Abstract
Three upland soils from Thailand, a natural forest, a 16-year-old reforested site, and an agricultural field, were studied with regard to methane uptake and the community composition of methanotrophic bacteria (MB). The methane uptake rates were similar to rates described previously for forest and farmland soils of the temperate zone. The rates were lower at the agricultural site than at the native forest and reforested sites. The sites also differed in the MB community composition, which was characterized by denaturing gradient gel electrophoresis (DGGE) of pmoA gene fragments (coding for a subunit of particulate methane monooxygenase) that were PCR amplified from total soil DNA extracts. Cluster analysis based on the DGGE banding patterns indicated that the MB communities at the forested and reforested sites were similar to each other but different from that at the farmland site. Sequence analysis of excised DGGE bands indicated that Methylobacter spp. and Methylocystis spp. were present. Sequences of the “forest soil cluster” or “upland soil cluster α,” which is postulated to represent organisms involved in atmospheric methane consumption in diverse soils, were detected only in samples from the native forest and reforested sites. Additional sequences that may represent uncultivated groups of MB in the Gammaproteobacteria were also detected.
The current atmospheric mixing ratio of the greenhouse gas methane (CH4) is 1.75 ppm by volume (8). An estimated 30 Tg of CH4 year−1 is consumed via microbiological oxidation in upland soils, accounting for about 6% of the global methane sink (17). Atmospheric methane oxidation has been detected in many different upland soils, including arctic and subarctic tundra soils, grasslands and arable soils of the temperate zone, tropical forest soils, savannah soils, and even arid desert soils (25, 31). Most studies have been performed to estimate the methane uptake capacity of upland soils of the temperate zone, and few data are available for tropical and subtropical soils (10, 21, 22, 27, 28). Most of these data indicate that the methane uptake rates of tropical and subtropical forest soils are comparable to those of forest soils of the temperate zone, but higher oxidation rates have been reported for some tropical soils (29).
Among upland soils, forest soils are much more efficient methane sinks than cultivated soils (1, 3). Changes in land use, especially cultivation of formerly undisturbed soils, reduce the sink strength for atmospheric methane by 60 to 90% (30, 31). Such reductions have been reported for tropical soils as well (10, 21). However, the methane oxidation rate seems to recover much faster after abandonment of agricultural activities (22) compared to systems in the temperate zone.
Methane oxidation in upland soils is mediated by methanotrophic bacteria (MB). Seven recognized genera of MB belong to the group containing the type I methanotrophs (Gammaproteobacteria), while the group containing the type II methanotrophs consists of four genera of MB (Alphaproteobacteria). The enzyme methane monooxygenase catalyzes the first step in methane oxidation. With the exception of Methylocella (6, 7, 9), all MB possess the particulate form of this enzyme (particulate methane monooxygenase [pMMO]). The pmoA gene, encoding the active site subunit of the pMMO, is therefore a specific and almost universal marker gene for methanotrophs and has often been targeted by cultivation-independent methods to characterize the MB communities in different soils. Sequences closely related to the sequences of seven genera of MB have been detected by these methods in upland soils (4, 12, 15, 29). The presence of some pmoA sequences indicated that uncultivated MB are present in these soils. Uncultivated organisms that harbor pmoA sequences of upland soil cluster α (USCα) or upland soil cluster γ are most likely involved in atmospheric methane oxidation in upland soils (16, 23). With the exception of one soil sample from a rainforest in Brazil, in which USCα was detected (16), characterization of soil methanotrophic communities has been restricted to upland soils from the temperate zone.
In the present study, we compared the methane oxidation activities and methanotrophic community compositions of tropical soils under different land uses, including a natural forest site, a reforestation site, and a cornfield. The methane oxidation activities and community compositions of these soils were analyzed and compared with other data available for temperate soils.
MATERIALS AND METHODS
Sampling sites.
