Abstract
Lactoperoxidase is an enzyme that contributes to the antimicrobial defense in secretory fluids and that has attracted interest as a potential biopreservative for foods and other perishable products. Its antimicrobial activity is based on the formation of hypothiocyanate (OSCN−) from thiocyanate (SCN−), using H2O2 as an oxidant. To gain insight into the antibacterial mode of action of the lactoperoxidase enzyme system, we generated random transposon insertion mutations in Escherichia coli MG1655 and screened the resultant mutants for an altered tolerance of bacteriostatic concentrations of this enzyme system. Out of the ca. 5,000 mutants screened, 4 showed significantly increased tolerance, and 2 of these had an insertion, one in the waaQ gene and one in the waaO gene, whose products are involved in the synthesis of the core oligosaccharide moiety of lipopolysaccharides. Besides producing truncated lipopolysaccharides and displaying hypersensitivity to novobiocin and sodium dodecyl sulfate (SDS), these mutants were also shown by urea-SDS-polyacrylamide gel electrophoresis analysis to have reduced amounts of porins in their outer membranes. Moreover, they showed a reduced degradation of p-nitrophenyl phosphate and an increased resistance to ampicillin, two indications of a decrease in outer membrane permeability for small hydrophilic solutes. Additionally, ompC and ompF knockout mutants displayed levels of tolerance to the lactoperoxidase system similar to those displayed by the waa mutants. These results suggest that mutations which reduce the porin-mediated outer membrane permeability for small hydrophilic molecules lead to increased tolerance to the lactoperoxidase enzyme system because of a reduced uptake of OSCN−.
Lactoperoxidase (LP) is a heme peroxidase that catalyzes the oxidation by hydrogen peroxide of a wide range of substrates. It is found in secretions such as milk, tears, saliva, and airway mucus from humans and other mammals and, together with several other related heme peroxidases, has an important function in the host defense against microbial invaders (8, 17, 46). The major physiological substrate for LP is thiocyanate (SCN−), which is present in milk, for example, at concentrations ranging from 1 to 15 ppm and which is oxidized to hypothiocyanate (OSCN−), a reactive oxidant, and to some other minor reaction products. Bromide and iodide anions may also be oxidized by LP, but unlike, for instance, chloroperoxidase and myeloperoxidase, LP is unable to oxidize the chloride ion (7). The antimicrobial activity of peroxidase enzyme systems is based on the oxidative power of their reaction products, and enzymes and other proteins in the bacterial cell membrane that have exposed thiol groups (-SH) are believed to be the major targets of the LP/SCN− system, as they are converted to disulfides. As a result, exposed cells would accumulate membrane damage, resulting in a loss of the pH gradient, K+ leakage, and an inhibition of respiration or the transport of solutes such as amino acids and glucose (8, 35, 41). However, other cellular thiol compounds, such as glutathione (20), can also be oxidized, at least in vitro, but it remains to be demonstrated whether these oxidations take place in vivo and whether LP activity destabilizes the cellular redox homeostasis in this way. When LP is applied at physiological concentrations, the effect of the LP system is mainly bacteriostatic, which may be related to the fact that thiol-disulfide conversions are reversible. At higher concentrations or when cells are appropriately sensitized, for example, by heat (48) or hydrostatic pressure treatment (10), the LP system can have a bactericidal effect. This bactericidal effect may be related to the production of small amounts of oxygen radicals by the LP system (4), which cause irreversible protein oxidation by the formation of carbonyl groups.
Natural antimicrobial systems such as the lactoperoxidase system have gained considerable interest as “natural” preservatives of foods and industrial, cosmetic, and health care products, and the potential of the LP/H2O2/SCN− system to extend the shelf life of various foods has been amply demonstrated (17, 39, 43). It is assumed that the use of the LP system in foods presents little or no toxicological concern provided that the physiological exposure is not exceeded, since LP is an endogenous enzyme in humans that, in addition, is also naturally present in many foods, such as milk (7). Furthermore, several studies reporting a protective effect of LP in the diet against enteropathogens have stimulated the development of LP-containing nutraceuticals which claim health-promoting effects (13).
The cellular defense mechanisms and stress response of microorganisms against the LP system are poorly known, as opposed to the defense mechanisms against H2O2 and superoxide (O2°−), two other forms of oxidative stress, which have been described in detail for Escherichia coli. We recently demonstrated that the LP system induces a specific and unique stress response in E. coli (40). Here we report the isolation and characterization of random transposon insertion mutants of E. coli MG1655 with increased tolerance to the LP system.
MATERIALS AND METHODS
Growth media and chemicals.
The standard growth medium was Luria-Bertani (LB) medium (10 g/l tryptone, 5 g/l yeast extract, 5 g/l NaCl, 10 g/l agar for solid medium). Antibiotics (Sigma-Aldrich, Bornem, Belgium) were added as appropriate at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; and chloramphenicol, 30 μg/ml. Tryptone soy broth (TSB) (Oxoid, Basingstoke, England) was used as the growth medium for evaluations of the bacteriostatic activity of the lactoperoxidase system. Stock solutions of lactoperoxidase (10 mg/ml) and of glucose oxidase (100 units/ml) (Sigma-Aldrich) were stored at −18°C in a 50% glycerol solution in phosphate-buffered saline (2.87 mM KH2PO4, 7.12 mM K2HPO4, 0.151 M NaCl, pH 6.0). Potassium thiocyanate (KSCN) (Acros Organics, Geel, Belgium) was stored at 4°C as a 25 mM stock solution. Filter-sterilized glucose was stored at room temperature as a 20% stock solution.
