Abstract
In Escherichia coli, three additional proteins having l-cysteine desulfhydrase activity were identified as O-acetylserine sulfhydrylase-A, O-acetylserine sulfhydrylase-B, and MalY protein, in addition to tryptophanase and cystathionine β-lyase, which have been reported previously. The gene disruption for each protein was significantly effective for overproduction of l-cysteine and l-cystine. Growth phenotype and transcriptional analyses suggest that tryptophanase contributes primarily to l-cysteine degradation.
l-cysteine is an important amino acid in terms of its applications in the pharmaceutical, food, and cosmetic industries. However, due to feedback inhibition by l-cysteine of serine acetyltransferase (SAT; EC 2.3.1.30), which catalyzes the formation of O-acetyl-l-serine from acetyl-coenzyme A (CoA) and l-serine (8, 9, 16), high-level production of l-cysteine from glucose has not been successfully achieved in microorganisms. In order to obtain l-cysteine producers, we previously constructed Escherichia coli cysE genes that encode altered SATs. These genes were genetically desensitized to the feedback inhibition by l-cysteine through site-directed or random mutagenesis (21, 32). We found that, in the recombinant E. coli cells expressing the altered cysE gene, there was a marked production of l-cysteine plus l-cystine.
In the same investigation (21), it was demonstrated that proteins with l-cysteine desulfhydrase (CD) activity play an important role in l-cysteine degradation in E. coli cells. In order to further improve l-cysteine production, a host strain having a lower level of CD activity must be constructed. CD is known to catalyze the degradation of l-cysteine to pyruvate, ammonia, and sulfide by the following reaction: HSCH2CH(NH2)COOH + H2O → CH3COCOOH + H2S + NH3. This type of enzyme activity has been demonstrated to be present in several mammalian tissues (15) and in bacteria, such as Salmonella enterica serovar Typhimurium (6, 17) and E. coli (2, 11). In E. coli, cystathionine-β-lyase (CBL) (17) encoded by metC, which catalyzes mainly the conversion of cystathionine to homocysteine, pyruvate, and ammonia (9), as well as tryptophanase (TNase) encoded by tnaA, which primarily degrades l-tryptophan to indole, pyruvate, and ammonia (22), has been shown to exhibit CD activity in vitro (9, 23). We have previously reported that CBL and TNase catalyzed the CD reaction and acted on l-cysteine degradation in E. coli cells by analyses with CD activity staining and gene disruption (2). However, the double CD gene-disrupted mutant still had a low level of CD activity, suggesting that unknown CD proteins remain to be identified. Thus, we report here further identification and characterization of the CDs involved in l-cysteine degradation in E. coli.
Identification of CDs in E. coli.
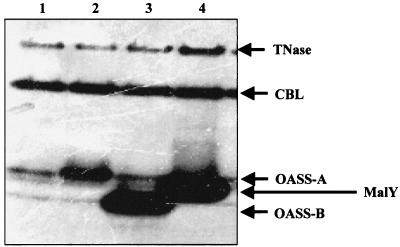
An E. coli cell has 4,388 kinds of open reading frame (ORF), including the function-unknown and -deduced genes. The library was constructed by placing each ORF under the lacZ promoter in vector pCA24N (20). E. coli wild-type strain JM39 was independently transformed with nine libraries, each consisting of 480 kinds of plasmids, and then approximately 1,200 colonies appeared from each library on Luria-Bertani (LB) medium (27) containing chloramphenicol (100 μg/ml). As a whole, more than 10,000 independent E. coli clones were obtained. The transformed cells from each library were mixed and grown at 37°C in 5 ml of LB medium containing chloramphenicol (100 μg/ml). When the absorbance at 600 nm reached 0.5, isopropyl-β-d-thiogalactopyranoside was added to the culture medium to a final concentration of 0.01 mM to induce gene expression. After cultivation for 4 h at 37°C, cell extracts were separated by native polyacrylamide