Three adjacent sampling sites were located in Amphur Wang Nham Keaw, Nakorn Ratchasrima province in Thailand. The forest site (site SK) was a natural dry evergreen forest within the Sakerat Experimental Station. The dominant tree species were Hopea ferrea, Pterocarpus marcrocarpus, Xylia xylocarpar, Dalbergia cochinchinensis, Lagerstroemia duppereana, and Shorea henryana. The reforestation site (site AC) was located about 5 km from site SK. It was planted with fast-growing, nitrogen-fixing tree species in 1988. The studies were performed on a plantation of Acacia mangium. Before reforestation, this site was used as farmland (corn and cassava). The third sampling site was a cornfield (site CF) located adjacent to the restoration area (2 km away). It was deforested more than 40 years ago, and corn has been continuously cultivated at this site for the last 16 years. The soils at sites SK and AC were highly acidic (pH 4.2, 1:1 in water), while the pH at site CF was 5.6. The soil texture at site SK is clay (clay content, 51%), and at sites AC (clay content, 30%) and CF (clay content, 21%) the texture is sandy clay loam. Soil preparation at the CF site started in mid-June 2003, and seeds of corn were sown on 10 July 2003. On this date, chemical fertilizer (16-20-0, N-P2O5-K2O) was applied at a rate of 50 kg ha−1. Soil samples for molecular analysis were taken from a depth of 10 to 20 cm on 27 and 28 July 2003.
Methane oxidation activity.
The net methane flux across the soil surface was determined in March and July 2003 using a closed static chamber method. The chambers consisted of a top made of transparent acrylic glass (30 cm wide by 30 cm long by 15 cm high) that could be attached to a fixed collar made of stainless steel. The collar was inserted 10 to 15 cm deep into the soil and remained there during the whole study period. The chambers were 10 to 15 m from each other. During measurement, the chamber top was inserted into a 1-cm-deep water-filled channel of the fixed collar. Using 30-ml syringes inserted through silicon stoppers in the chamber lids, 6 to 10 25- to 30-ml gas samples were taken from each chamber during the 1 h after capping. Methane mixing ratios of the gas samples were determined with a Shimadzu gas chromatograph (model 14B) equipped with a flame ionization detector and an Unibead C packed column (injector temperature, 120°C; oven temperature, 100°C; detector temperature, 300°C). The first-order uptake rate constant was estimated from the exponential decrease in methane over time. The methane oxidation rate was calculated from the first-order uptake rate constant by multiplication with the initial methane mixing ratio in the chamber headspace.
DNA extraction.
DNA was extracted from 0.5 g of soil using a Fast DNA SPIN kit (Bio 101, La Jolla, CA) according to the manufacturer's instructions, with modifications. Cell lysis was performed with a cell disruptor (FP 120 FastPrep; Savant Instruments Inc., Farmingdale, NY) operating at 6.5 m s−1 by using two 45-s cycles. After centrifugation (20,000 × g, 10 min) the supernatant was collected, and the pellet containing soil and beads was resuspended in buffers supplied with the kit for a second DNA extraction. Proteins and cell debris were precipitated as described in the manufacturer's instructions, and 1 ml of binding matrix was added to the supernatant. For purification, the matrix-bound DNA was washed three times with 1 ml of a 5.5 M guanidine isothiocyanate solution (modified as described by Yeates and Gillings [33]). After the last centrifugation step (14,000 × g, 1 min) the supernatant was removed, and the matrix-bound DNA was suspended in 0.6 ml of a guanidine isothiocyanate solution and loaded onto SPIN filters supplied with the kit. Final elution of the DNA from the filters was performed twice with 100 μl of DNase-free water. DNA extracts were purified with polyvinylpolypyrrolidone and a QIAquick PCR purification kit (QIAGEN, Hilden, Germany) (23).
PCR amplification and denaturing gradient gel electrophoresis (DGGE).
A partial fragment of the pmoA gene was PCR amplified with primer A189 with a GC clamp (12) in combination with either primer A682 (15) or primer mb661 (5). A touchdown PCR program (12) with annealing temperatures decreasing from 64 to 57°C was used for 35 cycles. A fragment of the mmoX gene encoding the active site subunit of soluble methane monooxygenase (sMMO) was amplified with a newly designed forward primer, mmoXf945 (5′-TGG GGY GNA ATC TGG AT-3′), and reverse primer mmoXB1401 of Auman et al. (2). A GC clamp described by Iwamoto et al. (18) was attached to the 5′ end of primer mmoXf975. The PCR program consisted of an initial denaturation step of 5 min at 94°C, followed by 35 cycles of 94°C for 1 min, 63 to 56°C for 1 min, and 72°C for 1 min and then a final extension step of 72°C for 7 min.