Construction of MG1655 ompC and MG1655 ompF.
The one-step inactivation method of Datsenko and Wanner (5) was used to make a mutant with an insertional mutation in ompC. First, the kanamycin resistance gene (kan) from plasmid pKD4 was amplified with the primers PompmutC1 and PompmutC2 (Table 1). In addition to a priming sequence of 19 or 20 nucleotides at their 3′ ends, these primers have an ∼50-nucleotide extension homologous to ompC that will allow an exchange of the PCR product with the genomic ompC gene. The PCR product was then electroporated into MG1655 carrying the pKD46 plasmid, which expresses the arabinose-inducible λ Red recombinase (29), to promote recombination. Kanamycin-resistant transformants were selected and cured of pKD46 by growth at 37°C, which is a nonpermissive temperature for replication of this plasmid. The ompC insertional mutant was verified by PCR with the primers PompC1 and PompC2 (Table 1) and was designated LMM-PDS1.
TABLE 1.
Primers used for this study
| Primer | 5′-3′a |
|---|---|
| Pcat1 | AAGCACCGCCGGACATC |
| Pcat2 | CTCCCAGAGCCTGATAA |
| PompmutC1 | AAGGCATATAACAGAGGGTT AATAACATGAAAGTTAAAG TACTGTgtgtaggctggagctgcttc |
| PompmutC2 | CCAGACCCAGAGCTACGATG TTATCAGTGTTGATGCCAG CGTCACcatatgaatatcctcctta |
| PompC1 | TGATCGCAACCAACAAAGAA |
| PompC2 | GAATGGACTTGCCGACTGAT |
| PompF1 | TCAAACATGACGAGGTTCCA |
| PompF2 | GCAGTGGCAGGTGTCATAAA |
| Pkan | CAGTCATAGCCGAATAGCCT |
Sequences in lowercase were needed to amplify the kan gene from the pKD4 plasmid. Sequences in uppercase are homologous to the ompC gene.
The ompF::Tn5 allele from E. coli BE strain BZB1107 (2) was transduced with phage P1 into MG1655 (26). Direct transduction between these two strains did not succeed, probably because the E. coli BE DNA was degraded by the K12 restriction-modification system. Therefore, we first transduced the ompF::Tn5 allele into the restriction-negative E. coli K12 strain MT102, and from there to MG1655, to yield strain LMM-PDS2. Since the location and orientation of the transposon in ompF are unknown, the correct replacement of ompF by ompF::Tn5, which contained the kan gene, was verified by PCR with the primer couples PompF1-PompF2, Pkan-PompF1, and Pkan-PompF2 in separate reactions (Table 1).
Screening for transposon insertion mutants with increased tolerance to the LP system.
Transposon knockout mutants of E. coli MG1655 were constructed by using λNK1324, which carries a mini-Tn10 transposon with a chloramphenicol resistance gene, according to the protocol described by Kleckner et al. (15). Approximately 5,000 mutants were then screened for tolerance to the LP system by recording their growth curves in a Bioscreen C automatic growth analyzer (Thermo Electron Corporation, Vantaa, Finland) in 300 μl of TSB medium in the presence of (i) lactoperoxidase (5 μg/ml), (ii) KSCN (0.25 mM), (iii) glucose (0.4%), and (iv) glucose oxidase (0.1 units/ml). The function of the glucose oxidase enzyme and of glucose was to generate H2O2 in situ, which subsequently served as a substrate for the lactoperoxidase enzyme. Cells were inoculated at an initial density of approximately 104 CFU/ml by diluting cultures grown at 37°C in LB medium in microplates for 21 h. Incubation was done at 25°C for 44 to 64 h, and the optical density was measured every 15 min by use of a wide-band filter (405 to 600 nm). Under these conditions, the LP system caused complete growth inhibition of E. coli MG1655 for at least 40 h, while at 37°C, growth inhibition was achieved for no longer than 2 h. Mutants showing growth within <40 h were retested in five replicates with the same test, and when their phenotype was confirmed, their mini-Tn10-induced mutation was transduced via P1 into MG1655 and the LP tolerance of the transductants was examined once more.
Cloning and identification of gene knockouts.
Mini-Tn10-containing genomic DNA fragments were isolated by random cloning of ca. 40-kb large genomic DNA fragments from LP-tolerant mutants into the pWEB cosmid cloning vector (Biozym, Landgraaf, The Netherlands) and the selection of chloramphenicol-resistant transformants of E. coli EPI305 (Biozym). The genomic DNA flanking the transposon was then sequenced from the cosmid template by the use of primers complementary to the respective ends of the cat gene (32) (Pcat1 and Pcat2; Table 1). Sequence analysis was performed by MWG-Biotech (Ebersberg, Germany).
Isolation and electrophoretic separation of LPS.
For the isolation of lipopolysaccharide (LPS), a protocol described by Moller et al. (28) was used. LPS was separated by 18% Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (33) and visualized by silver staining (12).
Isolation and electrophoretic separation of outer membrane proteins.
Outer membrane proteins were isolated by freeze-thawing of cell suspensions in the presence of lysozyme and subsequent washing with 0.5% sarcosyl as described by Mecsas et al. (25). Air-dried (3 h at 37°C) pellets were weighed, dissolved in urea sample buffer, and loaded in a urea-SDS-PAGE gel, ensuring that equivalent amounts of outer membrane material (on a weight basis) were loaded in each lane. Urea was present in the sample buffer and the SDS-PAGE gel at a concentration of 8% because this improves the separation of OmpC and OmpF (45). Gels were stained with Coomassie blue.
