DSM 266), 16S rRNA, pos. 441-464 | AAATCGGGATATTCTTCCTCCAC (20%) | 34 |
| Chlc190 | Chlorobium clathratiforme (DSM 5477), 16S rRNA, pos. 190-211 | GGCAGAACAACCATGCGATTGT (20%) | This study |
pos., position.
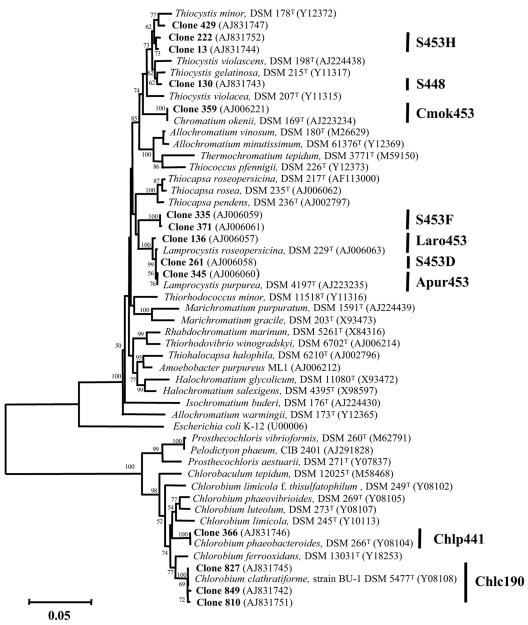
Cultures of resuspended colonies that did not hybridize with any of our probes were initially identified by comparative 16S rRNA gene sequence analysis. Nucleic acids were extracted from these cultures using the MagNA Pure LC automated extractor (Roche Molecular Biochemicals, Indianapolis, Ind.) and the DNA isolation extraction kit produced by the same manufacturer. 16S rRNA genes were amplified and amplicons purified as described previously (31) and sequenced with an ABI PRISM Ready Reaction dye deoxy terminator cycle sequencing kit and an ABI Prism 310 automated sequencer (Perkin-Elmer, Rodgau-Jügesheim, Germany). Sequences were aligned with a subset of bacterial 16S rRNA gene sequences obtained from the Ribosomal Database Project (22) by using the CLUSTAL W service at the European Bioinformatics Institute (16). Sequences from a 16S rRNA gene clone library of Lake Cadagno (7, 31, 32) as well as sequences of other purple and green sulfur bacteria were included in the phylogenetic analysis. Phylogenetic relationships were estimated using MEGA, version 2.0 (19) (Fig. 1).
FIG. 1.
Neighbor-joining tree depicting the phylogenetic positions of specific clones from a 16S rRNA gene clone library from the chemocline of Lake Cadagno and from enrichments of phototrophic sulfur bacteria within the purple and green sulfur bacteria. Acronyms represent specific probes, and bars represent their respective targets (see also Table 1).
Probe design and evaluation.
Based on comparative analysis of sequences retrieved from resuspended cultures and those of reference organisms, oligonucleotide probes S453H, S448, and Chlc190 were designed (Fig. 1; Table 1). Probe specificity with reference to available 16S rRNA sequences was checked with the ARB program (20) and in the EMBL/GenBank databases using FASTA (24). Clones prepared as described for the clone-fluorescent in situ hybridization technique (28), pure cultures of C. okenii DSM 169, Chromatium vinosum DSM 180, L. roseopersicina DSM 229, L. purpurea DSM 4197, Amoebobacter roseus DSM 235, Burkholderia cepacia DSM 50181, Brevundimonas diminuta DSM 1635, and Campylobacter jejuni DSM 4688 and of water samples from the chemocline of Lake Cadagno were used to test probe specificity and to establish appropriate in situ hybridization conditions for the specific detection.