All PCR mixtures contained each primer at a concentration of 1.0 μM, 5 μl of Accutaq-LA 10× buffer (Sigma-Aldrich, Taufkirchen, Germany), 3.5 mM MgCl2, 25 μg of bovine serum albumin (Roche Diagnostics GmbH, Mannheim, Germany), each deoxynucleoside triphosphate (Promega, Mannheim, Germany) at a concentration of 200 μM, 2.5 U of REDAccuTaq LA DNA polymerase (Sigma-Aldrich), 1 μl of template DNA, and enough sterile water (Q-Biogene, Heidelberg, Germany) to bring the volume to 50 μl. Mixed pmoA PCR products were separated by DGGE. Visible bands were excised and reamplified. PCR products from excised DGGE bands were purified and sequenced exactly as described by Knief et al. (23).
Phylogenetic sequence analysis.
Phylogenetic tree reconstructions were based on deduced amino acid sequences of partial pmoA and amoA sequences and were prepared with the ARB software package (24). The original tree construction included sequences of all DGGE bands, all available pmoA sequences in the GenBank database (July 2004), and selected public domain amoA sequences. Then 22 representative sequences from this study plus 38 public domain sequences were selected for analysis.
Cluster analysis.
A cluster analysis was performed based on the presence or absence of 21 different bands visible in DGGE gels of PCR products amplified with primers A189-GC and A682 and with primers A189-GC and mb661. Jaccard similarity coefficients were calculated, and a hierarchical tree was constructed based on average linkage and Euclidean distances using SYSTAT, version 10.2 (SPSS Inc., Richmond, CA).
Isolation of methanotrophic bacteria.
Diluted nitrate mineral salts medium at a slightly acid pH (pH 5.8) was used to enrich for MB as described by Dunfield et al. (9) with soil samples from forested sampling sites SK and AC.
Nucleotide sequence accession numbers.
Representative pmoA nucleotide sequences obtained during this study have been deposited in the GenBank, EMBL, and DDBJ nucleotide sequence databases under accession numbers AJ868266 to AJ868287.
RESULTS
In situ atmospheric methane oxidation.
Methane flux was measured in March 2003 and in July 2003 (on the day that soil samples were taken for molecular studies). Ten chambers were used for flux estimates at site SK, while five chambers were used at sites AC and CF. Only data from chambers with significant correlation of the measurement points (the Pearson correlation coefficient of log-transformed flux data versus time was significantly more than zero at the P = 0.05 level) were taken into account for calculation of the average methane flux. Thus, the average values given below for site SK were based on the data from only five chambers in March and six chambers in July, and the average values for sites AC and CF were based on four chambers in March and three chambers in July. Net atmospheric methane consumption was observed at sites SK and AC. At site CF net oxidation of methane was observed only in March. The methane uptake rates obtained from different chambers at the same sampling site showed large variations. Not all chambers at a site showed net consumption of methane. Likewise, in some chambers neither net emission nor net consumption was observed. The net methane fluxes at site SK ranged from −4.4 to −0.6 mg CH4 m−2 day−1 in March and from −0.2 to −1.2 mg CH4 m−2 day−1 in July. At site AC, the net methane fluxes ranged from −0.9 to −1.8 mg CH4 m−2 day−1 in March and from 2.7 to −5.6 mg CH4 m−2 day−1 in July; and at site CF the values ranged from −1.4 to 1.4 mg CH4 m−2 day−1 in March and from 0.3 to 1.4 mg CH4 m−2 day−1 in July. A negative flux indicates net consumption, while a positive flux indicates net emission. To test for significant differences in the net methane fluxes for the sampling sites, the nonparametric Mann-Whitney test was used. For this comparison, the flux data for both forested sites for both months were combined and compared to the data for the farmland site. The results of this analysis indicate that the net methane fluxes at the two forested sites were significantly different (more negative) than the flux at the CF site (P < 0.01).
Characterization of the methanotrophic communities.
Using cultivation-independent methods, the MB communities were characterized by using soil samples taken from a depth of 10 to 20 cm during the second half of the wet season. Duplicate soil samples were taken from each sampling site. The pmoA gene fragment, which was used as a functional marker for characterization of the methanotrophic communities, was successfully PCR amplified from all soil DNA extracts. A fragment of the mmoX gene could not be amplified from any sample. Two different reverse primers were used for amplification of the pmoA gene fragment. Primer A682 is the less specific of the two, since it was designed to amplify both pmoA and amoA gene fragments (15). Primer mb661 was designed to amplify pmoA gene fragments specifically and did not amplify amoA gene fragments (5).
