Abstract
Here we describe a quantitative PCR-based approach to estimating the relative abundances of major taxonomic groups of bacteria and fungi in soil. Primers were thoroughly tested for specificity, and the method was applied to three distinct soils. The technique provides a rapid and robust index of microbial community structure.
The diversity of the bacterial and fungal communities in soil is extraordinary (8, 27, 29). High levels of bacterial and fungal diversity make quantifying and characterizing soil microbial communities a daunting task. In recent years, quantitative PCR (qPCR, also referred to as real-time PCR) has emerged as a promising tool for studying soil microbial communities (10, 12, 20, 28). qPCR is based on the real-time detection of a reporter molecule whose fluorescence increases as PCR product accumulates during each amplification cycle (23). The qPCR approach is somewhat unique among methods of community analysis in that it allows for a relatively rapid yet quantitative assessment of the abundances of specific phylogenetic groups of microorganisms in soil.
In this paper, we describe a qPCR-based approach to assessing soil microbial community structure at broad taxonomic levels. We developed and tested nine individual qPCR assays to quantify the abundances of the dominant groups of bacteria and fungi found in the soil environment.
qPCR assays were conducted in polypropylene 96-well plates on an ABI Prism 7000 sequence detection system (Applied Biosystems). Each 25-μl reaction contained the following: 12.5 μl of ABsolute qPCR Master Mix (ABgene), 1.25 μl of each primer (10 μM; Invitrogen), 2.5 μl bovine serum albumin (10 mg ml−1; Promega), 1.0 μl SYBRGreen dye (1,600-fold dilution in H2O; Molecular Probes, Eugene, OR), 1.0 μl ROX dye (80-fold dilution in H2O; ABgene), 0.5 μl H2O, and 5 μl template DNA (0.5 ng μl−1). PCR conditions were 15 min at 95°C, followed by 40 cycles of 95°C for 1 min, 30 s at the annealing temperature, and 72°C for 1 min. Annealing temperatures (see Table 2) were experimentally optimized to maximize the group specificity of the amplification. Each plate included triplicate reactions per DNA sample and the appropriate set of standards. Melting curve analysis of the PCR products was conducted following each assay to confirm that the fluorescence signal originated from specific PCR products and not from primer-dimers or other artifacts.
TABLE 2.
Primers used for qPCR assays, annealing temperatures, target regions, and the specificity of the amplicons cloned from qPCR assays with soil DNAa
| Target group | Forward primer | Reverse primer | Approximate amplicon length (bp) | Annealing temp (°C) | % of soil clones belonging to the target groupc |
|---|---|---|---|---|---|
| All Bacteria | Eub338 | Eub518 | 200 | 53 | 100 |
| α-Proteobacteria | Eub338 | Alf685 | 365 | 60 | 75 |
| β-Proteobacteria | Eub338 | Bet680 | 360 | 60 | 96 |
| Actinobacteria | Actino235 | Eub518 | 300 | 60 | 60 |
| Firmicutes | Lgc353 | Eub518 | 180 | 60 | 100 |
| Bacteroidetes | Cfb319 | Eub518 | 220 | 65 | 100 |
| Acidobacteria | Acid31 | Eub518 | 500 | 50 | 100 |
| All Fungi | 5.8s | ITS1f | 300b | 53 | 100 |
| Basidiomycota | ITS4b | 5.8sr | 500b | 55 | 100 |
The bacterial qPCR assays target 16S rRNA genes, while the fungal assays target the internal transcribed spacer region found in rRNA genes.
The length of the targeted ITS region can vary significantly between different fungal strains.
The percentage includes only those clones that were nonchimeric and exceeded the 80% confidence threshold for taxonomic assignment using the RDP classifier program.
A plasmid standard containing the target region was generated for each primer set using DNA extracted from the appropriate positive control strain (see below). The amplified products were run on a 1.5% agarose gel to confirm the specificity of the amplification, and products were cloned using the TOPO TA cloning kit (Invitrogen). Plasmids were isolated using the Qiaprep Plasmid Miniprep kit (QIAGEN, Valencia, CA) with DNA concentrations determined by PicoGreen fluorometry (Molecular Probes). Standard curves were generated using triplicate 10-fold dilutions of plasmid DNA. We used at least three nonzero standard concentrations per assay, and the plasmid DNA concentrations ranged from 5 × 10−7 to 5 × 10−3 ng of DNA reaction−1. Target copy numbers for each reaction were calculated from the standard curves (22), assuming that the average molecular mass of a double-stranded DNA molecule is 660 g mol−1.