gel electrophoresis (PAGE) and the gel was stained by gently shaking in the solution containing l-cysteine, pyridoxal 5′-phosphate (PLP), and BiCl3 as described previously (2, 35). When the gel was kept at 4°C for 1.5 h, which was a shorter time than in the previous condition (3 h), three CD proteins were newly detected in JM39 in addition to CBL and TNase (Fig. 1): O-acetylserinesulfhydrylase-A (OASS-A), encoded by cysK (lane 2), O-acetylserinesulfhydrylase-B (OASS-B), encoded by cysM (lane 3), and MalY, encoded by malY (lane 4). OASS-A primarily catalyzes the synthesis of l-cysteine from O-acetylserine and sulfide along the l-cysteine biosynthetic pathway (3, 7, 19). OASS-B is considered to be an isomer of OASS-A (30, 31), but its function(s) is not yet clearly understood. Because both OASS-A and -B require PLP for CD activity in a similar manner as CBL, we think that these enzymes can catalyze the analogous reaction. MalY protein is also a PLP-dependent enzyme with the activity of the carbon-sulfur bond cleavage (βC-S lyase) of cystathionine (35), as well as a transcriptional regulator in mal gene expression (5, 26, 28). The secondary and tertiary structures of MalY are highly homologous to those of CBL (4, 5, 9, 18). Consequently, the five CD proteins in E. coli were identified as TNase, CBL, OASS-A, OASS-B, and MalY.
FIG. 1.
Detection of OASS-A, MalY, and OASS-B by CD activity staining. Preparation of native PAGE gel, cell extracts, and CD activity staining were carried out according to the method described previously (2). Lane 1, wild-type JM39 harboring pBluescript II SK+ (vector control; Toyobo Biochemicals, Osaka, Japan); lane 2, JM39 harboring pcysK (cysK plasmid; OASS-A is overexpressed); lane 3, JM39 harboring pcysM (cysM plasmid; OASS-B is overexpressed); lane 4, JM39 harboring pmalY (malY plasmid; MalY is overexpressed).
Effect of gene disruption on total CD activities and l-cysteine production in E. coli.
For the construction of cysK-, cysM-, and malY-disrupted strains, internal fragments of the cysK, cysM, and malY genes of strain JM39 were amplified by PCR and cloned in pEL3, which has a thermosensitive replicon (1). The primers used were 5′-CGC CGC GGA TCC CAA TCT ACC GGT TAT TTT GAT AAC C-3′ and 5′-CGC CGC GGA TCC CAA GCT GGC ATT ACT GTT GCA ATT C-3′ for cysK, 5′-GCG GCG GGA TCC TAG GTT GAG TGA ATG TTA AAC GCC C-3′ and 5′-GCG GCG GGA TCC ATA CTG CAT TTG TCG GCA GCA ACA-3′ for cysM, and 5′-ATC CAG TCG ATG ATC GAT ACC GGG ATC C-3′ and 5′-CGC GGG ATC CTT AAC GAA CAG CGC GGA TGG CGT TA-3′ for malY. Gene disruption was performed as described previously (2). The ampicillin-sensitive strains obtained were characterized and designated as JM39ΔcysK, JM39ΔcysM, and JM39ΔmalY. JM39ΔtnaA ΔmetC ΔcysM ΔmalY and JM39ΔtnaA ΔmetC ΔcysK ΔcysM ΔmalY were constructed from JM39ΔtnaA ΔmetC (2) in the same manner as described above. CD gene disruption was confirmed by PCR and by CD activity staining. In each CD gene-disrupted mutant, the corresponding CD protein bands disappeared on the activity staining gel. No CD bands were detected in the quintet mutant, JM39ΔtnaA ΔmetC ΔcysK ΔcysM ΔmalY (data not shown). Total CD activities of the cell extracts prepared from wild-type JM39 and CD gene-disrupted strains cultured in LB medium were measured as described previously (2) (Table 1). CD activity was measured by colorimetric assay of the sulfide formed from l-cysteine by the enzyme sources (2, 29). The total CD activities of all mutants were lower than that of wild-type JM39. Interestingly, even the quintet mutant still had a low level of CD activity. The reason for the residual activity remains unclear; however, the presence of another CD protein(s), which fails to be separated by native PAGE or is inactivated by oxygen during native PAGE, is possible.
