Abstract
Anthocyanins are red, purple, or blue plant pigments that belong to the family of polyphenolic compounds collectively called flavonoids. Their demonstrated antioxidant properties and economic importance to the dye, fruit, and cut-flower industries have driven intensive research into their metabolic biosynthetic pathways. In order to produce stable, glycosylated anthocyanins from colorless flavanones such as naringenin and eriodictyol, a four-step metabolic pathway was constructed that contained plant genes from heterologous origins: flavanone 3β-hydroxylase from Malus domestica, dihydroflavonol 4-reductase from Anthurium andraeanum, anthocyanidin synthase (ANS) also from M. domestica, and UDP-glucose:flavonoid 3-O-glucosyltransferase from Petunia hybrida. Using two rounds of PCR, each one of the four genes was first placed under the control of the trc promoter and its own bacterial ribosome-binding site and then cloned sequentially into vector pK184. Escherichia coli cells containing the recombinant plant pathway were able to take up either naringenin or eriodictyol and convert it to the corresponding glycosylated anthocyanin, pelargonidin 3-O-glucoside or cyanidin 3-O-glucoside. The produced anthocyanins were present at low concentrations, while most of the metabolites detected corresponded to their dihydroflavonol precursors, as well as the corresponding flavonols. The presence of side product flavonols is at least partly due to an alternate reaction catalyzed by ANS. This is the first time plant-specific anthocyanins have been produced from a microorganism and opens up the possibility of further production improvement by protein and pathway engineering.
Among the natural pigments in plants, anthocyanins are the largest water-soluble group, found in most fruits, flower petals, and leaves. These fascinating compounds can exist in many structural forms, both simple and complex, governed by physiological regulations and chemical modifications which have profound effects on their stability and colors (13). Among the variety of biological roles, anthocyanins are utilized for the recruitment of pollinators and seed dispersers and in UV protection. Initial interest in the practical applications of brightly colored anthocyanins has stemmed from their potential as replacements for banned dyes because they have no apparent adverse effects on human health (2, 5, 24). Recently, however, much attention has been drawn to anthocyanin-derived plant products due to their general antioxidant properties (10, 17, 22) and a consistent association between the consumption of diets rich in fruits and vegetables and a lower risk of chronic diseases, including cancer and cardiovascular disease (6, 14). As a result, anthocyanins, currently produced industrially as mixtures from various plant extracts, are becoming attractive targets for fermentation production from well-characterized microbial hosts such as Escherichia coli.
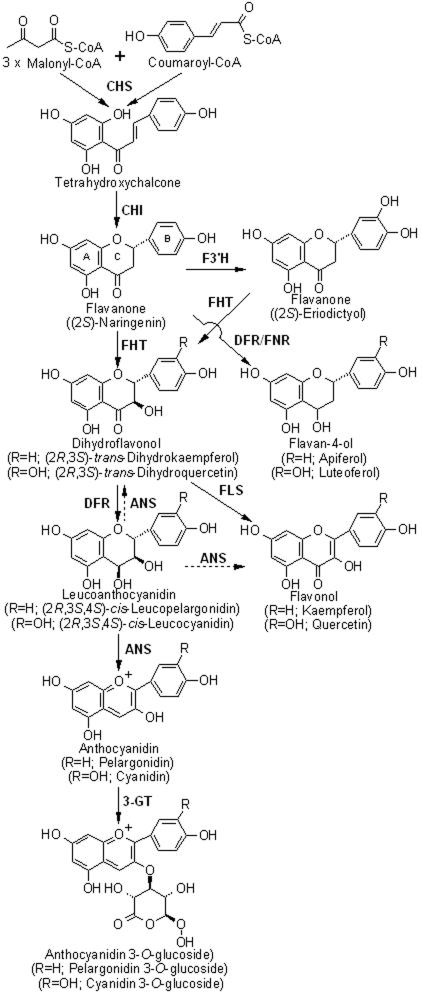
Six major classes of anthocyanidins, the aglycon forms of anthocyanins, exist: pelargonidin, cyanidin, delphinidin, peonidin, malvidin, and petunidin. The basic structure of an anthocyanin is a glycosylated form of polyhydroxy and polymethoxy derivatives of 2-phenylbenzopyrylium or flavylium salts (13). Differences between individual anthocyanins are the number of hydroxyl groups, the nature and number of sugars attached to the molecule and the position of the attachment, and the nature and number of aliphatic or aromatic acids attached to the sugars on the molecule. Biosynthesis of anthocyanins proceeds via the pathway chalcone → flavanone → dihydroflavonol → anthocyanidin → anthocyanin (Fig. 1) and has only recently been completely elucidated (16). The cDNA sequences of a large number of enzymes involved in the anthocyanin biosynthesis pathway from various plant species are now available.
FIG. 1.
Anthocyanin 3-O-glucoside biosynthetic pathway in plants. Compounds in parentheses represent the starting, intermediate, and final metabolites used or produced in the present study. The dashed arrows represent the leucoanthocyanidin oxidation activities that ANS demonstrates. Abbreviations: CHS, chalcone synthase; CHI, chalcone isomerase; FNR, flavanone reductase; FLS, flavonol synthase; F3′H, flavonoid 3′ hydroxylase; CoA, coenzyme A.