For all of the qPCR assays, there was a linear relationship between the log of the plasmid DNA copy number and the calculated threshold cycle value across the specified concentration range (R2 > 0.95 in all cases; data not shown). Amplification efficiencies, calculated using the methods described by Pfaffl (22), varied from 1.7 to 2.1 across the nine qPCR assays; these values are consistent with those reported in other studies (10, 25, 28).
The selected primer sets target the major phylogenetic groups of soil microorganisms and have been tested for specificity previously (Table 1). The bacterial primer sets were also tested in silico using the Probe Match software (5) and were found to be specific for the target groups (>90% matches to target group). The specificity of the qPCR assays was further tested with DNA extracted from bacterial and fungal strains representative of the target and nontarget phylogenetic groups. The following cultures were obtained from the German Culture Collection (DSMZ, Braunschweig, Germany) and cultivated using the recommended procedures: Acidobacterium capsulatum (DSMZ 11244), Arthrobacter crystallopoietes (DSMZ 20117), Bacillus subtilis (DSMZ 10), Flavobacterium aquatile (DSMZ 1132), Hyphomicrobium facile (DSMZ 1565), Myxococcus xanthus (DSMZ 435), Variovorax paradoxus (DSMZ 645), Saccharomyces cerevisiae (DSMZ 1334), and Halobacterium salinarum (DSMZ 670). Strains of Pseudomonas aeruginosa and Amanita rubescens were obtained from Patricia Holden at the University of California—Santa Barbara and Jeri Parrent at Duke University, respectively. DNA was extracted from each of the cultures using the UltraClean Microbial DNA kit (MoBio Laboratories, Solana Beach, CA), and each qPCR assay was tested for specificity using DNA from each of the 11 strains. Except for the α-Proteobacteria qPCR assay, which also amplified DNA from M. xanthus, a δ-proteobacterium, all of the assays were specific for the target group, and there was no appreciable amplification of DNA from strains representative of the nontarget groups (data not shown).
TABLE 1.
A description of the group-specific primers used for the qPCR assaysa
| Domain | Target group | Primer sequence (5′-3′) | Primer name | Reference |
|---|---|---|---|---|
| Bacteria | All groups | ACT CCT ACG GGA GGC AGC AG | Eub338 | 13 |
| All groups | ATT ACC GCG GCT GCT GG | Eub518 | 18 | |
| α-Proteobacteriab | TCT ACG RAT TTC ACC YCT AC | Alf685 | 13 | |
| β-Proteobacteriab | TCA CTG CTA CAC GYG | Bet680 | 21 | |
| Actinobacteriac | CGC GGC CTA TCA GCT TGT TG | Actino235 | 26 | |
| Firmicutesd | GCA GTA GGG AAT CTT CCG | Lgc353 | 17 | |
| Bacteroidetese | GTA CTG AGA CAC GGA CCA | Cfb319 | 15 | |
| Acidobacteria | GAT CCT GGC TCA GAA TC | Acid31 | 2 | |
| Eucarya (Fungi) | All groups | TCC GTA GGT GAA CCT GCG G | ITS1f | 7 |
| All groups | CGC TGC GTT CTT CAT CG | 5.8s | 31 | |
| Basidiomycota | CAG GAG ACT TGT ACA CGG TCC AG | ITS4b | 7 | |
| Basidiomycota | TCG ATG AAG AAC GCA GCG | 5.8sr | 31 |
The bacterial taxonomy follows that described in the latest edition of Bergey's Manual of Systematic Bacteriology.
This class is within the phylum Proteobacteria.
Commonly labeled the Actinomycete or high-GC-content gram-positive group.
Commonly labeled the low-GC-content gram-positive group.
Commonly labeled the Cytophaga-Flavobacteria or Cytophaga-Flexibacteria-Bacteroides group.