In situ hybridization with fluorescent (Cy3-labeled) oligonucleotide probes was performed on aliquots (1 μl) of paraformaldehyde-fixed bacteria or water samples (n = 3) spotted onto gelatin-coated slides [0.1% gelatin, 0.01% KCr(SO4)2] (13). Hybridizations were performed in 9 μl of hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl, 5 mM EDTA, 0.01% sodium dodecyl sulfate [SDS]; pH 7.2) in the presence of 10 to 40% formamide depending on the probe (Table 1), 1 μl of the probe (25 ng μl−1), and 1 μl of a solution of DAPI (4′,6′-diamidino-2-phenylindole; 200 ng μl−1) at 46°C for 2 h (37). After hybridization, the slides were washed in buffer containing 20 mM Tris-HCl, pH 7.2; 10 mM EDTA; 0.01% SDS; and either 450, 318, 159, 112, 80, or 56 mM NaCl depending on the formamide concentration during hybridization (10, 20, 25, 30, 35, and 40%, respectively) for 20 min at 48°C. They were subsequently rinsed with distilled water and air dried (37). The slides were mounted with Citifluor AF1 (Citifluor Ltd., London, United Kingdom) and examined by epifluorescence microscopy using filter sets F31 (AHF Analysentechnik, Tübingen, Germany; D360/40, 400DCLP, and D460/50 for DAPI) and F41 (AHF Analysentechnik; HQ535/50, Q565LP, and HQ610/75 for Cy3). Microorganisms were counted at 1,000× magnification in 40 fields covering an area of 0.01 mm2 each (11). Numbers were expressed as means ± standard errors.
Collection and in situ analysis of chemocline samples.
For chemical and microbial analyses, water samples were collected from the chemocline of Lake Cadagno over the center of the lake, at its deepest point (21 m), at the end of the summer season (October) before partial water mixing and surface freezing over a period of 10 years (1994 to 2003, except for 1996, when no samples were taken). The chemocline and the bacterial plume in Lake Cadagno were located at each sampling date using temperature, conductivity, pH, dissolved oxygen, turbidity, and redox potential measurements with a YSI 6000 profiler (Yellow Springs Inc., Yellow Springs, OH) (31, 32). In addition, photosynthetically available light transmission conditions were determined down to the chemocline in steps of 0.1 m using two LI-193SA spherical quantum sensors and a LI-COR 1000 data logger (LI-COR Ltd., Lincoln, NE). These measurements were used to calculate vertical attenuation coefficients (Kd) of photosynthetically available radiation (8) for the mixolimnion and the bacterial layer. The YSI 6000 profiler and LI-COR sensors were fixed at the lowest part of a thin-layer pneumatic multisyringe sampler (3, 4, 31, 32), which was used to take 20 simultaneous samples of 100 ml over a total depth of 2 m.
From the water samples, 11-ml subsamples were immediately transferred to screw-cap tubes containing 0.8 ml of a 4% zinc acetate solution that were stored on ice and used to determine sulfide concentrations colorimetrically (12) using a Merck (Switzerland) Spectroquant kit (31, 32). Fifteen-milliliter subsamples were filtered immediately after sampling through 0.22-μm polycarbonate membrane filters (25-mm diameter; Millipore, Volketswil, Switzerland) (13). Bacteria were fixed by overlaying the filters with 4% paraformaldehyde in phosphate-buffered saline (PBS; 0.13 M NaCl, 7 mM Na2HPO4, 3 mM NaH2PO4, pH 7.2) for 30 min at room temperature (1). The filters were subsequently rinsed twice with PBS by vacuum filtration and transferred into plastic bags with 1 ml of 50% ethanol in PBS. In sealed bags, the bacterioplankton was released from filters and resuspended by slightly massing the filter with thumb and forefinger (18). The complete release of the bacteria from filters was checked microscopically after DAPI staining. Resuspended bacterial cells were than transferred into Eppendorf tubes and stored in 50% ethanol in PBS at −20°C until further use (1, 31). Potential long-term storage effects on the detection of bacterial cells were ruled out after samples obtained and first analyzed 1994 were reanalyzed after DAPI staining and in situ hybridization. Equivalents of mixed samples from each depth over the whole chemocline were pooled, and the cell numbers obtained were averaged over the whole chemocline.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences obtained in this study were deposited in the EMBL/GenBank databases with accession numbers AJ831742 to AJ831747, AJ831751, and AJ831752.