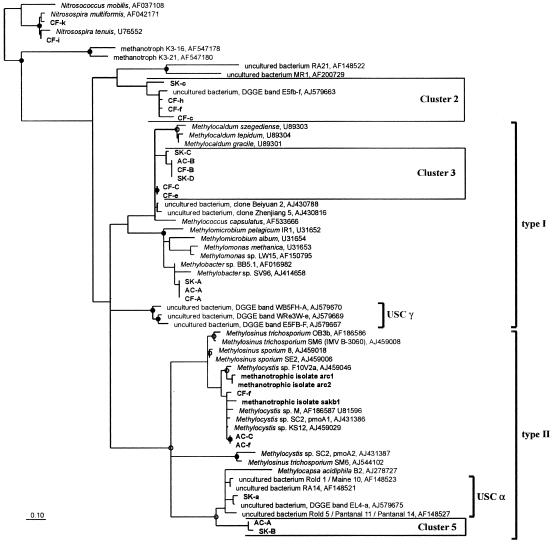
The recovery of PmoA sequences very closely related to sequences from isolates of Methylocystis and Methylobacter indicated that well-known cultivated genera of MB were present at all sites (>96% amino acid identity) (Table 1). Sequences of the USCα, which probably represented uncultivated MB, were detected at sites SK and AC (98% identity) but not at site CF. Likewise, pmoA sequences belonging to a group that was a closely related to but clearly distinct from previously detected USCα sequences (cluster 5) were detected in these two soils but not at site CF. Cluster 5 sequences AC-A1, AC-A2, and SK-B in the phylogenetic tree (Fig. 1) exhibited maximum levels of identity of 81% to USCα sequences, represented by EL4-a in Fig. 1. While USCα sequences were obtained only when primer A682 was used in PCRs, the novel sequence type was detected only when primer mb661 was used.
TABLE 1.
PmoA sequence types detected at the different sampling sitesa
| Detected taxon | Site SK
|
Site AC
|
Site CF
|
|||
|---|---|---|---|---|---|---|
| A682b | mb661 | A682b | mb661 | A682b | mb661 | |
| Ammonia oxidizers (AmoA) | + | |||||
| Cluster 2 (PmoA/AmoA) | + | + | ||||
| Methylobacter | + | + | + | + | ||
| Cluster 3 | + | + | + | + | ||
| Methylocystis | + | + | + | + | ||
| USCα | + | + | ||||
| Cluster 5 | + | + | ||||
Phylogenetic positions of the different taxa are shown in Fig 1.
FIG. 1.
Phylogenetic tree based on deduced amino acid sequences of partial pmoA and amoA sequences, showing the relationship of sequences detected at the different Thailand sampling sites to sequences of pure MB cultures and sequences detected in other cultivation-independent studies. Sequences from this study are indicated by boldface type and are designated according to the sites (SK, native forest; AC, reforestation; CF, cornfield). Sequences recovered from PCR products that were amplified using the A682 reverse primer are indicated by lowercase letters, while uppercase letters indicate sequences from PCR products that were obtained using the mb661 reverse primer. PmoA sequences of two methanotrophic isolates obtained from soils at sites SK and AC (sak1b and arc2) were also included. The tree was calculated based on 146 amino acid positions using the Treepuzzle algorithm with the Jones-Taylor-Thornton evolutionary model (20). The positions of recovered PmoA and AmoA sequences in the tree were confirmed by the topology of a neighbor-joining tree. The solid circles indicate branches that were present in 90% of 25,000 reconstructed Treepuzzle trees, and the open circles indicate branches that were present in 80% of the trees. AmoA sequences of ammonia-oxidizing bacteria were used as an outgroup. Bar = 0.10 change per amino acid position.
Another PmoA sequence type that was indicative of another taxon of uncultivated MB was detected at all sampling sites. PmoA sequences of this “cluster 3” formed two separate branches, which were distantly related to sequences of MB in the Gammaproteobacteria. The levels of amino acid identity of PmoA sequences in these branches were 93% to each other, 93 to 97% to sequences of the uncultivated bacteria represented by “clone Zhenjiang 5” and “clone Beijyuan 2,” and 87 to 93% to sequences of cultivated methanotrophs of the genera Methylococcus and Methylocaldum. Since the amino acid identity of these novel sequences was approximately equal to that observed for PmoA sequences from members of both genera (Methylococcus and Methylocaldum), the phylogenetic relationship could not be resolved and is represented by a multifurcation in the tree (Fig. 1).