The qPCR assays were further tested and optimized using DNA extracted from three distinct soils: a tallgrass prairie at the Konza Prairie Biological Station in Manhattan, Kansas (39°05′N, 96°35′W), a desert shrubland in the Mojave Desert, California (34°54′N, 115°36′W), and a coniferous forest near Durham, North Carolina (35°58′N, 79°5′W). All soil samples were collected from the 0- to 5-cm depth and were sieved to 4 mm, homogenized, and frozen at −80°C within 4 days of collection. DNA was extracted using the UltraClean Mega Soil DNA kit (MoBio Laboratories), with an aliquot of the extracted DNA further purified on a Sepharose 4B (Sigma-Aldrich, St. Louis, MO) column, as described by Jackson et al. (9).
To test the specificity of the qPCR assays with soil DNA templates, qPCR products were selected at random and cloned using the procedure described above. For each of the nine qPCR assays, 20 to 25 positive clones were reamplified using plasmid-specific primers, purified using the QIAquick 96 PCR purification kit (QIAGEN), and sequenced on an Applied Biosystems 3700 automated sequencer using the BigDye Terminator kit (v3.0). Sequences were proofread using Sequencher 4.2 (Gene Codes, Ann Arbor, MI) and screened for chimeras using Chimera_Check (version 2.7) software (http://rdp.cme.msu.edu) before being assigned to taxonomic groups using the RDP classifier program (5). For the fungal qPCR assays, sequences were identified by BLAST against the National Center for Biotechnology Information's nucleotide database (1). The short length of many of the cloned amplicons limited the number of clones that could be identified with confidence (10 to 20 per qPCR assay). With the exception of the Actinobacteria and α-Proteobacteria qPCR assays, more than 95% of the cloned qPCR amplicons that could be identified belonged to the correct target group (Table 2). All of the nontarget clones identified from the α-Proteobacteria and Actinobacteria qPCR assays were identified as belonging to the δ-Proteobacteria and Verrucomicrobia groups, respectively. Without further testing, these two qPCR assays cannot be considered specific for the targeted groups.
We tested the sensitivity of the qPCR assays to changes in target DNA concentrations by spiking soil DNA (0.25 ng DNA from the prairie soil) with various amounts of DNA from the representative target strain (a 2× dilution series ranging from 2.5 to 0.16 ng DNA). We compared the known amount of pure culture DNA added to each reaction to the estimated copy number calculated from the standard curve for each qPCR assay. For all nine assays, the R2 value for the linear regression relating the amount of target DNA added (ng of DNA reaction−1) to the target copy number was ≥0.93, indicating that the assays were quantitative across the range of DNA concentrations tested. Variability in the slope of the linear regression between assays (from 2 × 106 to 8 × 106) is likely a result of differences in amplification efficiencies, rRNA gene copy numbers, or both.
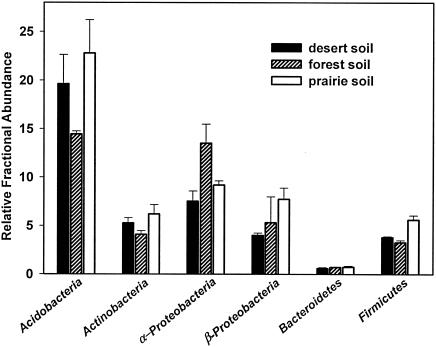
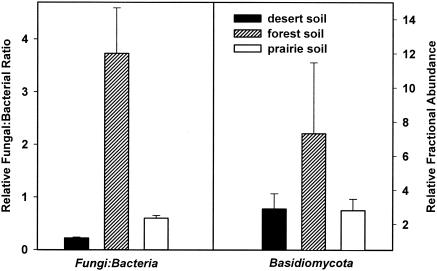
Figures 1 and 2 show the estimated relative abundances of the six targeted bacterial groups, the basidiomycete fungi, and the overall fungal/bacterial ratio in the three soils. The relative abundance of each bacterial group is calculated as the ratio between the measured copy number for each group-specific qPCR assay and the “all Bacteria” assay. The relative fungal/bacterial ratio is the ratio of copy numbers measured with the “all Fungi” and “all Bacteria” qPCR assays. The relative abundance of Basidiomycota is estimated from the ratio of copy numbers measured with the “Basidiomycota” and “all Fungi” qPCR assays. Fractional copy numbers provide a more accurate index of target abundances, since the efficiency of PCR amplification can vary across DNA samples (25).