RESULTS AND DISCUSSION
Sequence retrieval and probe design.
Sequence analysis of clones harboring PCR-amplified, almost-complete 16S rRNA gene fragments from enrichment cultures that did not hybridize with probes previously used to characterize the population dynamics of purple and green sulfur bacteria retrieved two groups of sequences clustering with those of purple sulfur bacteria and one group clustering with green sulfur bacteria (Fig. 1). Within the purple sulfur bacteria, clone 130 was closely related to Thiocystis gelatinosa while clones 13, 222, and 429 were related to Thiocystis minor. Clones 810, 827, and 849 represented the green sulfur bacterium Chlorobium clathratiforme (Fig. 1). Sequences related to these bacteria have not been found in the original screening of the library, which initially only retrieved sequences of C. okenii and four populations of small-celled purple sulfur bacteria related to the genus Lamprocystis (31), and later that of C. phaeobacteroides (34). The failure to detect these potential members of the community of phototrophic sulfur bacteria might be due to two methodological constraints: (i) restriction fragment analysis (ARDRA) that was used in the initial screening to reduce sequence redundancy and to identify representative clones in the library (9) might have resulted in the omission of potentially new sequences, and (ii) samples used for the generation of the gene clone library, the initial in situ analysis, and the enrichment cultures were obtained in different years and seasons (June 1994, October 1998, and October 1999 and 2003, respectively) which could have resulted in large differences in the abundance of specific populations as indicated previously (34). Consequently, analyses of microbial diversity based on sequence information require linking this information to the organisms and an evaluation of their significance in the same environmental sample or studies on multiple samples taken at comparable times in the season.
Probe design and evaluation.
Oligonucleotide probe S453H targeted clones 13 and 222 related to T. minor but not the closely related clone 429 and T. minor (17) that displayed two and four mismatches to this probe, respectively. Other rRNA gene sequences exhibiting fewer mismatches were not retrieved from the EMBL database. A similarly strong specificity was obtained for probe S448, targeting clone 130, related to T. gelatinosa (Fig. 1). Database analysis showed one mismatch to Thiobaca trueperi (accession no. AJ404007) and Geobacter sulfurreducens (accession no. AE017215) and five mismatches to T. gelatinosa (14, 17). Probe Chlc190, designed to detect clones 810, 827, and 849 and their cultured relatives Pelodictyon phaeoclathratiforme and C. clathratiforme, also did not display any significant similarity to other rRNA gene sequences in the database.
In situ hybridization of clones 13, 130, and 810, prepared as described for the clone-fluorescent in situ hybridization technique (28), indicated that formamide concentrations of 10, 25, and 20%, respectively, were optimal for Cy3-labeled oligonucleotide probes S453H, S448, and Chlc190. While none of the pure cultures hybridized under these conditions, cells with morphologies resembling those of either small-celled purple sulfur bacteria (probes S453H and S448) or green sulfur bacteria (probe Chlc190) were detected with bright signals in the environmental samples (data not shown). Significant numbers of these cells, however, were not retrieved by any of these probes in the sample analyzed (October 1999), suggesting that the respective bacteria did not represent prominent populations in the chemocline at this time.
In situ analysis of chemocline samples.