We present here the construction of an artificial gene cluster that contains the four plant-derived genes of the lower anthocyanin biosynthesis pathway that converts the flavanones naringenin and eriodictyol into the first two colored and stable anthocyanins, pelargonidin 3-O-glucoside and cyanidin 3-O-glucoside. This is the first time that plant-specific anthocyanin molecules have been synthesized through microbial fermentation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
E. coli TOP10F (Invitrogen, Carlsbad, Calif.) was used for DNA manipulations, while E. coli JM109 was used for shake flask experiments. Plasmids pTrcHis2-TOPO (Invitrogen) and pK184 were used for cloning.
Chemicals.
Callistephin chloride (pelargonidin 3-O-glucoside chloride), kuromanin chloride (cyanidin 3-O-glucoside chloride), cyanidin, and pelargonidin standards were purchased from ExtraSynthase. Naringenin was purchased from Sigma-Aldrich (St. Louis, Mo.), and eriodictyol was purchased from Indofine. Both trans-dihydrokaempferol and trans-dihydroquercetin were enzymatically synthesized from (2S)-naringenin and (2S)-eriodictyol, respectively, using flavanone 3β-hydroxylase from Malus domestica heterologously expressed in E. coli, as previously described for Petunia (4). The cis and trans dihydroflavonol epimers formed during the anthocyanidin synthase (ANS) in vitro assays were separated by high-performance liquid chromatography (HPLC) and verified by nuclear magnetic resonance (NMR) measurements. Natural (2R,3S,4S)-cis-leucoanthocyanidins and unnatural (2R,3S,4R)-trans-leucoanthocyanidins were synthesized from dihydroflavonols by chemical synthesis as previously described by Tanner et al. (25).
DNA manipulations.
All DNA manipulations were performed according to standard procedures (21). Restriction enzymes, calf intestine alkaline phosphatase, and T4 DNA ligase were purchased from New England Biolabs and Promega. All PCRs and reverse transcription (RT)-PCRs were performed using Roche's Expand High Fidelity PCR system. MdF3H and MdANS cDNAs from M. domestica were kind gifts from Chikako Honda (National Institute of Fruit and Tree Science, Japan) (7). PGT8 cDNA from Petunia hybrida and dfr from Anthurium andraeanum were cloned in our lab based on DNA sequences available in GenBank (accession numbers AB027454 for PGT8 and AY232494 for dfr). The QIAGEN RNeasy MiniKit was used for total RNA isolation from P. hybrida corolla or A. andraeanum red spadix. Reverse transcription for the cDNA generation was performed using SuperScript II (Invitrogen). In all cases, after PCR or RT-PCR amplification, the absence of undesired mutations was verified by direct nucleotide sequencing.
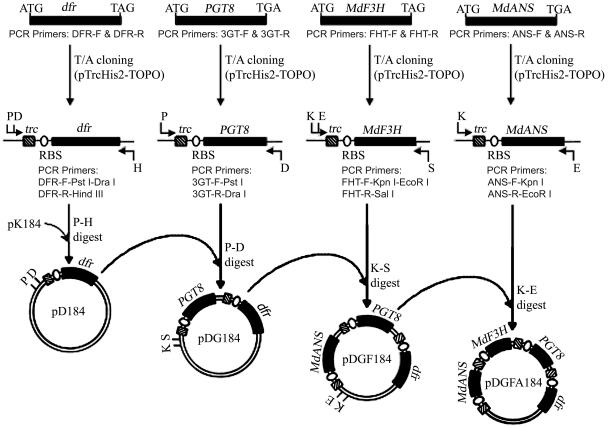
Construction of plasmid pDGFA184.
Plasmid pDGFA184 was constructed through two rounds of PCR for each one of the four genes cloned, as depicted in Fig. 2. In the first round of PCR, each of the four structural genes (from the ATG start codon to the stop codon) was amplified either from a plasmid provided or from total RNA, as previously described. After adding an A overhang to the PCR products using Taq polymerase (Fisher Scientific), each structural gene was individually cloned under the strong trc promoter by T/A cloning using pTrcHis2-TOPO as the cloning vector. The trc promoter is a hybrid E. coli promoter, which is induced by isopropyl-β-d-thiogalactopyranoside (IPTG). Two PstI sites and one SalI site in the A. andraeanum dfr gene were removed by introducing silent mutations using the QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, Calif.).
FIG. 2.
Schematic representation of the strategy used for constructing vector pDGFA184. Abbreviations used for restriction enzymes: P, PstI; H, HindIII; D, DraI; S, SalI; K, KpnI; E, EcoRV. By performing a first round of PCR or RT-PCR, the MdF3H, dfr, MdANS, or PGT8 gene was placed under the control of the E. coli trc promoter and an E. coli RBS derived from cloning vector pTrcHis2-TOPO. In a second round of PCR, each gene was amplified together with the trc promoter and RBS and placed sequentially into E. coli cloning vector pK184. The PCR and RT-PCR primer sequences used are presented in Table 1.