FIG. 1.
The relative abundances of the six bacterial groups in three different soils, as estimated using the qPCR assays. Error bars are the standard errors of the mean for the three replicates.
FIG. 2.
The estimated fungal/bacterial ratios and the relative abundances of basidiomycete fungi (as a fraction of total fungal abundance) in the three soil samples. Error bars are the standard errors of the mean for the three replicates.
Figures 1 and 2 show important differences in microbial community structure between the three soils, particularly with regard to the fungal/bacterial ratios. The results agree well with published data. Acidobacteria and Proteobacteria are generally the numerically dominant phyla in soil, with members of the Bacteroidetes and Firmicutes phyla being less common (3, 6, 14, 32). We would also expect the fungal/bacterial ratio to be higher in the coniferous forest soil than in the other two soils (4, 24). The high relative abundance of basidiomycete fungi in the forest soil (Fig. 2) has been observed in fungal clone libraries constructed from soil collected at the site (19).
A significant limitation of our method is that the estimated abundances of the different microbial groups may not equal the true percentages of these groups in the soil samples. There are a number of reasons for this: DNA extraction bias may alter the estimated abundances of certain groups (16), heterogeneity in ribosomal operon number (11, 30) may affect relative estimates of group abundances, and the tested qPCR assays do not necessarily amplify rRNA genes belonging to all members of each targeted group. In addition, we have not designed qPCR assays for those microbial groups (such as the Ascomycota, Verrucomicrobia, and Planctomycetes) that may be numerically important in soil. The exclusion of certain bacterial groups or members within a group may explain why the fractional percentages shown in Fig. 1 add up to only 40 to 52% for each soil. These limitations need to be considered when using this approach for the assessment of microbial community structure.
In this study, we examined microbial community structure at the coarsest level of taxonomic resolution. However, the qPCR approach described here can be adapted to provide more comprehensive and detailed assessments of soil microbial community structure. To do so requires designing the appropriate set of oligonucleotide primers, testing the primer sets, and experimentally optimizing the qPCR reaction conditions. The flexibility, ease of use, and quantitative nature of the qPCR technique make it a valuable tool for characterizing soil microbial communities.
Acknowledgments
We thank Rachel Steinberger, Wyatt Hartman, Jeri Parrent, Tim James, Patricia Holden, and Matt Wallenstein for their help.
Financial support was provided by a National Science Foundation postdoctoral fellowship in microbial biology awarded to N.F. and by NSF, NIGEC, and Mellon Foundation grants to R.B.J.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barns, S. M., S. L. Takala, and C. R. Kuske. 1999. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 65:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow, M. L., C. C. Radomski, J. M. McDermott, J. Davies, and P. E. Axelrood. 2002. Molecular characterization of bacterial diversity in Lodgepole pine (Pinus contorta) rhizosphere soils from British Columbia forest soils differing in disturbance and geographic source. FEMS Microbiol. Ecol. 42:347-357. [DOI] [PubMed] [Google Scholar]
- 4.Christensen, M. 1981. Species diversity and dominance in fungal communities, p. 201-232. In D. Wicklow and G. Carroll (ed.), The fungal community: its organization and role in the ecosystem. Marcel Dekker, New York, N.Y.
- 5.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardes, M., and T. D. Bruns. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 8.Hawksworth, D. L. 2001. The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol. Res. 105:1422-1432. [Google Scholar]
- 9.Jackson, C. R., J. P. Harper, D. Willoughby, E. E. Roden, and P. F. Churchill. 1997. A simple, efficient method for the separation of humic substances and DNA from environmental samples. Appl. Environ. Microbiol. 63:4993-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kabir, S., N. Rajendran, T. Amemiya, and K. Itoh. 2003. Quantitative measurement of fungal DNA extracted by three different methods using real-time polymerase chain reaction. J. Biosci. Bioeng. 96:337-343. [DOI] [PubMed] [Google Scholar]
- 11.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolb, S., C. Knief, S. Stubner, and R. Conrad. 2003. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl. Environ. Microbiol. 69:2423-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lane, D. 1991. 16s/23s rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, West Sussex, United Kingdom.