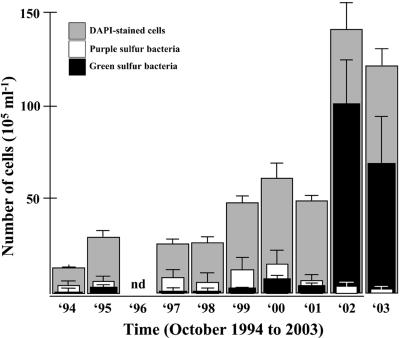
Seasonal analyses of water samples of the chemocline of Lake Cadagno between 1994 and 2003 revealed large shifts in abundance between purple and green sulfur bacteria and among different populations and a continuous increase in numbers of DAPI-stained cells. During the first 8 years, the increase was slow and about fourfold from (13 ± 1) × 105 ml−1 in October 1994 to (49 ± 2) × 105 ml−1 in October 2001 with a twofold increase in 1999 (Fig. 2). The latter numbers increased about threefold for years 2002 and 2003, with maximum numbers reaching to (140 ± 14) × 105 ml−1 in October 2002 (Fig. 2). The initial increase in cell numbers was accompanied by a concomitant slight increase in numbers of both purple and green sulfur bacteria, with numbers of purple sulfur bacteria generally higher than those of green sulfur bacteria. During the following 2 years, however, numbers of purple sulfur bacteria decreased by about 1 order of magnitude from their highest number in October 2000 with (15 ± 7) × 105 ml−1 to (6 ± 1) × 105 and (2 ± 1) × 105 ml−1 in 2002 and 2003, respectively (Fig. 2), while concomitantly numbers of green sulfur bacteria increased by more than 1 order of magnitude from (7 ± 1) × 105 ml−1 in October 2000 to (100 ± 23) × 105 ml−1 in October 2002 (Fig. 2). During the first 8 years, purple sulfur bacteria thus represented the most prominent group of phototrophic sulfur bacteria, accounting for between 70 and 95% of the phototrophic sulfur bacteria. The dominance of purple sulfur bacteria over green sulfur bacteria in the chemocline was reversed in the following 2 years of the analysis, with the percentage of purple sulfur bacteria being reduced to about 5% and that of green sulfur bacteria increased to up to 95% of the phototrophic sulfur bacteria (Fig. 2 and 3).
FIG. 2.
Numerical abundance of all cells (i.e., DAPI-stained cells, grey bars), and of purple (white bars) and green sulfur bacteria (black bars) in samples from October 1994 to October 2003 averaged over the whole chemocline of Lake Cadagno. Numbers were expressed as means ± standard errors.
FIG. 3.
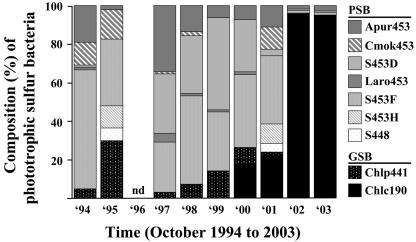
Relative abundance of different populations of phototrophic sulfur bacteria (%) for samples from October 1994 to October 2003 averaged over the whole chemocline of Lake Cadagno. Acronyms represent specific probes for the detection of purple sulfur bacteria (PSB; Cmok453, targeting Chromatium okenii; Apur453, Laro453, S453D, and S453F, targeting Lamprocystis purpurea, L. roseopersicina, and populations D and F, not yet represented by cultured relatives, respectively; and S453H and S448, targeting two populations within the genus Thiocystis) and green sulfur bacteria (GSB; Chlc190, targeting Chlorobium clathratiforme, and Chlp441, targeting C. phaeobacteroides).
In the initial 8 years, purple sulfur bacteria were represented by up to seven different populations (C. okenii, L. purpurea, L. roseopersicina, populations D and F, and the two populations related to T. minor and T. gelatinosa) each year (Fig. 3). The most stable population with respect to abundance was population F, which accounted for between 25 and 60% of the phototrophic sulfur bacteria (Fig. 3). C. okenii and all other populations of the genus Lamprocystis were usually present, except for L. roseopersicina and population D in 1995, but abundance fluctuated significantly (Fig. 3). The newly discovered populations related to the genus Thiocystis were only detected in significant numbers in 1995 and 2001, where they represented between 5 and 10% of the phototrophic sulfur bacteria. Green sulfur bacteria were initially entirely represented by C. phaeobacteroides, which, however, was replaced by bacteria related to C. clathratiforme within 2 years at the end of the 8-year period. In the last 2 years of the analysis, green sulfur bacteria and thus up to 95% of the phototrophic sulfur bacteria were represented by C. clathratiforme (Fig. 3).