In the second round of cloning, the dfr cDNA, together with the trc promoter and the ribosome binding site (RBS), was amplified by PCR using a forward primer hybridizing to a vector DNA region that lies upstream of the trc promoter and a reverse primer hybridizing to the vector DNA region immediately after the stop codon. Two restriction sites, PstI and DraI, were introduced into the forward primer, and a HindIII restriction site was introduced into the reverse primer. The resulting PCR fragment was digested with PstI and HindIII and inserted into E. coli vector pK184 digested with the same enzymes, yielding plasmid pD184. Similarly, the PGT8 cDNA was amplified together with the trc promoter and RBS using a forward primer hybridizing upstream of the trc promoter and carrying the restriction site PstI, while the reverse primer, designed immediately downstream from the stop codon, contained the restriction site DraI. The amplified PCR fragment was digested with PstI and DraI and inserted into plasmid pD184 digested with the same enzymes, generating plasmid pDG184. Next, the MdF3H cDNA was amplified together with the trc promoter and RBS with a forward primer carrying restriction sites KpnI and EcoRV and a reverse primer carrying restriction site SalI. The resulting PCR fragment was digested with KpnI and SalI and inserted into plasmid pDG184 digested with the same enzymes, yielding plasmid pDGF184. Finally, the MdANS cDNA was amplified together with the trc promoter and RBS using a forward primer carrying the restriction site KpnI and a reverse primer carrying restriction site EcoRV. The resulting PCR fragment was digested with KpnI and EcoRV and inserted into plasmid pDGF184 digested with the same enzymes. This resulted in the final plasmid pDGFA184. All the PCR primer sequences used for the first and second rounds of PCR are presented in Table 1.
TABLE 1.
Primers used in this study and referred to in Fig. 1
| Primer | Sequence (5′-3′)a |
|---|---|
| DFR-F | GATGATGCACAAGGGCACCGTGTGC |
| DFR-R | CTGAATGGCCGTTGTCTTGCCCGGTG |
| 3GT-F | CATGACTACTTCTCAACTTCACATTGC |
| 3GT-R | TCAAGTAAGCTTGTGACATTTAACTAGCTC |
| FHT-F | ATGGCTCCTCCTGCTACTACGC |
| FHT-R | CTAAGCAAATATGTCGTCCG |
| ANS-F | ATGGTGAGCTCTGATTCAGTGA |
| ANS-R | TCACTTGGGGAGCAAAGCCTCT |
| DFR-F-PstI-DraI | GGGGCTGCAGGGGTTTAAACCGACATCATAACGGTTCTG |
| DFR-R-HindIII | CCCCAAGCTTCCCCTGAATGGCCGTTGTCTTGCCCGGTG |
| 3GT-F-PstI | GGGGCTGCAGCCGACATCATAACGGTTCTG |
| 3GT-R-DraI | CCCCTTTAAACCCTCAAGTAAGCTTGTGACATTTAACTAGCTC |
| FHT-F-KpnI-EcoRI | GGGGGGTACCGGGGATATCCCGACATCATAACGGTTCTG |
| FHT-R-SalI | CCCCGTCGACCCCCTAAGCAAATATGTCGTCCGCTGGC |
| ANS-F-KpnI | GGGGGGTACCCCGACATCATAACGGTTCTG |
| ANS-R-EcoRI | CCCCGATATCCCCTCACTTGGGGAGCAAAGCCTCTT |
Underlining indicates restriction enzyme cleavage sites corresponding to the primer description. Boldface indicates the start codon, and italics indicate the stop codon.
Recombinant protein expression.
E. coli JM109 harboring plasmid pDGFA184 or plasmid pK184 (control) was preinoculated into Luria-Bertani (LB) liquid medium (3 ml) containing 50 μg/ml kanamycin and incubated at 37°C overnight with shaking. The following day, 1 ml of preinoculum was added to 200 ml LB liquid medium (also containing 50 μg/ml kanamycin) and the culture was left to grow at 37°C with shaking until the A600 reached approximately 0.6. At that point, IPTG was added to the culture to a final concentration of 1 mM and the culture was incubated at room temperature (with shaking) for 5 h. The cells were harvested by centrifugation, washed twice with washing buffer (0.9% NaCl), and resuspended in lysis buffer (Tris-HCl at 20 mM, NaCl at 200 mM, EDTA at 1 mM, β-mercaptoethanol at 1 mM, pH 7.0). Cell disruption was performed using glass beads, and soluble protein was obtained by centrifugation. Total protein was estimated using the bicinchoninic acid assay (Pierce Chemicals) and was used either for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (21) or for enzymatic assays.
Assay of ANS enzymatic activity.
Recombinant E. coli TOP10F carrying MdANS cDNA cloned into the pTrcHis2-TOPO vector was grown, induced, and lysed as described in the previous section, except that ampicillin (100 μg/ml) was used for antibiotic selection. The standard mixture for the ANS reaction (200 μl) was prepared by mixing 10 μl of 15 mM cis- or trans-leucoanthocyanidin, 4 μl of 500 mM sodium ascorbate, 2 μl of 500 mM ferrous sulfate, and 2 μl of 500 mM 2-oxoglutaric acid with 100 to 500 μg of total protein preparation. The reaction mixture was incubated at 30°C for 30 min, and the reaction was terminated by extraction with 400 μl ethyl acetate. The extract was dried by speed vacuum and then dissolved in 3 μl dimethyl sulfoxide, 27 μl water, and 1 μl concentrated HCl. The products were analyzed by HPLC using an Agilent 1100 series instrument and a reverse-phase ZORBAX SB C18 column (4.6 by 150 mm) maintained at 25°C. The compounds produced were separated by elution with an acetonitrile-water gradient, both containing 0.1% formic acid, at a flow rate of 1.0 ml/min. The HPLC conditions were as follows: 10 to 40% acetonitrile for 10 min and 40 to 10% acetonitrile for an additional 5 min. The A515 of pelargonidin and cyanidin was monitored. The retention times under these HPLC conditions for the standard authentic samples are presented in Table 2.