TABLE 1.
Effect of CD gene disruption on total CD activities and L-cysteine plus L-cystine production in E. coli
| Strain | Growth (OD610)a after 15 h of cultivation in LB plus 30 mM l-cysteine | CD activity (mU/mg of protein)b in the absence and presence of l-cysteine
|
Concn of l-cysteine plus l-cystine after 72 h of cultivation in C1 + TS medium (mg/liter/OD562)c | |
|---|---|---|---|---|
| No l-cysteine | 10 mM l-cysteine | |||
| JM39 (wild type) | 4.0 | 20.6 ± 0.1 | 27.6 ± 0.1 | 590 ± 160 |
| JM39ΔtnaA | 2.2 | 15.7 ± 0.1 | 14.1 ± 0.1 | 1,290 ± 70 |
| JM39ΔmetC | 3.5 | 15.0 ± 0.1 | 27.6 ± 0.5 | 1,240 ± 100 |
| JM39ΔcysK | 3.5 | 18.2 ± 0.5 | 29.9 ± 0.1 | NTd |
| JM39ΔcysM | 3.6 | 17.9 ± 0.5 | 27.8 ± 0.1 | 1,100 ± 70 |
| JM39ΔmalY | 3.6 | 15.3 ± 0.1 | 27.1 ± 0.5 | 1,360 ± 90 |
| JM39ΔtnaA ΔmetC ΔcysM ΔmalY | NT | 9.1 ± 0.5 | 19.6 ± 0.1 | 1,080 ± 10 |
| JM39ΔtnaA ΔmetC ΔcysK ΔcysM ΔmalY | NT | 8.7 ± 0.5 | 11.5 ± 0.1 | NT |
The values are means from three independent experiments. The variations in the values were less than 10%.
One unit of activity was defined as the amount of enzyme required to produce 1 μmol of sulfide per min from l-cysteine at 37°C. The values are means ± standard deviations from three independent experiments.
The values are means ± standard deviations from three independent experiments.
NT, not tested since cysK-disrupted cells showed l-cysteine auxotrophy.
In order to confirm whether CDs were involved in l-cysteine degradation in E. coli cells, we analyzed the l-cysteine productivities of wild-type JM39 and each CD gene-disrupted mutant harboring pEAS-m (24, 33), which carries the cDNA encoding feedback-insensitive SAT from Arabidopsis thaliana. It should be noted that two mutants, JM39ΔcysK and JM39ΔtnaA ΔmetC ΔcysK ΔcysM ΔmalY, were not tested, because OASS-A encoded by cysK is cysteine synthetase, which is essential for l-cysteine biosynthesis. In contrast, it was found that OASS-B encoded by cysM was not an isomer of OASS-A based on the fact that JM39ΔcysM grew on a medium lacking l-cysteine (data not shown). For the production of l-cysteine and l-cystine, a loopful of cells cultured for 24 h on LB solid medium containing ampicillin (50 μg/ml) at 30°C was inoculated into 20 ml of C1 plus TS medium in a 500-ml flask and cultured at 30°C on a reciprocal shaker maintained at 120 strokes per min (33). Growth was measured by optical density at 562 nm (OD562) of culture broth after appropriate dilution with 0.1 N HCl. The amounts of l-cysteine and l-cystine were determined by microbioassay with Pediococcus acidilactici IFO3076, as described by Tsunoda et al. (34). As l-cysteine in the culture fluid was easily oxidized to l-cystine, which was slightly soluble in water, the culture fluids were assayed after l-cystine was dissolved with 0.5 N HCl. As shown in Table 1, the production of l-cysteine and l-cystine by these mutants was higher than that observed in the case of JM39. The amounts of l-cysteine and l-cystine produced after 72 h of cultivation increased by a factor of 1.8 to 2.3. These results clearly indicate that CD proteins played important roles in l-cysteine degradation in E. coli cells and that the corresponding gene disruption was effective in the production of l-cysteine and l-cystine by E. coli cells. However, the amounts of l-cysteine and l-cystine decreased significantly after 96 h of cultivation in all the strains, probably because of the remaining CD enzyme(s) (data not shown).