SDS, novobiocin, and ampicillin sensitivity tests.
SDS and novobiocin sensitivities were tested as described by Moller et al. (28). Twofold serial dilutions of SDS (200 to 0.1 mg/ml) and novobiocin (200 to 1.6 μg/ml) in LB broth were made in microplates, inoculated in a final volume of 200 μl with approximately 105 CFU/ml from an overnight culture, and incubated on a rotary shaker (200 rpm) at 37°C, and after 15 h of growth, their optical densities (at 600 nm) were measured with a microplate reader (Multiskan RC; Thermo Electron Corporation). Growth was scored as positive for optical densities of ≥0.1.
For tests of ampicillin sensitivity, overnight cultures were resuspended to approximately 107 CFU/ml in honeycomb microplates containing LB broth with various concentrations of ampicillin, and growth at 37°C was monitored with a Bioscreen C automatic growth analyzer using a wide-band filter.
Evaluation of outer membrane permeability for small hydrophilic solutes.
Measurements of outer membrane permeability were based on the membrane diffusion barrier model described by Martinez et al. (22). The whole-cell alkaline phosphatase assay of Wang et al. (47) was applied to assess the outer membrane permeability. In this assay, the hydrolysis of p-nitrophenyl phosphate (pNPP) by an E. coli cell suspension under controlled incubation conditions is measured by the increase in absorption at 420 nm and then corrected for cell density (A420/A600). Since the test is conducted at a low substrate concentration and with cells transformed with the plasmid pIV26 (21) to overexpress PhoA in the periplasm, product formation will be controlled by the diffusion of pNPP through the outer membrane, and thus A420/A600 is a valid measure of outer membrane permeability.
RESULTS
Screening for mutants with increased tolerance to the LP antimicrobial system.
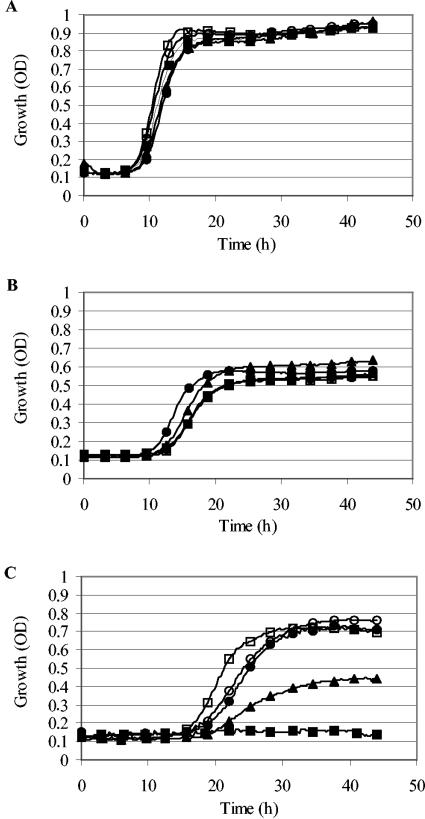
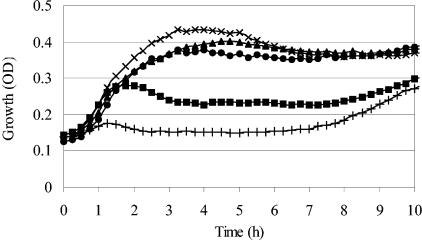
In several preparatory experiments, we determined suitable screening conditions for the detection of mutants with an increased tolerance to the LP system. The parameters that were varied include the growth medium, growth temperature, initial cell number, and component concentrations of the LP system. The conditions that were finally chosen (see Materials and Methods) ensured the complete growth inhibition of E. coli MG1655 for at least 40 h. Out of ca. 5,000 random mini-Tn10 mutants, 12 showed an increased tolerance to the LP system after a first screen, and for 4 of these, the phenotype could be confirmed with a second test. These mutants were cotransduced with mini-Tn10 by P1 transduction. The resulting strains were designated LMM-PDS3, LMM-PDS4, LMM-PDS5, and LMM-PDS6. Growth curves for E. coli MG1655 and the four selected mutants in the absence of additives, in the presence of the H2O2-generating glucose oxidase/glucose enzyme system and KSCN, and in the presence of the entire LP system (glucose oxidase, glucose, KSCN, and lactoperoxidase) are shown, respectively, in Fig. 1A, B, and C. The LP system completely inhibited the growth of E. coli MG1655 for at least 40 h. The mutants all grew in the presence of the LP system within <40 h, but compared to the growth of the wild-type strain in TSB without additives, they exhibited an extended lag phase, a lower exponential growth rate, and a lower stationary-phase cell density. Exposure to H2O2 stress from the glucose oxidase/glucose enzyme system in the absence of the LP enzyme (Fig. 1B) also resulted in partial growth inhibition, but interestingly, this inhibition was the same for the wild-type strain and the mutants, indicating that the mutants had acquired tolerance specifically to the LP system.
FIG. 1.
Growth curves for E. coli strains MG1655 (▪), LMM-PDS3 (waaO) (•), LMM-PDS4 (waaQ) (▴), LMM-PDS5 (ulaA) (○), and LMM-PDS6 (lrp) (□) incubated at 25°C. (A) Growth in TSB. (B) Growth in TSB with the addition of KSCN, glucose, and glucose oxidase. (C) Growth in TSB with the addition of KSCN, glucose, glucose oxidase, and lactoperoxidase. Only a few measurement points are labeled with symbols.