TABLE 2.
Retention times of standard (authentic) compounds used in the present study
| Compound | Retention timea (min) |
|---|---|
| Naringenin | 11.7 |
| Eriodictyol | 10.7 |
| cis-Dihydrokaempferol | 9.7 |
| trans-Dihydrokaempferol | 10.1 |
| cis-Dihydroquercetin | 8.3 |
| trans-Dihydroquercetin | 8.4 |
| Kaempferol | 11.9 |
| Quercetin | 10.8 |
| cis-Leucocyanidin | 3.9 |
| trans-Leucocyanidin | 4.8 |
| Pelargonidin | 7.8 |
| Cyanidin | 6.7 |
| Pelargonidin 3-O-glucoside (callistephin chloride) | 5.1 |
| Cyanidin 3-O-glucoside, (kuromanin chloride) | 4.4 |
Retention times were estimated according to the reverse-phase HPLC conditions described in Materials and Methods.
E. coli shake flask experiments.
Compounds produced from recombinant E. coli strain JM109 carrying plasmid pDGFA184 were identified as follows. Three milliliters of Luria broth rich medium containing 50 μg/ml kanamycin was inoculated with the recombinant strain and incubated at 37°C with vigorous shaking. The next day, 200 ml of LB liquid medium containing 50 μg/ml kanamycin was inoculated with 1 ml of an overnight culture and the culture was incubated at 37°C with vigorous shaking at 300 rpm to an A600 of approximately 0.6. Next, the inducer IPTG was added to the culture to a final concentration of 1 mM. In order to avoid the formation of inclusion bodies, after the addition of the inducer the culture was incubated at room temperature with vigorous shaking overnight. Cells were then harvested by centrifugation and washed twice with M9 minimal medium. The pellets were resuspended in 200 ml M9 minimal medium containing 50 μg/ml kanamycin, 5 mM UDP-glucose, 1 mM IPTG, and various concentrations of naringenin or eriodictyol. In order to produce growth curves, the cultures were left to grow at room temperature with vigorous shaking until the late stationary phase was reached. For flavonoid extraction experiments, the cultures were left to grow at room temperature for 65 h with 0.25 mM naringenin or 0.1 mM eriodictyol, again with vigorous horizontal shaking.
Flavonoid extraction.
After completion of the shake flask experiments, the supernatant was separated from the cells by centrifugation. Flavonoids were extracted from the supernatant with an equal volume (approximately 200 ml) of ethyl acetate for 2 h at room temperature. The organic layer was evaporated to dryness by lyophilization or rotary evaporation, and the resulting orange powder was dissolved in 2 ml dimethyl sulfoxide. The compounds produced were separated by HPLC using the same method previously described for the ANS in vitro assay. Anthocyanins and anthocyanidins were detected and quantified by monitoring A515, quercetin and kaempferol were detected and quantified by monitoring A360, and flavanones, dihydroflavonols, and leucoanthocyanidin epimers were detected and quantified by monitoring A290. The quantitative calibration curves were obtained with standard anthocyanin, anthocyanidin, flavanone, dihydroflavonol, flavonol, and leucoanthocyanidin solutions.
MS.
Mass spectrometry (MS) analysis was performed using a Thermo Finnigan LCQ Advantage system. Dried, HPLC-purified compounds or standard powder was dissolved in methanol and then diluted with water containing 0.1% formic acid and 80% acetonitrile before injection into the MS system. For MS analysis, the mass charge was adjusted at 433. Further tandem MS analysis of ions with m/z = 433 was performed at 35% relative energy.
NMR spectrometry.
The identity of dihydroflavonol epimers was determined by NMR measurements. For that purpose, 1H NMR data were obtained in CD3OD (Aldrich) (using 3.31 ppm for reference of residual CD3OD) at 500 MHz using a Varian instrument. Signals were compared to published data (1, 11, 19).
RESULTS
The presence of naringenin in the medium results in E. coli growth reduction.
We tested the effect of naringenin on E. coli growth by incubating cultures of JM109 carrying plasmid pK184 in M9 minimal medium (50 μg/ml kanamycin) in the presence of different concentrations of naringenin (0 mM, 0.5 mM, and 1 mM). Naringenin had an inhibitory effect on E. coli growth; the final dry cell weight concentration dropped from 0.586 mg/ml in the control (no naringenin) to 0.492 mg/ml in the presence of 1 mM naringenin in the minimal medium, both after 13 h of cell growth. This 16% reduction in the final dry cell weight could be a direct result of naringenin uptake by E. coli.
Biochemical characterization of M. domestica ANS.