The observations for the initial 8 years essentially reflect those from many studies performed during the last century that describe purple sulfur bacteria as the typical and characteristic bacterial group in the chemocline of Lake Cadagno (see references 25 and 33 for reviews). As in this study, the most abundant populations were found to belong to the genus Lamprocystis (31, 33, 34). In contrast to Thiocystis, all populations of the genus Lamprocystis form aggregates with up to 900 cells in the chemocline of Lake Cadagno (31, 34). In these aggregates, they are associated with sulfate-reducing bacteria that are also able to grow by disproportionation of inorganic sulfur compounds (26). Aggregates can provide distinct but relatively stable microenvironmental conditions (29) and thus a growth advantage in a habitat with rapid changes of environmental conditions (e.g., intensity of light and sulfide concentrations) (9, 15, 21). Potential limitation for sulfide in these aggregates could be overcome by the synergistic association with the sulfate-reducing bacteria that presumably resembles a source-sink relationship for sulfide between the sulfate-reducing and the purple sulfur bacteria (26). Fluctuations in populations of purple sulfur bacteria between years most likely reflect small variations in physicochemical conditions in the chemocline since purple sulfur bacteria of the genera Chromatium and Lamprocystis have been found microstratified in the chemocline of Lake Cadagno with large shifts in the significance of specific populations analyzed over an annual cycle (34).
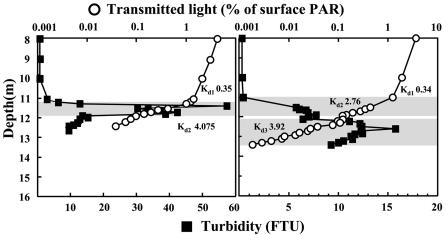
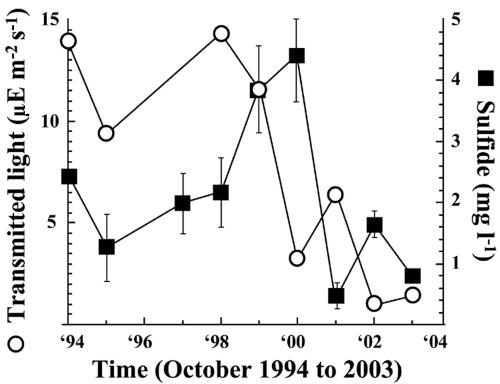
The most prominent observation of this study is the pronounced shift in dominance from purple sulfur to green sulfur bacteria and between populations of green sulfur bacteria from C. phaeobacteroides to C. clathratiforme. This shift becomes evident in samples of the year 2000 for the first time, suggesting changes in environmental conditions in the chemocline before 2000. Basic environmental conditions, such as temperature, conductivity, pH, dissolved oxygen, and redox potential, used to localize the chemocline, however, were similar at all times (data not shown). Kd values for photosynthetically available radiation were also similarly high in the mixolimnion and chemocline at all times, indicating low absorption and thus good light transmission in the mixolimnion and large absorption in the bacterial plume of the chemocline (Fig. 4). Profiles of turbidity and Kd values in the chemocline, however, differed before and after 2000. Before 2000, both turbidity and Kd profiles indicate the presence of a well-defined bacterial plume with a fast and steady reduction in light transmission with depth (Fig. 4). Profiles in and after 2000 show a turbidity profile with lower values that is expanding into deeper parts of the chemocline. Light transmission, however, was already reduced above the major turbidity peak to values found below the major turbidity peak before 2000 (Fig. 4). As a consequence, about 10-fold-lower light intensities reached the surface of this plume. Concomitant with the population shifts a reduction in transmitted light reaching the chemocline from usually above 10 μE m−2 s−1 before 2000 to below 5 μE m−2 s−1 after 2000 was observed (Fig. 5). Sulfide concentrations (mean values over the chemocline) increased between 1998 and 2000, but decreased afterwards and remained low between 2001 and 2003 (Fig. 5).