Identification of mini-Tn10 insertion sites and initial characterization of mutants LMM-PDS3 and LMM-PDS4.
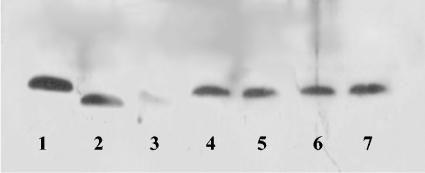
Mini-Tn10-containing fragments from the four mutants were cloned, and a short sequence of genomic DNA flanking the transposons was determined. The insertions were located in waaO, waaQ, ulaA, and lrp for mutants LMM-PDS3, LMM-PDS4, LMM-PDS5, and LMM-PDS6, respectively, and their exact locations are presented in Table 2. The finding that two insertions were in the waa (formerly rfa) operon, which is involved in core oligosaccharide synthesis of lipopolysaccharide (LPS) molecules, suggested a role for the LPS layer in the tolerance to the LP system and prompted us to investigate these two mutants in more detail. Since mutations in the waa operon result in truncated LPS molecules and, as a result, can cause hypersensitivity to SDS and hydrophobic antibiotics such as novobiocin (49), we isolated LPS, subjected it to SDS-PAGE analysis, and tested the SDS and novobiocin sensitivities of the mutants. Both waa knockout mutants (LMM-PDS3 and LMM-PDS4) did indeed have LPS molecules that were reduced in size (Fig. 2, lanes 2 and 3), while the LPS molecules of both other mutants were indistinguishable from wild-type LPS. Furthermore, both waa mutants also showed increased sensitivities to SDS and novobiocin. The MICs of SDS and novobiocin, determined by a 15-h growth test described in Materials and Methods, were reduced 2-fold for the waaO mutant and 500-fold and 12-fold, respectively, for the waaQ mutant compared to the wild-type strain MG1655 (Table 3). The LMM-PDS5 and LMM-PDS6 mutants had no altered sensitivity to SDS or novobiocin (data not shown).
TABLE 2.
Locations of transposon insertions in four lactoperoxidase-tolerant mutants
| Strain | Knocked-out gene | Coordinates of gene open reading framea | Position of transposon on coding strand | Orientation of transposon-encoded resistance marker relative to gene coding strand |
|---|---|---|---|---|
| LMM-PDS3 | waaO | 3801081-3800062 | 3801072 | Same |
| LMM-PDS4 | waaQ | 3806121-3805087 | 3805205 | Same |
| LMM-PDS5 | ulaA | 4417946-4419400 | 4418348 | Opposite |
| LMM-PDS6 | lrp | 0931818-0932312 | 0932016 | Same |
From Escherichia coli K-12 MG1655 complete genome (GenBank accession number U00096).
FIG. 2.
LPS profiles of wild-type MG1655 (lane 1), LMM-PDS3 (waaO) (lane 2), LMM-PDS4 (waaQ) (lane 3), LMM-PDS5 (ulaA) (lane 4), LMM-PDS6 (lrp) (lane 5), LMM-PDS1 (ompC) (lane 6), and LMM-PDS2 (ompF) (lane 7).
TABLE 3.
MICs of SDS and novobiocin for E. coli MG1655, LMM-PDS3 (waaO), and LMM-PDS4 (waaQ)
| Strain | MIC
|
|
|---|---|---|
| SDS (mg/ml) | Novobiocin (μg/ml) | |
| MG1655 | 100 | 100 |
| LMM-PDS3 | 50 | 50 |
| LMM-PDS4 | 0.2 | 12.5 |
Outer membrane protein content and permeability to hydrophilic molecules.
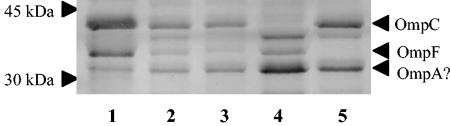
Since truncated LPS molecules can result in reduced amounts of proteins, particularly porins, being incorporated into the outer membrane (1, 16), we conducted an electrophoretic analysis of the outer membrane proteins of the waa mutants and of ompC and ompF knockout mutants for comparison. The latter allowed us to identify the OmpC and OmpF proteins on the gel. For the same amount of outer membrane material, both mutants showed reduced quantities of the OmpC and OmpF proteins compared to those in strain MG1655 (Fig. 3, lanes 2 and 3). The amount of a third protein of approximately 35 kDa that is present in outer membrane extracts of MG1655 and that has been proposed to be OmpA in other studies (3, 19) was slightly increased in both waa mutants.
FIG. 3.
SDS-PAGE gel of Coomassie blue-stained outer membrane proteins. Lanes: 1, MG1655; 2, LMM-PDS3 (waaO); 3, LMM-PDS4 (waaQ); 4, LMM-PDS1 (ompC); and 5, LMM-PDS2 (ompF). OmpC and OmpF bands were identified by comparisons to the corresponding knockout reference strains. The OmpA band was presumptively identified based on its position relative to OmpC and OmpF and by comparison with the results of Brissette et al. (3) and Liu and Ferenci (19).