ANS is a member of the nonheme ferrous and 2-oxoglutarate-dependent oxygenase family with wide substrate specificity (27, 28). In one of the most thorough investigations, it was shown that ANS from Arabidopsis thaliana was able to convert both naturally occurring (2R,3S,4S)-cis-leucocyanidin and unnatural (2R,3S,4R)-trans-leucocyanidin to the corresponding anthocyanidins, albeit with very low efficiency (26). Only 2% of the final products when (2R,3S,4S)-cis-leucocyanidin was used as a substrate and only 4% when unnatural (2R,3S,4R)-trans-leucocyanidin was used as a substrate corresponded to cyanidins. The rest of the product was identified as cis-dihydroquercetin, trans-dihydroquercetin, and quercetin.
In an effort to understand some of the biochemical properties of the recombinant ANS cloned from M. domestica, we performed the in vitro ANS assay using (2R,3S,4S)-cis-leucocyanidin, (2R,3S,4R)-trans-leucocyanidin, (2R,3S,4S)-cis-leucopelargonidin, and (2R,3S,4R)-trans-leucopelargonidin. All compounds were accepted as substrates, with the unnatural trans-leucoanthocyanidins catalyzed more efficiently than the natural cis epimers, in accordance with the previous report by Turnbull et al. (26). HPLC analysis of the in vitro ANS reaction indicated that dihydroquercetin and quercetin (in the case of leucocyanidin as the substrate) or dihydrokaempferol and kaempferol (in the case of leucopelargonidin) were the major products; only 1% of the final ANS products corresponded to cyanidin or pelargonidin. The majority of the product, 82%, corresponded to dihydroflavonols (dihydrokaempferol or dihydroquercetin), while the rest corresponded to flavonols (kaempferol or quercetin). This result was consistent, regardless of the leucoanthocyanidin epimer used as a substrate. The observed product distributions from M. domestica ANS incubations are presented in Table 3.
TABLE 3.
Observed product distributions from M. domestica incubations (30 min) with the cis and trans epimers of leucocyanidina
| Substrate | % cis-DHQ | % trans-DHQ | % Cyanidin | % Quercetin |
|---|---|---|---|---|
| (2R,3S,4S)-cis-LCD | 56 | 26 | 1 | 17 |
| (2R,3S,4R)-trans-LCD | 40 | 42 | 1 | 17 |
The enzymatic assay was performed as described in Materials and Methods. Reaction products were quantified by integrating peak areas in the HPLC profiles and comparing them with standard curves of authentic samples. Cyanidin was monitored at 515 nm, quercetin at 360 nm, and dihydroquercetin at 290 nm. Results shown are the averages of two independent experiments. Abbreviations: LCD, leucocyanidin; DHQ, dihydroquercetin.
Expression of the anthocyanin biosynthesis pathway in E. coli.
We designed a gene cluster in pK184 that would allow flavanones such as naringenin or eriodictyol to be converted into the first stable anthocyanin, pelargonidin 3-O-glucoside (from naringenin) or cyanidin 3-O-glucoside (from eriodictyol). For that purpose, four genes of plant origin, MdF3H from M. domestica, dfr from A. andraeanum, MdANS from M. domestica, and PGT8 from P. hybrida, were selected to be heterologously expressed in E. coli. Extensive past research had shown that flavanone 3β-hydroxylase (FHT), ANS, and UDP-glucose:flavonoid 3-O-glucosyltransferase (3-GT) can be efficiently expressed in E. coli (3, 4). However, this has only recently been demonstrated for dihydroflavonol 4-reductase (DFR) (18, 30). One particular aspect of DFR enzymes is that some of them accept only dihydrokaempferol or dihydroquercetin as a substrate (32). We have recently shown in our lab that A. andraeanum DFR, heterologously expressed in E. coli, is able to catalyze very efficiently the reduction of both dihydrokaempferol and dihydroquercetin to the corresponding leucoanthocyanidins (E. J. Leonard et al., unpublished data). Based on this evidence, we decided to use the dfr cDNA gene isolated from A. andraeanum for our cloning purposes.
All four genes were placed individually under the control of the strong trc promoter in the low-copy-number E. coli vector pK184. We chose a low-copy-number vector in order to avoid the likely high transcription and translation levels that would result from higher-copy-number plasmids and could lead to deleterious effects on the cell. An RBS (AGAGG) and a reinitiation RBS (AAGGAG) located after a minicistron sequence, both derived from cloning vector pTrcHis2-TOPO, were present 46 and 16 bp, respectively, from the start codon of each gene. Even though the same untranslated region was present in front of each one of the four genes, no recombination events were observed and the vectors proved stable in E. coli JM109 even after 65 h of growth.
For assessing the expression levels of the recombinant proteins, we grew the recombinant E. coli cultures at suboptimal (room) temperature in order to avoid inclusion body formation. When the total protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, only a slight increase at the level of protein production was observed in the region of 40 kDa (where ANS, with a molecular mass of 40,470 Da; DFR, with a molecular mass of 39,417 Da; and FHT, with a molecular mass of 41,137 Da, migrate) while protein expression was not evident in the region of 50 kDa (where 3-GT, with a molecular mass of 49,706 Da, migrates). This result is in accordance with similar results previously reported for the flavanone biosynthetic pathway heterologously expressed in E. coli using a medium-copy-number vector (9).
Analysis of fermentation products.