TNase contributes primarily to l-cysteine degradation in E. coli.
Although the mechanisms are not yet understood, it has previously been observed that the growth of E. coli cells is inhibited by excess l-cysteine (12-14, 25). Therefore, the effect of CD gene disruption on the growth of E. coli cells in the presence of l-cysteine was examined. All of the strains were grown in LB plus 30 mM l-cysteine at 37°C, and cell growth was measured by optical density at 610 nm. As shown in Table 1, the growth of the tnaA disruptant JM39ΔtnaA was significantly inhibited, while all of the strains showed the same level of growth when cultured in LB medium (data not shown). This result suggests that TNase is a key enzyme in l-cysteine degradation in E. coli cells.
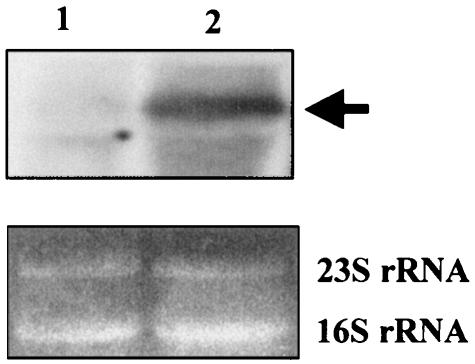
We examined the role of CD enzymes on l-cysteine degradation. Wild-type JM39 and CD gene-disrupted mutants were cultivated in LB plus 10 mM l-cysteine, and total CD activities were measured (Table 1). All of the single mutants, except for JM39ΔtnaA, showed a prominent increase in CD activity, ranging from 38% to 84% of that observed in the absence of l-cysteine. The CD activity from the tnaA-disrupted strain was virtually unchanged in the presence or absence of l-cysteine. Northern blot analysis for tnaA was carried out by using a Gene Images Random-Prime Labeling and Detection System (Amersham Pharmacia Biotech, Buckinghamshire, England). Strain JM39 was cultivated in LB medium or LB plus 10 mM l-cysteine, and total RNA was prepared using an RNeasy Protect Bacteria Mini kit (QIAGEN, Valencia, Calif.). As a DNA probe, the DNA fragment of tnaA was prepared by PCR with oligonucleotide primers 5′-CCG TTC CGC ATT CGT GTT AT-3′ and 5′-TGC GGT GAA GTG ACG CAA TA-3′. As shown in Fig. 2, the addition of l-cysteine resulted in an elevated level of the specific transcript (ca. 1.7 kb), while the basal level of expression in LB medium was fairly low. Transcription of the E. coli tna operon, consisting of two major structural genes, tnaA encoding tryptophanase and tnaB encoding tryptophan permease, have been studied in detail (10). This operon also contains a 319-bp transcribed leader regulatory region, tnaC, preceding tnaA and specifying a 24-residue leader peptide, TnaC, and expression of the tna operon is induced by l-tryptophan. Interestingly, the tnaA DNA probe detected a tnaC-tnaA transcript of ca. 1.7-kb in the presence of l-cysteine (lane 2), because no transcript was observed when the DNA fragment of tnaB was used as a probe (data not shown). These results indicate that TNase synthesis is induced by l-cysteine, in agreement with previous data on native PAGE (2). Our results may suggest that TNase contributes mainly to l-cysteine degradation and that a novel transcriptional regulation system is involved in tnaA expression.