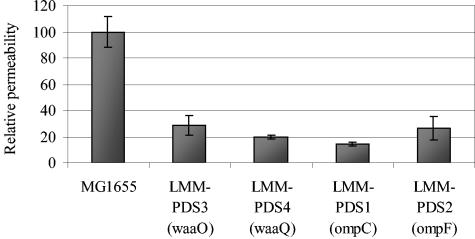
Since OmpC and OmpF form channels allowing the passive diffusion of small hydrophilic molecules through the outer membrane, we speculated that the increased tolerance of the waa mutants to the LP system might stem from a reduced uptake of OSCN−, the active component generated by the system. Therefore, we compared the outer membrane permeabilities of the different strains by using a pNPP assay and an ampicillin sensitivity test (see Materials and Methods). Figure 4 shows that compared to wild-type MG1655, both waa mutants exhibited a reduced uptake of pNPP, as was the case for porin knockout mutants of ompC and ompF which were included in the experiment as controls. Resistance to the hydrophilic antibiotic ampicillin was used as an alternative method to estimate the porin-dependent outer membrane permeability. Figure 5 shows that compared to wild-type E. coli MG1655, the waa mutants and the ompF mutant had an increased resistance to 5 μg/ml of ampicillin. In contrast, the ompC mutant showed a reduced resistance, probably because in this strain OmpF is still present and other proteins, including (the putative) OmpA, are overexpressed (Fig. 3).
FIG. 4.
Outer membrane permeability of mutant strains relative to that of MG1655. The height of each bar is correlated with the absorption at 420 nm and thus with the degradation of p-nitrophenyl phosphate by alkaline phosphatase. Each bar represents the average of three independent experiments, with the error bars corresponding to the standard deviation.
FIG. 5.
Growth curves for E. coli strains MG1655 (▪), LMM-PDS3 (waaO) (•), LMM-PDS4 (waaQ) (▴), LMM-PDS1 (ompC) (+), and LMM-PDS2 (ompF) (×) incubated at 37°C in LB supplemented with 5 μg/ml ampicillin. Each curve represents the averages of three independent experiments.
Tolerance of ompC and ompF knockout mutants to the LP system.
The tolerance of the ompC and ompF mutants to the LP system was analyzed and compared to that of the wild-type strain MG1655 and one of the isolated LP-tolerant waa mutants, using the same approach as that for the initial screening of the mutant collection. Both porin mutants showed an increased tolerance to the LP system, to a similar extent as that of the waaO mutant shown in Fig. 1C (data not shown).
DISCUSSION
For this work, we isolated four transposon insertion mutants of the Escherichia coli K12 strain MG1655 that showed increased tolerance to the antimicrobial effect of the lactoperoxidase system. Interestingly, this tolerance was specific to the lactoperoxidase system since no increased resistance to H2O2 was observed (Fig. 1). This suggests that hypothiocyanate, the presumed active antimicrobial component of the lactoperoxidase system when thiocyanate is used as a substrate, has a mode of action which is different from that of H2O2. This idea is further supported by the recent finding that the lactoperoxidase system elicits a distinct stress response in E. coli which is different from the responses induced by H2O2 and the superoxide generator plumbagin (40).
Two of the four mutants had an insertion in the central waa operon, which in E. coli K12 consists of 10 genes whose products are involved in the assembly and decoration of the core oligosaccharide of LPS (34). The LMM-PDS4 mutant has an insertion in waaQ, the first gene of the operon, whose gene product adds a heptose side chain to the last heptose of the inner core oligosaccharide. A nonpolar knockout mutation of this gene does not produce a particular phenotype (28, 49). Therefore, the LPS truncation (Fig. 2) and SDS and novobiocin sensitivity (Table 3) observed in our work suggest that the mini-Tn10 insertion in waaQ has a polar effect, knocking out the expression of the downstream genes in the operon and producing a phenotype reminiscent of deep rough mutants of E. coli (28, 49). Specifically, we can anticipate that LPS in the waaQ mutant will completely lack an outer core oligosaccharide chain due to the knockout of waaG and, additionally, will lack phosphoryl residues on both inner core heptoses due to the knockout of waaP and waaY. The waaO knockout mutant, on the other hand, still has functional waaG and waaP genes, and its LPS is thus predicted to retain an outer core oligosaccharide consisting of one glucose residue and to lack only one phosphoryl group on its inner core heptoses. The lack of an extra phosphoryl residue in the waaQ LPS may explain the extreme hypersensitivity of this mutant to the hydrophobic compounds SDS and novobiocin compared to the moderate hypersensitivity of the waaO strain. The hydrophobic properties of the waaQ LPS may also explain the difficulties that we encountered in the electrophoretic analysis of this LPS (Fig. 2).