FIG. 4.
Profiles of turbidity and Kd of photosynthetically available radiation (PAR) in the mixolimnion and the chemocline. The October 1998 profiles (left) are representative for profiles before 2000, and the October 2002 profiles (right) are representative for those in and after 2000.
FIG. 5.
Intensity values of transmitted light reaching the chemocline (μE m−2 s−1) and sulfide concentrations (mg liter−1) averaged over the whole chemocline of Lake Cadagno analyzed from October 1994 to October 2003.
Kd values at the upper chemocline border in October in and after 2000 are similar to those measured in the bacterial plume experienced during winter when the lake is covered by 2 m of ice and snow (34). At this time, numbers of purple sulfur bacteria were generally 1 order of magnitude lower than in October, indicating negative effects of reduced light conditions on purple sulfur bacteria (34). Since green sulfur bacteria require approximately one-fourth of the light intensity of the purple sulfur bacteria in order to grow at comparable growth rates (2) and show different light absorption optima than purple sulfur bacteria (6), their large increase in numbers under these conditions is a logical consequence. However, C. phaeobacteroides, the only detectable green sulfur bacterium before 2000, was only detected when light intensities reaching the chemocline were relatively high, that is, during summer and fall (34). At this time, the species was distributed over the whole chemocline and thus encountered a light intensity range from high to low and, similar to reports for other meromictic lakes (35), was present at the same depths as purple sulfur bacteria (34).
Since no further environmental data on Lake Cadagno are available, a causal relationship between population shifts and environmental factors cannot be established. Speculations, however, could be based on results obtained for two meromictic lakes in the same larger region as Lake Cadagno that identified unusually deep mixing events due to very cold winters in 1998 and 1999 as a reason for enhanced nutrient availability, especially that of phosphorus, resulting in a 10- to 20-fold increase in phytoplankton density between 1998 and 2000 (30). Although our data on both biotic and abiotic changes in the chemocline of Lake Cadagno support mixing events as the cause for the changes, the discussion about potential reasons for the change in populations and environmental data remains highly speculative. Future perspectives will therefore include studies on the relationship between population dynamics and nutrient availability during the season and the effect of nutrient availability on the potential reversal of the population shifts in the next years.
Acknowledgments
This work was supported by grants from the Swiss National Science Foundation (NF31-46855.96) and the canton of Ticino (Switzerland).
We are indebted to Nadia Ruggeri-Bernardi, AnnaPaola Caminada, Sandro Peduzzi, and Laura Pezzoni for technical support.