FIG. 2.
TNase induction by the addition of l-cysteine. Total RNA of JM39 was prepared from cells cultivated in LB medium (lane 1) and LB plus 10 mM l-cysteine (lane 2). Each lane was loaded with 20 μg of total RNA. The arrow indicates a tnaC-tnaA transcript of ca. 1.7 kb. The 23S and 16S rRNAs are total RNA-loading controls detected by UV spectrometer.
In conclusion, five CD enzymes were identified in E. coli cells, and the gene disruption for each protein was significantly effective for overproduction of l-cysteine and l-cystine. However, it is noteworthy that the quintet mutant JM39ΔtnaA ΔmetC ΔcysK ΔcysM ΔmalY in the presence of l-cysteine showed higher CD activity than that observed in the absence of l-cysteine (Table 1). It appears that other CDs, in addition to the five proteins identified, could be induced by l-cysteine in E. coli. We must further analyze the quintet mutant in order to investigate the mechanism. Through the CD activity staining described here, some faint bands were still seen in the gel and some proteins appeared not to be migrated into the gel. We therefore think that unidentified proteins with CD activity, which may be induced by l-cysteine, are still present. Development of alternative methods to detect the remaining CDs, including enzyme purification and CD activity staining, is necessary and is currently in progress. We will also analyze the genome-wide expression profile in each CD gene-disrupted mutant using DNA microarray technology. It is possible that one gene disruption would affect the expression of other CD proteins and the whole metabolic profile in E. coli.
Acknowledgments
The technical assistance of S. Yamada of our laboratory is greatly appreciated.
This work was supported in part by a grant-in-aid from Japan Society for the Promotion of Science for Young Scientists (no. 01978 to N.A.) and by a grant from Ajinomoto Co., Inc., to H.T.
REFERENCES
- 1.Armstrong, K. A., R. Acosta, E. Ledner, Y. Machida, M. Pancotto, M. McCormick, H. Ohtsubo, and F. Ohtsubo. 1984. A 37-103 molecular weight plasmid-encoded protein is required for replication and copy number control in the plasmid pSC101 and its temperature-sensitive derivative pHS1. J. Mol. Biol. 175:331-347. [DOI] [PubMed] [Google Scholar]
- 2.Awano, N., M. Wada, A. Kohdoh, T. Oikawa, H. Takagi, and S. Nakamori. 2003. Effect of cysteine desulfhydrase gene disruption on l-cysteine overproduction in Escherichia coli. Appl. Microbiol. Biotechnol. 62:239-243. [DOI] [PubMed] [Google Scholar]
- 3.Byrne, C. R., R. S. Monroe, K. A. Ward, and N. M. Kredich. 1987. DNA sequences of the cysK regions of Salmonella typhimurium and Escherichia coli and linkage of the cysK regions to ptsH. J. Bacteriol. 170:3150-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clausen, T., R. Huber, B. Laber, H. D. Pohlenz, and A. Messerschmidt. 1996. Crystal structure of the pyridoxal-5′-phosphate dependent cystathionine beta-lyase from Escherichia coli at 1.83 Å. J. Mol. Biol. 262:202-224. [DOI] [PubMed] [Google Scholar]