Anthocyanin biosynthesis was performed by culturing the recombinant E. coli strain in M9 minimal medium with glucose as the carbon source and supplemented with naringenin or eriodictyol at a concentration of 0.2 mM and UDP-glucose at a concentration of 5 mM. UDP-glucose was added to the medium since it is the glucose donor during the last step of anthocyanidin 3-O-glucoside biosynthesis, catalyzed by 3-GT. After the shake flask cultures were harvested, various polyphenolic compounds were extracted from the fermentation broth using ethyl acetate.
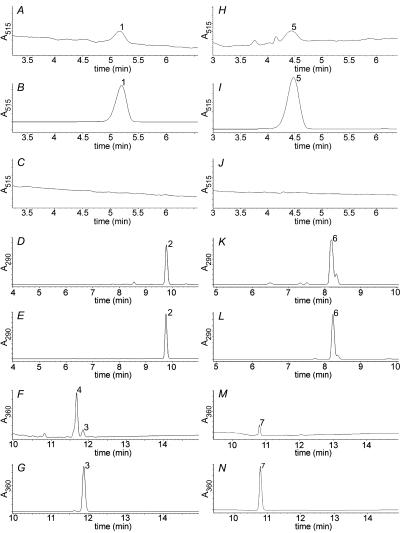
We first performed HPLC analysis of the fermentation broth obtained from both the recombinant and control cultures and using naringenin or eriodictyol as the precursor flavanone. For detection, we monitored A515. Only one compound appeared to be present (Fig. 3A and H), with the same retention time as the standard sample of pelargonidin 3-O-glucoside (Fig. 3B) or cyanidin 3-O-glucoside (Fig. 3I). No compounds absorbing at 515 nm appeared in the control cultures (Fig. 3C and J). In order to further verify the identity of at least one of the synthesized colored compounds, pelargonidin 3-O-glucoside was separated from the culture broth by HPLC and evaporated to dryness in order to be further analyzed by MS, in parallel with the pelargonidin 3-O-glucoside standard. Both samples produced peaks with an m/z ratio of 433 (the molecular weight of pelargonidin 3-O-glucoside), thus verifying the biosynthesis of authentic pelargonidin 3-O-glucoside from the recombinant E. coli strain.
FIG. 3.
HPLC analysis of shake flask supernatants of recombinant JM109 carrying plasmid pDGFA184 performed as described in Materials and Methods. A, pelargonidin 3-O-glucoside (peak 1) produced from the recombinant strain when fed with naringenin; B, standard callistephin chloride (peak 1); C, shake flask supernatant of JM109 carrying empty vector pK184 fed with naringenin (control); D, dihydrokaempferol (peak 2) produced from the recombinant strain when fed with naringenin; E, standard dihydrokaempferol (peak 2); F, side product kaempferol (peak 3) produced from the recombinant strain fed with naringenin; remaining starting material naringenin (peak 4) is shown; G, standard kaempferol (peak 3); H, cyanidin 3-O-glucoside (peak 5) produced from the recombinant strain fed with eriodictyol; I, standard kuromanin chloride (peak 5); J, shake flask supernatant of JM109 carrying empty vector pK184 fed with eriodictyol (control); K, dihydroquercetin (peak 6) produced from the recombinant strain fed with eriodictyol; L, standard dihydroquercetin (peak 6); M, side product quercetin (peak 7) produced by the recombinant strain fed with eriodictyol; N, standard quercetin (peak 7).
Besides anthocyanins, the recombinant E. coli expressing the lower anthocyanin pathway accumulated large amounts of the precursor metabolites dihydrokaempferol (5.56 mg/liter; Fig. 3D; authentic sample shown in Fig. 3E) or dihydroquercetin (8.8 mg/liter; Fig. 3K; authentic sample shown in Fig. 3L). Since ANS converts leucoanthocyanidins largely into dihydroflavonols in vitro, it is not clear whether the synthesized dihydroflavonols were mainly the result of high FHT or ANS activity. This is especially true since no leucoanthocyanidins were detected in the fermentation broth. The side product kaempferol (or quercetin) was also present in the fermentation broth at concentrations of 440 μg/liter (Fig. 3F; authentic sample shown in Fig. 3G) and 444 μg/liter (Fig. 3M; authentic sample shown in Fig. 3N), respectively. The biosynthesis of kaempferol is most likely a direct product of ANS, as its biosynthesis was clearly demonstrated during the in vitro ANS assays. No anthocyanidins (pelargonidin or cyanidin) and no apiferol or luteoforol (the products of the flavanone reductase activity the A. andraeanum DFR demonstrates) were detected in the fermentation broth (16).
The amounts of anthocyanins synthesized were relatively low; the pelargonidin 3-O-glucoside concentration in the fermentation broth was 5.6 μg/liter, while cyanidin 3-O-glucoside was present at a concentration of 6.0 μg/liter. In an attempt to increase production levels, we ran similar shake flask experiments using M9 minimal medium supplemented with cofactors such as 2-oxoglutarate (0.5 mM), sodium ascorbate (0.5 mM), and ferrous sulfate (0.5 mM). These are cofactors necessary in the general reaction scheme of 2-oxoglutarate, nonheme Fe(II)-dependent oxygenases such as ANS (20, 23, 31). A dramatic decrease in the concentration of anthocyanins was observed under these conditions, to levels below 1.0 μg/liter.