Sequences whose phylogenetic position was intermediate with respect to sequences belonging to clearly definable AmoA or PmoA sequence types were detected in soil samples from sites SK and CF. These “cluster 2” sequences exhibited 88 to 97% identity to a sequence previously recovered by cultivation-independent methods from a temperate forest soil, represented by DGGE band E5FB-f (23). Sequences that were phylogenetically most closely related to amoA genes of ammonia-oxidizing bacteria were detected only at site CF.
Isolation of MB from forested sites.
The methanotrophic isolate sakb1 was isolated from site SK, while isolate arc2 was obtained from site AC. Based on the partial pmoA gene sequences, these isolates were identified as Methylocystis spp. (Fig. 1). This result was confirmed by 16S rRNA sequence analysis and by the morphology of the isolates. The pmoA sequences of the isolates from site AC were not identical to the pmoA sequences that were detected in this soil by cultivation-independent methods. At site SK, pmoA sequences that indicated the presence of Methylocystis spp. were not detected.
Relationship between sampling site and community composition of methane-oxidizing bacteria.
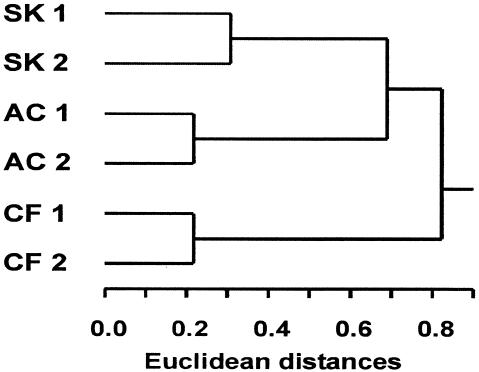
The DGGE banding pattern of PCR products obtained with both backward primers indicated that there were clear differences in the community compositions of methane-oxidizing bacteria at the different sampling sites. The duplicates analyzed from each sampling site were very similar to each other, as demonstrated by the results of a cluster analysis performed based on the DGGE banding pattern (Fig. 2). The duplicates from all soil samples formed separate clusters. The MB community similarity between sites SK and AC was greater than the similarity of either site SK or site AC to site CF.
FIG. 2.
Cluster analysis based on the presence or absence in DGGE gels of 21 bands from PCR products obtained with primers A189-GC and A682 and with primers A189-GC and mb661. Jaccard similarity coefficients were calculated, and a hierarchical tree was constructed based on average linkage and Euclidean distances.
DISCUSSION
In recent years, several studies have examined the methane oxidation activity together with the structure of the methanotrophic community of diverse upland soils (13, 16, 19, 23, 26). With the exception of a Brazilian rainforest soil (16), all data on methanotrophic community structure are data for upland soils of the temperate zone of the Northern Hemisphere. Because some tropical sites have much higher rates of methane oxidation than temperate sites (27, 28, 29), it is of interest to investigate the MB community composition in these areas.
The measured methane uptake rates of the Thailand soils (up to −5.6 mg CH4 m−2 day−1 at site AC) were similar to rates published previously in several other studies of upland soils (up to −7 mg CH4 m−2 day−1) but were not as high as the highest rates reported for some tropical forest soils (up to −14 mg CH4 m−2 day−1) (27, 28). Both forested sites (sites SK and AC) showed net methane uptake, but only weak methane uptake or even net methane emission was observed at site CF. This indicates that some parts of the soil profile might have been saturated with water to the level that methanogenesis developed and overloaded the total methane oxidation potential. Although the study was not blocked in order to rule out spatial effects, this general trend agrees well with previous studies indicating that the conversion of natural forest sites to farmland leads to a 60 to 90% reduction in the atmospheric methane uptake rate but that the activity recovers at reforested sites (21, 29, 32). In addition, it has been found that the oxidation rates of tropical soils recover much faster after abandonment of agricultural activities than the oxidation rates of temperate systems recover (22, 29). This may explain why the methane oxidation rate at reforestation site AC was comparable to the rate at site SK, although site AC was reforested only 16 years ago.