- 14.Lipson, D. A., and S. K. Schmidt. 2004. Seasonal changes in an alpine soil bacterial community in the Colorado Rocky Mountains. Appl. Environ. Microbiol. 70:2867-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 16.Martin-Laurent, F., L. Philippot, S. Hallet, R. Chaussod, J. C. Germon, G. Soulas, and G. Catroux. 2001. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl. Environ. Microbiol. 67:2354-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meier, H., R. Amann, W. Ludwig, and K. H. Schleifer. 1999. Specific oligonucleotide probes for in situ detection of a major group of Gram-positive bacteria with low DNA G+C content. Syst. Appl. Microbiol. 22:186-196. [DOI] [PubMed] [Google Scholar]
- 18.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Brien, H., J. Parrent, J. Jackson, J. Moncalvo, and R. Vilgalys. Fungal community analysis by large-scale ITS sequencing of environmental samples. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 20.Okano, Y., K. R. Hristova, C. M. Leutenegger, L. E. Jackson, R. F. Denison, B. Gebreyesus, D. Lebauer, and K. M. Scow. 2004. Application of real-time PCR to study effects of ammonium on population size of ammonia-oxidizing bacteria in soil. Appl. Environ. Microbiol. 70:1008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Overmann, J., M. J. L. Coolen, and C. Tuschak. 1999. Specific detection of different phylogenetic groups of chemocline bacteria based on PCR and denaturing gradient gel electrophoresis of 16S rRNA gene fragments. Arch. Microbiol. 172:83-94. [DOI] [PubMed] [Google Scholar]
- 22.Pfaffl, M. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:2002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raeymaekers, L. 2000. Basic principles of quantitative PCR. Mol. Biotechnol. 15:115-122. [DOI] [PubMed] [Google Scholar]
- 24.Read, D. 1985. Mycorrhizal mycelia and nutrient cycling in plant communities, p. 193-217. In A. Fitter, D. Atkinson, and D. Read (ed.), Ecological interactions in soil: plants, microbes, and animals. Blackwell Scientific Publications, Oxford, United Kingdom.
- 25.Smits, T. H. M., C. Devenoges, K. Szynalski, J. Maillard, and C. Holliger. 2004. Development of a real-time PCR method for quantification of the three genera Dehalobacter, Dehalococcoides, and Desulfitobacterium in microbial communities. J. Microbiol. Methods 57:369-378. [DOI] [PubMed] [Google Scholar]
- 26.Stach, J. E. M., L. A. Maldonado, A. C. Ward, M. Goodfellow, and A. T. Bull. 2003. New primers for the class Actinobacteria: application to marine and terrestrial environments. Environ. Microbiol. 5:828-841. [DOI] [PubMed] [Google Scholar]
- 27.Straatsma, G., F. Ayer, and S. Egli. 2001. Species richness, abundance, and phenology of fungal fruit bodies over 21 years in a Swiss forest plot. Mycol. Res. 105:515-523. [Google Scholar]
- 28.Stubner, S. 2002. Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreen (TM) detection. J. Microbiol. Methods 50:155-164. [DOI] [PubMed] [Google Scholar]
- 29.Torsvik, V., L. Ovreas, and T. F. Thingstad. 2002. Prokaryotic diversity: magnitude, dynamics, and controlling factors. Science 296:1064-1066. [DOI] [PubMed] [Google Scholar]
- 30.Tourova, T. P. 2003. Copy number of ribosomal operons in prokaryotes and its effect on phylogenetic analyses. Microbiology 72:389-402. [PubMed] [Google Scholar]
- 31.Vilgalys, R., and M. Hester. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172:4238-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou, J. Z., B. C. Xia, H. S. Huang, D. S. Treves, L. J. Hauser, R. J. Mural, A. V. Palumbo, and J. M. Tiedje. 2003. Bacterial phylogenetic diversity and a novel candidate division of two humid region, sandy surface soils. Soil Biol. Biochem. 35:915-924. [Google Scholar]