DISCUSSION
In the present study, we demonstrate for the first time the expression of an artificial gene cluster in E. coli of four plant-derived genes involved in the lower anthocyanin biosynthetic pathway and the biosynthesis of the first two stable anthocyanins, pelargonidin 3-O-glucoside and cyanidin 3-O-glucoside.
In the past, engineering entire metabolic pathways on a single operon has proven to be successful in E. coli (12). However, we decided to proceed with the use of individual promoters and RBS for each one of the four genes after a recent study demonstrated that the construction of the flavanone biosynthetic pathway on a single operon did not lead to efficient flavanone production yields (9). Despite following a similar approach, the amount of anthocyanins produced from our recombinant strain was relatively small, about 2 orders of magnitude lower than the amount of flavanones produced in the presence of tyrosine (9).
The increase in the anthocyanin 3-O-glucoside production yield would involve a more thorough understanding of the biochemical properties of some critical enzymes identified in the present study, such as DFR and, most important, ANS. We chose to use the A. andraeanum DFR based on in vitro enzymatic assay data that demonstrated its higher specific activity compared to DFR enzymes isolated from other plant sources (Leonard et al., unpublished). Still, the large amount of dihydroflavonols detected in the fermentation broth demonstrates that DFR remains a potential rate-limiting step in the linear reaction pathway that we constructed. In addition, it is now evident that ANS demonstrates a low activity toward anthocyanidin formation when expressed in E. coli, with most of its various leucoanthocyanidin substrates being converted to their corresponding dihydroflavonols and flavonols (26, 27). It is possible that the elucidation of the role of enzyme complexes formed by anthocyanin biosynthetic enzymes, as previously suggested (26, 29), would allow better pathway engineering in the future.
Another significant step to follow is the expression of the gene cluster constructed in the present study together with an artificial gene cluster that would allow the conversion of phenylalanine or tyrosine to naringenin, such as the one previously described (9). Such simultaneous expression would permit the conversion of the two amino acids that are native to E. coli to anthocyanins and later to other polyphenolic compounds that are considered specific to plants. It is important to note here that certain metabolic steps involved in flavonoid biosynthesis in general and anthocyanin biosynthesis in particular are catalyzed by cytochrome P450 monooxygenases, such as flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase, whose functional expression has proven to be challenging in the past (8). In that respect, utilization of other, eukaryotic systems as production platforms for at least some types of anthocyanins may prove to be a wiser selection and a better alternative.
Whether E. coli or any other host is utilized as a production platform, the possibilities of metabolically engineering high-value anthocyanin compounds are tremendous due to the natural coloration that these compounds provide at low pH values. This coloration would provide an easy and low-cost screening method for anthocyanin producers derived from the application of various protein engineering and combinatorial techniques. Such techniques could be applied toward improvement of enzyme function, alteration of substrate specificities, and introduction of novel catalytic activities and would result not only in better production of natural compounds but also in the generation of novel anthocyanins with unique structures and functions.
Acknowledgments
This work was supported by a research grant from the U.S. National Science Foundation (BES-0331404) to M. A. G. Koffas.
We thank Alice Bergmann and Cheng Zhao for the mass spectrometer analysis, which was supported by NSF award CHE0091977. We thank Amalia Koffas for providing expertise in plant cultivation.
REFERENCES
- 1.Baderschneider, B., and P. Winterhalter. 2001. Isolation and characterization of novel benzoates, cinnamates, flavonoids, and lignans from Riesling wine and screening for antioxidant activity. J. Agric. Food Chem. 49:2788-2798. [DOI] [PubMed] [Google Scholar]
- 2.Boyd, W. 2000. Natural colors as functional ingredients in healthy foods. Cereal Foods World. 45:221-222. [Google Scholar]
- 3.Britsch, L., J. Dedio, H. Saedler, and G. Forkmann. 1993. Molecular characterization of flavanone 3β-hydroxylases. Consensus sequence, comparison with related enzymes and the role of conserved histidine residues. Eur. J. Biochem. 217:745-754. [DOI] [PubMed] [Google Scholar]
- 4.Britsch, L., B. Ruhnau-Brich, and G. Forkmann. 1992. Molecular cloning, sequence analysis, and in vitro expression of flavanone 3β-hydroxylase from Petunia hybrida. J. Biol. Chem. 267:5380-5387. [PubMed] [Google Scholar]
- 5.Brouillard, R. 1982. Chemical structure of anthocyanins, p. 1-40. In P. Markakis (ed.), Anthocyanins as food colors. Academic Press, Inc., New York, N.Y.