Fragments of the pmoA gene were amplified from all soil DNA extracts, but mmoX genes were not recovered from the soil samples. This is in accordance with results obtained for upland soils of central Europe (23). Either the dominant MB in upland soils do not possess mmoX genes or this assay has a higher detection limit than the pmoA assay. The former possibility is reasonable considering that MB in upland soils have to live on very low substrate concentrations. Under these oligotrophic conditions it should be preferable to use pMMO rather than sMMO, since it has a lower energy demand for the oxidation of methane to methanol. MB expressing pMMO have higher growth yields and show a higher affinity for methane than MB expressing sMMO (11).
The only known genera of MB detected in the Thailand soil samples were Methylocystis and Methylobacter. While Methylocystis has been detected in several different upland soils (4, 16, 23, 26, 30), Methylobacter has been detected only in an acidic heathland soil (4). Some Methylocystis spp. were also isolated from the soils, although interestingly, based on PmoA phylogeny the strains were not the same strains detected by cultivation-independent methods in the soil samples. This result emphasizes the fact that both molecular and cultivation methods are subject to inherent biases.
Unusual sequences obtained from several DGGE bands indicated that different taxa of uncultivated putative methanotrophs were also present in the soil samples. Organisms harboring pmoA sequences of USCα have been detected previously in several upland soils, including acidic tropical forest soils (16). Such sequences were present at sampling sites SK and AC, as were sequences that represent cluster 5, which is closely related to USCα. Cluster 3 sequences represent a group of uncultivated type I methanotrophs related to Methylocaldum and Methylococcus. A further cluster of PmoA sequences within this broad Methylococcus-Methylocaldum group (type X MB) is formed by sequences that were detected in different rice field soils (clone Beiyuan 2 and clone Zhenijang 5) (14). Thus, there seem to be diverse uncultivated MB taxa which have (on the PmoA level) a phylogenetic position between Methylocaldum spp. and Methylococcus spp. So far, the detection of a third cluster representing uncultivated bacteria, cluster 2, has been restricted to upland soils. Since the sequences of this branch are only distantly related (<69%) to PmoA sequences of recognized genera of MB, the possibility that these sequences code for the AmoA of uncultivated ammonia-oxidizing bacteria cannot be excluded.
The different land uses at the sampling sites were reflected by different methane uptake rates and different soil pHs, as well as by different methanotrophic communities, especially for the cornfield soil. The pmoA sequences of USCα and cluster 5 were detected in samples from sites SK and AC but not in the soil samples from the cornfield. The absence of USCα at site CF is in agreement with results of other studies, in which the methanotrophic communities of some farmland soils were analyzed and in which USCα sequences were not detected, although they were present in adjacent grassland and forest soils (23). Although the organisms harboring USCα sequences appear to occur more frequently in acidic soils than in soils with a neutral pH, they do occur in soils at a wide pH range (pH 4.0 to 8.0) (23), so their absence from the farmland soil is probably not the result of a pH shift alone. Other sequences that led to the different community compositions at the different sampling sites were sequences of Methylocystis spp., cluster 2, and ammonia-oxidizing bacteria (Table 1). Sequences of Methylobacter and cluster 3 were detected at all sampling sites. A cluster analysis was used to verify the differences numerically. The resulting cluster diagram confirmed the high similarity of replicate samples from one sampling site and differences between the sampling sites (Fig. 2). The differences within the replicate samples from one site were due to faint DGGE bands visible in one sample but not in the other samples. Thus, pmoA sequences with abundance near the detection limit of the methods were mainly responsible for the dissimilarity of duplicate samples. The main purpose of this cluster analysis was to compare the different sampling sites. The community compositions at sites SK and AC were more similar to each other than either was to the community composition at site CF. This finding is in accordance with the methane uptake rates and pHs of the sampling sites, which also indicated a high degree of dissimilarity between site CF and sites SK and AC.
These data suggest that land use changes are connected not only to changes in the methane uptake capacity of soils but also to changes in the community composition of methane-oxidizing bacteria. The change in the community composition may be an important factor leading to the general decrease in the methane uptake rate of farmland soils. To understand the effects on land use and to confirm our findings, however, further studies on the relationship among land use, MB community composition, and the in situ methane uptake rate are needed.