- 5.Clausen, T., A. Schlegel, R. Peist, E. Schneider, C. Steegborn, Y.-S. Chang, A. Haase, G. P. Bourenkov, H. D. Bartunik, and W. Boos. 2000. X-ray structure of MalY from Escherichia coli: a pyridoxal 5′-phosphate-dependent enzyme acting as a modulator in mal gene expression. EMBO J. 19:831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, J. M., and K. J. Monty. 1973. The cysteine desulfhydrase of Salmonella typhimurium kinetic and catalytic properties. J. Biol. Chem. 248:5943-5949. [PubMed] [Google Scholar]
- 7.Cook, P. F. 2003. Alpha, beta-elimination reaction of O-acetylserine sulfhydrylase. Is the pyridine ring required? Biochim. Biophys. Acta 1647:66-69. [DOI] [PubMed] [Google Scholar]
- 8.Denk, D., and A. Böck. 1987. l-cysteine biosynthesis in Escherichia coli: nucleotide sequence and expression of the serine acetyltransferase (cysE) gene from the wild-type and cysteine excreting mutant. J. Gen. Microbiol. 133:515-525. [DOI] [PubMed] [Google Scholar]
- 9.Dwivedi, C. M., R. C. Ragin, and J. R. Uren. 1982. Cloning, purification, and characterization of β-cystathionase from Escherichia coli. Biochemistry 21:3064-3069. [DOI] [PubMed] [Google Scholar]
- 10.Gong, F., K. Ito, Y. Nakamura, and C. Yanofsky. 2001. The mechanism of tryptophan induction of tryptophanase operon expression: tryptophan inhibits release factor-mediated cleavage of TnaCpeptidyl-tRNAPro. Proc. Natl. Acad. Sci. USA 98:8997-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guarneros, G., and M. V. Ortega. 1970. Cysteine desulfhydrase activities of Salmonella typhimurium and Escherichia coli. Biochim. Biophys. Acta 198:132-142. [DOI] [PubMed] [Google Scholar]
- 12.Harris, C. L. 1981. Cysteine and growth inhibition of Escherichia coli: threonine-deaminase as the target enzyme. J. Bacteriol. 145:1031-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris, C. L., and L. Lui. 1981. Cysteine and growth inhibition of Escherichia coli: depression of the ilvGEDA operon. Biochem. Biophys. Res. Commun. 101:1145-1151. [DOI] [PubMed] [Google Scholar]
- 14.Kari, C., Z. Nagy, P. Kovacs, and F. Hernadi. 1971. Mechanism of the growth inhibitory effect of cysteine on Escherichia coli. J. Gen. Microbiol. 68:349-356. [DOI] [PubMed] [Google Scholar]
- 15.Kato, A., M. Ogura, H. Kimura, T. Kawai, and M. Suda. 1966. Control mechanism in the rat liver enzyme system converting L-methionine to L-cysteine. II. Crystallization of the serine dehydratase inhibitor-forming enzyme and its identity with cystathionase, homoserine dehydratase, cysteine desulfurase and cysteine desulfhydrase. J. Biochem. (Tokyo) 59:34-39. [DOI] [PubMed] [Google Scholar]
- 16.Kredich, N. M. 1983. Regulation of cysteine biosynthesis in Escherichia coli and Salmonella typhimurium, p. 115-132. In K. M. Herrmann and R. L. Sommerville (ed.), Amino acids: biosynthesis and genetic regulation. Addison-Wesley Publishing Company, London, United Kingdom.