Mutations leading to an altered LPS structure not only affect the sensitivity of E. coli to hydrophobic compounds but can also compromise the proper folding and membrane integration of outer membrane proteins such as porins and consequently alter the outer membrane's permeability for hydrophilic solutes (1, 6, 16, 18). The analysis shown in Fig. 3 confirmed that both waa mutants did indeed contain reduced amounts of OmpC and OmpF. Since we were not aware of a method to directly measure the uptake of hypothiocyanate ions (OSCN−), the presumed active antimicrobial species under our experimental conditions, through the outer membrane, we adopted two widely used methods for evaluating outer membrane permeability that make use of other hydrophilic solutes, assuming that these would provide a valid indication of OSCN− permeation. The first method uses p-nitrophenyl phosphate (pNPP), a hydrophilic molecule that enters the cell via porins (21, 24, 53) and that is converted by the periplasmic alkaline phosphatase into p-nitrophenol, a yellow compound that can be quantified by its absorption at 420 nm. At low substrate concentrations and in the presence of excess alkaline phosphatase (by phoA overexpression from the plasmid pIV26 [21]), the uptake of pNPP rather than the availability of alkaline phosphatase is the rate-limiting step (23), and the increase in absorption at 420 nm is a measure of the outer membrane permeability. Resistance to the hydrophilic antibiotic ampicillin was used as an alternative method to estimate the porin-dependent outer membrane permeability. The validity of this approach is based on the findings that ampicillin passes the outer membrane barrier through the porins, especially OmpF (30, 31), and that a reduction in porin content can increase the resistance to some antibiotics, including ampicillin (36, 37, 38). These methods demonstrated that the waaO and waaQ mutants had a lower uptake of the hydrophilic molecules pNPP and ampicillin (Fig. 4 and 5). We believe that this was caused by the lower porin content of these strains, since a similar reduction in pNPP permeability was apparent for OmpC and OmpF knockout strains and a similar ampicillin sensitivity was seen for an OmpF knockout strain. A knockout of OmpC did not affect the ampicillin sensitivity of E. coli, but this can be explained by the facts that ampicillin preferentially passes the outer membrane via OmpF (30, 31) and that the OmpC knockout strain still produces the OmpF porin (Fig. 3). Finally, the finding that both OmpF and OmpC mutants proved to be similarly resistant as the waa mutants to the lactoperoxidase system further strengthens the hypothesis that reduced outer membrane permeability for the hydrophilic OSCN− ion is the basis of the tolerance of the waa mutants to the LP system.
A reduced net uptake through the outer membrane is the basis of several emerging resistances to antimicrobial compounds in gram-negative bacteria (31, 44, 51). Pathogenic E. coli O157:H7 strains, for instance, exhibit an enhanced resistance to antimicrobials compared to other E. coli strains, which is believed to stem from differences in the permeative properties of their porins (24). Also, exposure to ampicillin results in the rapid emergence of E. coli mutants lacking OmpF (31). Although this mutation provides only a low level of resistance to ampicillin, it may allow for the temporary survival of a population that can subsequently acquire additional resistance mechanisms. Our work suggests that selection for this type of antibiotic resistance may even occur naturally in the absence of any antibiotic by selection for LP resistance in saliva, milk, tears, and airway mucus. Heme peroxidases are part of the nonspecific host defense mechanism of plants and animals. Besides lactoperoxidase, myeloperoxidase is another heme peroxidase that plays an important role in the killing of phagocytosed bacteria (11). However, its main substrate in phagocytes is Cl−, which is oxidized to HClO (hypochlorous acid). In view of the similar chemical structures and properties of the OSCN− and ClO− ions, it can be expected that a reduction of porin-mediated outer membrane permeability will also provide tolerance of the myeloperoxidase/Cl− enzyme system. Whether this will also increase the survival of porin-deficient mutants in phagocytes is unclear because the myeloperoxidase system is only one of multiple bactericidal mechanisms in these cells (11).
The development of a tolerance to OSCN− generated by the LP system and/or of ClO− is also relevant in the context of food preservation and disinfection. For example, the resistance of Salmonella to ClO− has been reported and several possible mechanisms have been suggested, but the role of porins was not investigated (27). Furthermore, besides studies of the potential of “natural” preservative systems such as the LP system to increase the safety and extend the shelf-life of perishable foods and other products, studies are also needed to address the potential emergence of LP tolerance. Finally, our research group has previously reported that a sublethal treatment with high hydrostatic pressure strongly sensitizes bacteria to the LP system, opening perspectives for the application of this combined treatment for mild food preservation (9, 10). The implications of the LP tolerance reported in the present work on the efficiency of this combined treatment should be further investigated.
In this work, we also identified two LP-resistant mutants (lrp and ulaA) that were not further investigated. Lrp is a global regulator that coordinates cellular metabolism with the nutritional state of the environment and, as such, affects the expression of multiple genes and operons, including some stress response genes (14, 42). Furthermore, Lrp-deficient mutants were found to possess a growth advantage during stationary phase (GASP phenotype) (54). UlaA is involved in the utilization of l-ascorbic acid (50, 52), an antioxidant that can neutralize OSCN− generated by the LP system. However, further work will be required to elucidate the precise mechanisms by which mutations in lrp and ulaA confer LP tolerance on E. coli.
Acknowledgments
This work was conducted in the framework of research projects financed by the K.U. Leuven Research Fund (OT/01/35) and the Fund for Scientific Research Flanders (F.W.O. G.0195.02).
REFERENCES
- 1.Ames, G. F., E. N. Spudich, and H. Nikaido. 1974. Protein composition of outer membrane of Salmonella typhimurium—effect of lipopolysaccharide mutations. J. Bacteriol. 117:406-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bainbridge, G., H. Mobasheri, G. A. Armstrong, E. J. A. Lea, and J. H. Lakey. 1998. Voltage-gating of Escherichia coli porin: a cystine-scanning mutagenesis study of loop 3. J. Mol. Biol. 275:171-176. [DOI] [PubMed] [Google Scholar]
- 3.Brissette, R. E., K. Tsung, and M. Inouye. 1992. Mutations in a central highly conserved non-DNA-binding region of OmpR, an Escherichia coli transcriptional activator, influence its DNA-binding ability. J. Bacteriol. 174:4907-4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, S. X., and P. Schopfer. 1999. Hydroxyl-radical production in physiological reactions—a novel function of peroxidase. Eur. J. Biochem. 260:726-735. [DOI] [PubMed] [Google Scholar]
- 5.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Cock, H., S. van Blokland, and J. Tommassen. 1996. In vitro insertion and assembly of outer membrane protein PhoE of Escherichia coli K-12 into the outer membrane—role of Triton X-100. J. Biol. Chem. 271:12885-12890. [DOI] [PubMed] [Google Scholar]
- 7.de Wit, J. N., and A. C. M. van Hooydonk. 1996. Structure, functions and applications of lactoperoxidase in natural antimicrobial systems. Neth. Milk Dairy J. 50:227-244. [Google Scholar]
- 8.Ekstrand, B. 1994. Lactoperoxidase and lactoferrin, p. 15-57. In V. M. Dillon and R. G. Board (ed.), Natural antimicrobial systems and food preservation. CAB International, Wallingford, United Kingdom.