It is noteworthy that the community composition at reforested site AC represents a kind of intermediate state between the communities at sites SK and CF. Since the community composition and the methane uptake activity at reforested site AC before the site was reforested are not known, we cannot conclude whether the community that is there today redeveloped or whether the community structure was not drastically changed after deforestation.
Acknowledgments
This work was supported by the Thailand Research Fund (grant MRG4580019) and the Deutsche Forschungsgemeinschaft (grant DU 377/1-1).
We thank Chongrak Watcharinrat of Kasetsart University for providing his experimental plots for soil sampling and Nathapol Lichaikul of the Joint Graduate School of Energy and Environment for sampling assistance and analysis. The Sakaerat Environmental Research Station (Ministry of Science and Technology) and the Re-Afforestation Research and Training Station (Ministry of Natural Resources and Environment) kindly permitted access to the experimental sites and other facilities.
REFERENCES
- 1.Ambus, P., and S. Christensen. 1995. Spatial and seasonal nitrous-oxide and methane fluxes in Danish forest-ecosystems, grassland-ecosystems, and agroecosystems. J. Environ. Qual. 24:993-1001. [Google Scholar]
- 2.Auman, A. J., S. Stolyar, A. M. Costello, and M. E. Lidstrom. 2000. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl. Environ. Microbiol. 66:5259-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Born, M., H. Dörr, and L. Ingeborg. 1990. Methane consumption in aerated soils of the temperate zone. Tellus Ser. B 42:2-8. [Google Scholar]
- 4.Bournes, D. G., I. R. McDonald, and J. C. Murrell. 2001. Comparison of pmoA PCR primer sets as tools for investigating methanotroph diversity in three Danish soils. Appl. Environ. Microbiol. 67:3802-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costello, A. M., and M. E. Lidstrom. 1999. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 65:5066-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dedysh, S. N., Y. Y. Berestovskaya, L. V. Vasylieva, S. E. Belova, V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, W. Liesack, and G. A. Zavarzin. 2004. Methylocella tundrae sp. nov., a novel methanotrophic bacterium from acidic tundra peatlands. Int. J. Syst. Evol. Microbiol. 54:151-156. [DOI] [PubMed] [Google Scholar]
- 7.Dedysh, S. N., W. Liesack, V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, J. D. Semrau, A. M. Bares, N. S. Panikov, and J. M. Tiedje. 2000. Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int. J. Syst. Evol. Microbiol. 50:955-969. [DOI] [PubMed] [Google Scholar]
- 8.Dlugokencky, E. J., S. Houweling, L. Bruhwiler, K. A. Masarie, P. M. Lang, J. B. Miller, and P. P. Tans. 2003. Atmospheric methane levels off: temporary pause or a new steady-state? Geophys. Res. Lett. 30:1-4. 8. [Google Scholar]
- 9.Dunfield, P. F., V. N. Khmelenina, N. E. Suzina, T. Y. A., and S. N. Dedysh. 2003. Methylocella silvestris sp. nov., a novel methanotrophic bacterium isolated from an acidic forest cambisol. Int. J. Syst. Evol. Microbiol. 53:1231-1239. [DOI] [PubMed] [Google Scholar]
- 10.Goreau, T. J., and W. Z. de Mello. 1988. Tropical deforestation: some effects on atmospheric chemistry. Ambio 17:275-281. [Google Scholar]
- 11.Hanson, R. S., and T. E. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henckel, T., M. Friedrich, and R. Conrad. 1999. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl. Environ. Microbiol. 65:1980-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henckel, T., U. Jäckel, S. Schnell, and R. Conrad. 2000. Molecular analyses of novel methanotrophic communities in forest soil that oxidize atmospheric methane. Appl. Environ. Microbiol. 66:1801-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann, T., H. P. Horz, D. Kemnitz, and R. Conrad. 2002. Diversity of the particulate methane monooxygenase gene in methanotrophic samples from different rice field soils in China and the Philippines. Syst. Appl. Microbiol. 25:267-274. [DOI] [PubMed] [Google Scholar]
- 15.Holmes, A. J., A. Costello, M. E. Lidstrom, and J. C. Murrell. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132:203-208. [DOI] [PubMed] [Google Scholar]
- 16.Holmes, A. J., P. Roslev, I. R. McDonald, N. Iversen, K. Henriksen, and J. C. Murrell. 1999. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl. Environ. Microbiol. 65:3312-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houghton, J. T., Y. Ding, D. J. Griggs, M. Noguer, P. J. van der Linden, and D. Xiaosu. 2001. IPPC climate change 2001: the scientific basis. Contribution of working group I to the third assessment report of the Intergovernmental Panel on Climate Change (IPCC). Cambridge University Press, Cambridge, United Kingdom.