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biebl, H., and N. Pfennig. 1978. Growth yields of green sulfur bacteria in mixed cultures with sulfur and sulfate reducing bacteria. Arch. Microbiol. 117:9-16. [Google Scholar]
- 3.Bosshard, P. P., Y. Santini, D. Grüter, R. Stettler, and R. Bachofen. 2000. Bacterial diversity and community composition in the chemocline of in the meromictic Lake Cadagno as revealed by 16S rDNA analysis. FEMS Microbiol. Ecol. 31:173-182. [DOI] [PubMed] [Google Scholar]
- 4.Bosshard, P. P., R. Stettler, and R. Bachofen. 2000. Seasonal and spatial community dynamics in the meromictic Lake Cadagno. Arch. Microbiol. 174:168-174. [DOI] [PubMed] [Google Scholar]
- 5.Camacho, A., J. Erez, A. Chicote, M. Florin, M. M. Squires, C. Lehmann, and R. Bachofen. 2001. Microbial microstratification, inorganic carbon photoassimilation and dark carbon fixation at the chemocline of the meromictic Lake Cadagno (Switzerland) and its relevance to the food web. Aquat. Sci. 63:91-106. [Google Scholar]
- 6.Del Don, C., K. W. Hanselmann, R. Peduzzi, and R. Bachofen. 2001. The meromictic alpine Lake Cadagno: orographical and biogeochemical description. Aquat. Sci. 63:70-90. [Google Scholar]
- 7.Demarta, A., M. Tonolla, A.-P. Caminda, N. Ruggeri, and R. Peduzzi. 1998. Phylogenetic diversity of the bacterial community from the anoxic layer of the meromictic Lake Cadagno. Doc. Ist. Ital. Idrobiol. 63:19-30. [Google Scholar]
- 8.Dubinsky, Z. 1980. Light utilization efficiency in natural phytoplankton communities, p. 83-97. In P. G. Falkowski (ed.), Primary productivity in sea. Plenum Press, New York, N.Y.
- 9.Egli, K., M. Wiggli, J. Klug, and R. Bachofen. 1998. Spatial and temporal dynamics of the cell density in a plume of phototrophic microorganisms in their natural environment. Doc. Ist. Ital. Idrobiol. 63:121-126. [Google Scholar]
- 10.Eichler, B., and N. Pfennig. 1988. A new purple sulfur bacterium from stratified freshwater lakes, Amoebobacter purpureus sp. nov. Arch. Microbiol. 149:395-400. [Google Scholar]
- 11.Fischer, K., D. Hahn, R. I. Amann, O. Daniel, and J. Zeyer. 1995. In situ analysis of the bacterial community in the gut of the earthworm Lumbricus terrestris L. by whole-cell hybridization. Can. J. Microbiol. 41:666-673. [Google Scholar]
- 12.Gilboa-Garber, N. 1971. Direct spectrophotometric determination of inorganic sulfide in biological materials and in other complex mixtures. Anal. Biochem. 43:129-133. [DOI] [PubMed] [Google Scholar]
- 13.Glöckner, F. O., R. I. Amann, A. Alfreider, J. Pernthaler, R. Psenner, K. Trebesius, and K.-H. Schleifer. 1996. An optimized in situ hybridization protocol for planktonic bacteria. Syst. Appl. Microbiol. 19:403-406. [Google Scholar]
- 14.Guyoneaud, R., R. Matheron, W. Liesack, J. Imhoff, and P. Caumette. 1997. Thiorhodococcus minus, gen. nov., and sp. nov., a new purple sulfur bacterium isolated from coastal lagoon sediments. Arch. Microbiol. 168:16-23. [DOI] [PubMed] [Google Scholar]
- 15.Hanselmann, K., and R. Hutter. 1998. Geomicrobiological coupling of sulfur and iron cycling in anoxic sediments of a meromictic lake: sulfate reduction and sulfide sources and sinks in Lake Cadagno. Doc. Ist. Ital. Idrobiol. 63:85-98. [Google Scholar]
- 16.Higgins, D., J. Thompson, T. Gibson, J. D. Thompson, D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imhoff, J. F., J. Süling, and R. Petri. 1998. Phylogenetic relationships among the Chromatiaceae, their taxonomic reclassification and description of the new genera Allochromatium, Halochromatium, Isochromatium, Marichromatium, Thiococcus, Thiohalocapsa, and Thermochromatium. Int. J. Syst. Bacteriol. 48:1129-1143. [DOI] [PubMed] [Google Scholar]
- 18.International Standards Organisation. 1998. Water quality—detection and enumeration of Legionella. Intern. Stand. 11731:1-16. [Google Scholar]