- 9.Garcia-Graells, C., C. Valckx, and C. W. Michiels. 2000. Inactivation of Escherichia coli and Listeria innocua in milk by combined treatment with high hydrostatic pressure and the lactoperoxidase system. Appl. Environ. Microbiol. 66:4173-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Graells, C., I. Van Opstal, S. C. M. Vanmuysen, and C. W. Michiels. 2003. The lactoperoxidase system increases efficacy of high-pressure inactivation of foodborne bacteria. Int. J. Food Microbiol. 81:211-221. [DOI] [PubMed] [Google Scholar]
- 11.Hampton, M. B., A. J. Kettle, and C. C. Winterbourn. 1998. Inside the neutrophil phagosome: oxidants, myeloperoxidase, and bacterial killing. Blood 92:3007-3017. [PubMed] [Google Scholar]
- 12.Heukeshoven, J., and R. Dernick. 1985. Simplified method for silver staining of proteins in polyacrylamide gels and the mechanism of silver staining. Electrophoresis 6:103-112. [Google Scholar]
- 13.Horton, B. S. 1995. Commercial utilization of minor milk components in the health and food industries. J. Dairy Sci. 78:2584-2589. [DOI] [PubMed] [Google Scholar]
- 14.Hung, S. P., P. Baldi, and G. W. Hatfield. 2002. Global gene expression profiling in Escherichia coli K12—the effects of leucine-responsive regulatory protein. J. Biol. Chem. 277:40309-40323. [DOI] [PubMed] [Google Scholar]
- 15.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 16.Koplow, J., and H. Goldfine. 1974. Alterations in outer membrane of cell-envelope of heptose-deficient mutants of Escherichia coli. J. Bacteriol. 117:527-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kussendrager, K. D., and A. C. M. van Hooijdonk. 2000. Lactoperoxidase: physico-chemical properties, occurrence, mechanism of action and applications. Br. J. Nutr. 84:S19-S25. [DOI] [PubMed] [Google Scholar]
- 18.Laird, M. W., A. W. Kloser, and R. Misra. 1994. Assembly of LamB and OmpF in deep rough lipopolysaccharide mutants of Escherichia coli K-12. J. Bacteriol. 176:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, X., and T. Ferenci. 1998. Regulation of porin-mediated outer membrane permeability by nutrient limitation in Escherichia coli. J. Bacteriol. 180:3917-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lovaas, E. 1992. Free-radical generation and coupled thiol oxidation by lactoperoxidase/SCN−/H2O2. Free Radic. Biol. Med. 13:187-195. [DOI] [PubMed] [Google Scholar]
- 21.Martinez, M. B., F. J. Schendel, M. C. Flickinger, and G. L. Nelsestuen. 1992. In vivo kinetic-studies of clustered enzymes using overexpression of alkaline-phosphatase in Escherichia coli. FASEB J. 6:A460. [Google Scholar]
- 22.Martinez, M. B., M. C. Flickinger, and G. L. Nelsestuen. 1996. Accurate kinetic modeling of alkaline phosphatase in the Escherichia coli periplasm: implications for enzyme properties and substrate diffusion. Biochemistry 35:1179-1186. [DOI] [PubMed] [Google Scholar]
- 23.Martinez, M. B., M. C. Flickinger, and G. L. Nelsestuen. 1999. Steady-state enzyme kinetics in the Escherichia coli periplasm: a model of a whole cell biocatalyst. J. Biotechnol. 71:59-66. [DOI] [PubMed] [Google Scholar]
- 24.Martinez, M. B., M. Flickinger, L. Higgins, T. Krick, and G. L. Nelsestuen. 2001. Reduced outer membrane permeability of Escherichia coli O157:H7: suggested role of modified outer membrane porins and theoretical function in resistance to antimicrobial agents. Biochemistry 40:11965-11974. [DOI] [PubMed] [Google Scholar]
- 25.Mecsas, J., P. E. Rouviere, J. W. Erickson, T. J. Donohue, and C. A. Gross. 1993. The activity of sigma(E), an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7:2618-2628. [DOI] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Mokgatla, R. M., P. A. Gouws, and V. S. Brozel. 2002. Mechanisms contributing to hypochlorous acid resistance of a Salmonella isolate from a poultry-processing plant. J. Appl. Microbiol. 92:566-573. [DOI] [PubMed] [Google Scholar]
- 28.Moller, A. K., M. P. Leatham, T. Conway, P. J. M. Nuijten, L. A. M. de Haan, K. A. Krogfelt, and P. S. Cohen. 2003. An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infect. Immun. 71:2142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy, K. C. 1998. Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nestorovich, E. M., C. Danelon, M. Winterhalter, and S. M. Bezrukov. 2002. Designed to penetrate: time-resolved interaction of single antibiotic molecules with bacterial pores. Proc. Natl. Acad. Sci. USA 99:9789-9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, J. T., D. Raychaudhuri, H. S. Li, S. Normark, and D. Mengin-Lecreulx. 1998. MppA, a periplasmic binding protein essential for import of the bacterial cell wall peptide l-alanyl-gamma-d-glutamyl-meso-diaminopimelate. J. Bacteriol. 180:1215-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pradel, E., and C. A. Schnaitman. 1991. Effect of RfaH (SfrB) and temperature on expression of rfa genes of Escherichia coli K-12. J. Bacteriol. 173:6428-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raetz, C. R. H., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reiter, B., and G. Harnulv. 1984. Lactoperoxidase antibacterial system—natural occurrence, biological functions and practical applications. J. Food Prot. 47:724-732. [DOI] [PubMed] [Google Scholar]