- 18.Iwamoto, T., K. Tani, K. Nakamura, Y. Suzuki, M. Kitagawa, M. Eguchi, and M. Nasu. 2000. Monitoring impact of in situ biostimulation treatment on groundwater bacterial community by DGGE. FEMS Microbiol. Ecol. 32:129-141. [DOI] [PubMed] [Google Scholar]
- 19.Jensen, S., A. J. Holmes, R. A. Olsen, and J. C. Murrell. 2000. Detection of methane oxidizing bacteria in forest soil by monooxygenase PCR amplification. Microb. Ecol. 39:282-289. [PubMed] [Google Scholar]
- 20.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 21.Keller, M., M. E. Mitre, and R. F. Stallard. 1990. Consumption of atmospheric methane in tropical soils of central Panama. Global Biogeochem. Cycles 4:21-27. [Google Scholar]
- 22.Keller, M., and W. A. Reiners. 1994. Soil-atmosphere exchange of nitrous oxide, nitric oxide, and methane under secondary succession of pasture to forest in the Atlantic lowlands of Costa Rica. Global Biogeochem. Cycles 8:399-409. [Google Scholar]
- 23.Knief, C., A. Lipski, and P. F. Dunfield. 2003. Diversity and activity of methanotrophic bacteria in different upland soils. Appl. Environ. Microbiol. 69:6703-6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckman, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mosier, A., D. Schimel, D. Valentine, K. Bronson, and W. Parton. 1991. Methane and nitrous oxide fluxes in native, fertilized and cultivated grasslands. Nature 350:330-332. [Google Scholar]
- 26.Reay, D. S., S. Radajewski, J. C. Murrell, N. McNamara, and D. B. Nedwell. 2001. Effects of land-use on the activity and diversity of methane oxidizing bacteria in forest soils. Soil Biol. Biochem. 33:1613-1623. [Google Scholar]
- 27.Singh, A. K., A. S. Raghubanshi, V. S. Reddy, S. Singh, and A. K. Kashyap. 1998. Methane flux from irrigated paddy and dryland rice fields, and from seasonally dry tropical forest and savanna soils of India. Soil Biol. Biochem. 30:135-139. [Google Scholar]
- 28.Singh, J. S., S. Singh, A. S. Raghubanshi, S. Singh, A. K. Kashyap, and V. S. Reddy. 1997. Effect of soil nitrogen, carbon and moisture on methane uptake by dry tropical forest soils. Plant Soil 196:115-121. [Google Scholar]
- 29.Smith, K. A., K. E. Dobbie, B. C. Ball, L. R. Bakken, B. K. Sitaula, S. Hansen, R. Brumme, W. Borken, S. Christensen, A. Prieme, D. Fowler, J. A. Macdonald, U. Skiba, L. Klemedtsson, A. Kasimir-Klemedtsson, A. Degorska, and P. Orlanski. 2000. Oxidation of atmospheric methane in northern European soils, comparison with other ecosystems, and uncertainties in the global terrestrial sink. Global Change Biol. 6:791-803. [Google Scholar]
- 30.Steinkamp, R., W. Zimmer, and H. Papen. 2001. Improved method for detection of methanotrophic bacteria in forest soils by PCR. Curr. Microbiol. 42:316-322. [DOI] [PubMed] [Google Scholar]
- 31.Whalen, R. T., and W. S. Reeburgh. 1990. Consumption of atmospheric methane by tundra soils. Nature 346:160-162. [Google Scholar]
- 32.Willison, T. W., C. P. Webster, K. W. T. Goulding, and D. S. Powlson. 1995. Methane oxidation in temperate soils: effects of land use and the chemical form of nitrogen fertilizer. Chemosphere 30:539-546. [Google Scholar]
- 33.Yeates, C., and M. R. Gillings. 1998. Rapid purification of DNA from soil for molecular biodiversity analysis. Lett. Appl. Microbiol. 27:49-53. [Google Scholar]