- 19.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 20.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Y. Kumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lüthy, L., M. Fritz, and R. Bachofen. 2000. In situ determination of sulfide turnover rates in a meromictic alpine lake. Appl. Environ. Microbiol. 66:712-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maidak, B. L., G. J. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. R. Woese. 1997. The RDP (Ribosomal Database Project). Nucleic Acids Res. 25:109-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Overmann, J., and N. Pfennig. 1989. Pelodictyon phaeoclathratiforme sp. nov., a new brown-colored member of the Chlorobiaceae forming net-like colonies. Arch. Microbiol. 152:401-406. [Google Scholar]
- 24.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peduzzi, R., R. Bachofen, and M. Tonolla. 1998. Lake Cadagno, a meromictic alpine lake. Doc. Ist. Ital. Idrobiol. 63:1-152. [Google Scholar]
- 26.Peduzzi, S., M. Tonolla, and D. Hahn. 2003. Isolation and characterization of aggregate-forming sulfate-reducing and purple sulfur bacteria from the chemocline of meromictic Lake Cadagno, Switzerland. FEMS Microbiol. Ecol. 45:29-37. [DOI] [PubMed] [Google Scholar]
- 27.Pfennig, N. 1978. Rhodocyclus purpureus gen. nov. and sp. nov., a ring-shaped, vitamin B12-requiring member of the family Rhodospirillaceae. Int. J. Syst. Bacteriol. 28:283-288. [Google Scholar]
- 28.Schramm, A., B. M. Fuchs, J. L. Nielsen, M. Tonolla, and D. Stahl. 2003. Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation; screening of clone libraries. Environ. Microbiol. 4:713-720. [DOI] [PubMed] [Google Scholar]
- 29.Schramm, A., C. M. Santegoeds, H. K. Nielsen, H. Ploug, M. Wagner, M. Pribyl, J. Wanner, R. I. Amann, and D. DeBeer. 1999. On the occurrence of anoxic microniches, denitrification, and sulfate reduction in aerated activated sludge. Appl. Environ. Microbiol. 65:4189-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simona, M. 2003. Winter and spring mixing depths affect the trophic status and composition of phytoplankton in the northern meromictic basin of Lake Lugano. J. Limnol. 62:190-206. [Google Scholar]
- 31.Tonolla, M., A. Demarta, R. Peduzzi, and D. Hahn. 1999. In situ analysis of phototrophic sulfur bacteria in the chemocline of meromictic Lake Cadagno (Switzerland). Appl. Environ. Microbiol. 65:1325-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tonolla, M., A. Demarta, S. Peduzzi, D. Hahn, and R. Peduzzi. 2000. In situ analysis of sulfate-reducing bacteria related to Desulfocapsa thiozymogenes in the chemocline of meromictic Lake Cadagno (Switzerland). Appl. Environ. Microbiol. 66:820-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tonolla, M., S. Peduzzi, A. Demarta, R. Peduzzi, and D. Hahn. 2004. Phototropic sulfur and sulfate-reducing bacteria in the chemocline of meromictic Lake Cadagno, Switzerland. J. Limnol. 63:157-166. [Google Scholar]
- 34.Tonolla, M., S. Peduzzi, D. Hahn, and R. Peduzzi. 2003. Spatio-temporal distribution of phototrophic sulfur bacteria in the chemocline of meromictic Lake Cadagno (Switzerland). FEMS Microbiol. Ecol. 43:89-98. [DOI] [PubMed] [Google Scholar]
- 35.Vila, X., C. A. Abella, J. B. Figueras, and J. P. Hurley. 1998. Vertical models of phototrophic bacterial distribution in the metalimnetic microbial communities of several freshwater North-American kettle lakes. FEMS Microbiol. Ecol. 25:287-299. [Google Scholar]
- 36.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Trüper, M. Dworking, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes. Springer Verlag, New York, N.Y.
- 37.Zarda, B., D. Hahn, A. Chatzinotas, W. Schoenhuber, A. Neef, R. I. Amann, and J. Zeyer. 1997. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch. Microbiol. 168:185-192. [Google Scholar]