- 6.Hannum, S. M. 2004. Potential impact of strawberries on human health: a review of the science. Crit. Rev. Food. Sci. Nutr. 44:1-17. [DOI] [PubMed] [Google Scholar]
- 7.Honda, C., N. Kotoda, M. Wada, S. Kondo, S. Kobayashi, J. Soejima, Z. L. Zhang, T. Tsuda, and T. Moriguchi. 2002. Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiol. Biochem. 40:955-962. [Google Scholar]
- 8.Hotze, M., G. Schroder, and J. Schroder. 1995. Cinnamate 4-hydroxylase from Catharanthus roseus, and a strategy for the functional expression of plant cytochrome P450 proteins as translational fusions with P450 reductase in Escherichia coli. FEBS Lett. 374:345-350. [DOI] [PubMed] [Google Scholar]
- 9.Hwang, E. I., M. Kaneko, Y. Ohnishi, and S. Horinouchi. 2003. Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster. Appl. Environ. Microbiol. 69:2699-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahkonen, M. P., and M. Heinonen. 2003. Antioxidant activity of anthocyanins and their aglycons. J. Agric. Food Chem. 51:628-633. [DOI] [PubMed] [Google Scholar]
- 11.Lundgren, L. N., and O. Theander. 1988. The constituents of conifer needles. 14. Cis-dihydroquercetin and trans-dihydroquercetin glucosides from needles of Pinus sylvestris. Phytochemistry 27:829-832. [Google Scholar]
- 12.Martin, V. J., D. J. Pitera, S. T. Withers, J. D. Newman, and J. D. Keasling. 2003. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21:796-802. [DOI] [PubMed] [Google Scholar]
- 13.Mazza, G., and E. Miniati. 1993. Anthocyanins in fruits, vegetables and grains. CRC Press, Inc., Boca Raton, Fla.
- 14.Middleton, E., Jr., C. Kandaswami, and T. C. Theoharides. 2000. The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease, and cancer. Pharmacol. Rev. 52:673-751. [PubMed] [Google Scholar]
- 15.Reference deleted.
- 16.Nakajima, J., Y. Tanaka, M. Yamazaki, and K. Saito. 2001. Reaction mechanism from leucoanthocyanidin to anthocyanidin 3-glucoside, a key reaction for coloring in anthocyanin biosynthesis. J. Biol. Chem. 276:25797-25803. [DOI] [PubMed] [Google Scholar]
- 17.Noda, Y., T. Kneyuki, K. Igarashi, A. Mori, and L. Packer. 2000. Antioxidant activity of nasunin, an anthocyanin in eggplant peels. Toxicology 148:119-123. [DOI] [PubMed] [Google Scholar]
- 18.Peters, D. J., and C. P. Constabel. 2002. Molecular analysis of herbivore-induced condensed tannin synthesis: cloning and expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides). Plant J. 32:701-712. [DOI] [PubMed] [Google Scholar]
- 19.Prescott, A. G., N. P. Stamford, G. Wheeler, and J. L. Firmin. 2002. In vitro properties of a recombinant flavonol synthase from Arabidopsis thaliana. Phytochemistry 60:589-593. [DOI] [PubMed] [Google Scholar]
- 20.Ryle, M. J., and R. P. Hausinger. 2002. Non-heme iron oxygenases. Curr. Opin. Chem. Biol. 6:193-201. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Satue-Gracia, M. T., I. M. Heinonen, and E. N. Frankel. 1997. Anthocyanins as antioxidants on human low-density lipoprotein and lecithin-liposome systems. J. Agric. Food Chem. 45:3362-3367. [Google Scholar]
- 23.Schofield, C. J., and Z. Zhang. 1999. Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes. Curr. Opin. Struct. Biol. 9:722-731. [DOI] [PubMed] [Google Scholar]
- 24.Stich, E., K. Kloos, P. Cortona, and S. Hake. 1999. Color me natural. Nutraceuticals World 2:64-70. [Google Scholar]
- 25.Tanner, G. J., A. R. Ashton, S. Arahams, J. M. Watson, P. J. Larkin, and K. T. Francki. 29 August 2002. Novel gene and uses therefor to modify pasture qualities of crops. International patent WO 2002-AU179.
- 26.Turnbull, J. J., M. J. Nagle, J. F. Seibel, R. W. Welford, G. H. Grant, and C. J. Schofield. 2003. The C-4 stereochemistry of leucocyanidin substrates for anthocyanidin synthase affects product selectivity. Bioorg. Med. Chem. Lett. 13:3853-3857. [DOI] [PubMed] [Google Scholar]
- 27.Turnbull, J. J., J. Nakajima, R. W. Welford, M. Yamazaki, K. Saito, and C. J. Schofield. 2004. Mechanistic studies on three 2-oxoglutarate-dependent oxygenases of flavonoid biosynthesis: anthocyanidin synthase, flavonol synthase, and flavanone 3β-hydroxylase. J. Biol. Chem. 279:1206-1216. [DOI] [PubMed] [Google Scholar]
- 28.Wilmouth, R. C., J. J. Turnbull, R. W. Welford, I. J. Clifton, A. G. Prescott, and C. J. Schofield. 2002. Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure (London) 10:93-103. [DOI] [PubMed] [Google Scholar]
- 29.Winkel, B. S. J. 2004. Metabolic channeling in plants. Annu. Rev. Plant Biol. 55:85-107. [DOI] [PubMed] [Google Scholar]
- 30.Xie, D. Y., L. A. Jackson, J. D. Cooper, D. Ferreira, and N. L. Paiva. 2004. Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula. Plant Physiol. 134:979-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, Z., J. Ren, D. K. Stammers, J. E. Baldwin, K. Harlos, and C. J. Schofield. 2000. Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase. Nat. Struct. Biol. 7:127-133. [DOI] [PubMed] [Google Scholar]
- 32.Zufall, R. A., and M. D. Rausher. 2004. Genetic changes associated with floral adaptation restrict future evolutionary potential. Nature 428:847-850. [DOI] [PubMed] [Google Scholar]