- 17.Kredich, N. M., L. J. Foote, and B. S. Kreenan. 1973. The stoichiometry and kinetics of the inducible cysteine desulfhydrase from Salmonella typhimurium. J. Biol. Chem. 248:6187-6196. [PubMed] [Google Scholar]
- 18.Martel, A., C. de la Tour Bouthier, and F. Le Goffic. 1987. Pyridoxal-5′-phosphate binding site of Escherichia coli beta cystathionase and cystathionine gamma synthase comparison of their sequences. Biochem. Biophys. Res. Commun. 147:565-571. [DOI] [PubMed] [Google Scholar]
- 19.Mino, K., T. Yamanoue, T. Sakiyama, N. Eisaki, A. Matsuyama, and K. Nakanishi. 2000. Effects of bioenzyme complex formation of cysteine synthetase from Escherichia coli on some properties and kinetics. Biosci. Biotechnol. Biochem. 64:1628-1640. [DOI] [PubMed] [Google Scholar]
- 20.Mori, H., K. Isono, T. Horiuchi, and T. Miki. 2000. Functional genomics of Escherichia coli in Japan. Res. Microbiol. 151:121-128. [DOI] [PubMed] [Google Scholar]
- 21.Nakamori, S., S. Kobayashi, C. Kobayashi, and H. Takagi. 1998. Overproduction of L-cysteine and L-cystine by Escherichia coli strains with a genetically altered serine acetyltransferase. Appl. Environ. Microbiol. 64:1607-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newton, W. A., and E. F. Snell. 1964. Catalytic properties of tryptophanase, a multifunctional pyridoxal phosphate enzyme. Proc. Natl. Acad. Sci. USA 51:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newton, W. A., Y. Morino, and E. F. Snell. 1965. Properties of crystalline tryptophanase. J. Biol. Chem. 240:1211-1218. [PubMed] [Google Scholar]
- 24.Noji, M., K. Inoue, N. Kimura, A. Gouda, and K. Saito. 1998. Isoformdependent differences in feedback regulation and subcellular localization of serine acetyltransferase involved in cysteine biosynthesis from Arabidopsis thaliana. J. Biol. Chem. 273:32739-32745. [DOI] [PubMed] [Google Scholar]
- 25.Park, S., and J. A. Imlay. 2003. High levels of intracellular cysteine promote oxidative DNA damage by driving the fenton reaction. J. Bacteriol. 185:1942-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reidl, J., and W. Boos. 1991. The malX malY operon of Escherichia coli encodes a novel enzyme II of the phosphotransferase system recognizing glucose and maltose and an enzyme abolishing the endogenous induction of the maltose system. J. Bacteriol. 173:4862-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Schlegle, A., A. Bohm, S. J. Lee, R. Peist, K. Decker, and W. Boos. 2002. Network regulation of Escherichia coli maltose system. J. Mol. Microbiol. Biotechnol. 4:301-307. [PubMed] [Google Scholar]
- 29.Siegel, L. M. 1965. A direct microdetermination for sulfide. Anal. Biochem. 11:126-132. [DOI] [PubMed] [Google Scholar]
- 30.Sirko, A., M. Hryniewicz, D. Hulanicka, and A. Böck. 1990. Sulfate and thiosulfate transport in Escherichia coli K-12: nucleotide sequence and expression of the cysTWAM gene cluster. J. Bacteriol. 172:3351-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sirko, A., M. Zatyka, and M. D. Hulanicka. 1987. Identification of Escherichia coli cysM gene encoding O-acetylserinesulfhydrylase B by cloning with mini-Mu-lac containing a plasmid replicon. J. Gen. Microbiol. 133:2719-2725. [DOI] [PubMed] [Google Scholar]
- 32.Takagi, H., C. Kobayashi, S. Kobayashi, and S. Nakamori. 1999. PCR random mutagenesis into Escherichia coli serine acetyltransferase: isolation of the mutant enzymes that cause overproduction of L-cysteine and L-cystine due to the desensitization to feedback inhibition. FEBS Lett. 452:323-327. [DOI] [PubMed] [Google Scholar]
- 33.Takagi, H., N. Awano, S. Kobayashi, C. Kobayashi, M. Noji, K. Saito, and S. Nakamori. 1999. Overproduction of L-cystine and L-cystine by expression of genes for feedback inhibition-insensitive serine acetyltransferase from Arabidopsis thaliana in Escherichia coli. FEMS Microbiol. Lett. 179:453-459. [DOI] [PubMed] [Google Scholar]
- 34.Tsunoda, T., S. Eguchi, and K. Narumi. 1961. On the bioassay of amino acids. II. Determination of arginine, aspartic acid and cysteine. Amino Acids 3:7-13. [Google Scholar]
- 35.Zdych, E., R. Peist, J. Reidl, and W. Boos. 1995. MalY of Escherichia coli is an enzyme with the activity of a βC-S lyase (cystathionase) J. Bacteriol. 177:5035-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]