- 36.Roantree, R. J., T. T. Kuo, and D. G. Macphee. 1977. Effect of defined lipopolysaccharide core defects upon antibiotic resistances of Salmonella typhimurium. J. Gen. Microbiol. 103:223-234. [DOI] [PubMed] [Google Scholar]
- 37.Ruiz, N., T. Montero, J. Hernandez-Borrell, and M. Vinas. 2003. The role of Serratia marcescens porins in antibiotic resistance. Microb. Drug Resist. 9:257-264. [DOI] [PubMed] [Google Scholar]
- 38.Sanderson, K. E., T. MacAlistor, J. W. Costerton, and K. J. Cheng. 1974. Permeability of lipopolysaccharide-deficient (rough) mutants of Salmonella typhimurium to antibiotics, lysozyme, and other agents. Can. J. Microbiol. 20:1135-1145. [DOI] [PubMed] [Google Scholar]
- 39.Seifu, E., E. M. Buys, and E. F. Donkin. 2004. Quality aspects of Gouda cheese made from goat milk preserved by the lactoperoxidase system. Int. Dairy J. 14:581-589. [Google Scholar]
- 40.Sermon, J., K. Vanoirbeek, P. De Spiegeleer, R. Van Houdt, A. Aertsen, and C. W. Michiels. 2005. Unique stress response to the lactoperoxidase-thiocyanate enzyme system in Escherichia coli. Res. Microbiol. 156:225-232. [DOI] [PubMed] [Google Scholar]
- 41.Shin, K., H. Hayasawa, and B. Lonnerdal. 2001. Inhibition of Escherichia coli respiratory enzymes by the lactoperoxidase-hydrogen peroxide-thiocyanate antimicrobial system. J. Appl. Microbiol. 90:489-493. [DOI] [PubMed] [Google Scholar]
- 42.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Matthews. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tenovuo, J. 2002. Clinical applications of antimicrobial host proteins lactoperoxidase, lysozyme and lactoferrin in xerostomia: efficacy and safety. Oral Dis. 8:23-29. [DOI] [PubMed] [Google Scholar]
- 44.Thiolas, A., C. Bornet, A. vin-Regli, J. M. Pages, and C. Bollet. 2004. Resistance to imipenem, cefepime, and cefpirome associated with mutation in Omp36 osmoporin of Enterobacter aerogenes. Biochem. Biophys. Res. Commun. 317:851-856. [DOI] [PubMed] [Google Scholar]
- 45.Uemura, J., and S. Mizushima. 1975. Isolation of outer membrane proteins of Escherichia coli and their characterization on polyacrylamide-gel. Biochim. Biophys. Acta 413:163-176. [DOI] [PubMed] [Google Scholar]
- 46.van Hooijdonk, A. C. M., K. D. Kussendrager, and J. M. Steijns. 2000. In vivo antimicrobial and antiviral activity of components in bovine milk and colostrum involved in non-specific defense. Br. J. Nutr. 84:S127-S134. [DOI] [PubMed] [Google Scholar]
- 47.Wang, Y. P., U. Ha, L. Zeng, and S. G. Jin. 2003. Regulation of membrane permeability by a two-component regulatory system in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 47:95-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolfson, L. M., and S. S. Sumner. 1994. Antibacterial activity of the lactoperoxidase system against Salmonella typhimurium in Trypticase soy broth in the presence and absence of a heat-treatment. J. Food Prot. 57:365-368. [DOI] [PubMed] [Google Scholar]
- 49.Yethon, J. A., D. E. Heinrichs, M. A. Monteiro, M. B. Perry, and C. Whitfield. 1998. Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J. Biol. Chem. 273:26310-26316. [DOI] [PubMed] [Google Scholar]
- 50.Yew, W. S., and J. A. Gerlt. 2002. Utilization of l-ascorbate by Escherichia coli K-12: assignments of functions to products of the yjf-sga and yia-sgb operons. J. Bacteriol. 184:302-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yigit, H., G. J. Anderson, J. W. Biddle, C. D. Steward, J. K. Rasheed, L. L. Valera, J. E. McGowan, and F. C. Tenover. 2002. Carbapenem resistance in a clinical isolate of Enterobacter aerogenes is associated with decreased expression of OmpF and OmpC porin analogs. Antimicrob. Agents Chemother. 46:3817-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, Z. G., M. Aboulwafa, M. H. Smith, and M. H. Saier. 2003. The ascorbate transporter of Escherichia coli. J. Bacteriol. 185:2243-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zimmermann, W., and A. Rosselet. 1977. Function of outer membrane of Escherichia coli as a permeability barrier to beta-lactam antibiotics. Antimicrob. Agents Chemother. 12:368-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zinser, E. R., and R. Kolter. 2000. Prolonged stationary-phase incubation selects for lrp mutations in Escherichia coli K-12. J. Bacteriol. 182:4361-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]