Abstract
Oxalate oxidase is thought to be involved in the production of hydrogen peroxide for lignin degradation by the dikaryotic white rot fungus Ceriporiopsis subvermispora. This enzyme was purified, and after digestion with trypsin, peptide fragments of the enzyme were sequenced using quadrupole time-of-flight mass spectrometry. Starting with degenerate primers based on the peptide sequences, two genes encoding isoforms of the enzyme were cloned, sequenced, and shown to be allelic. Both genes contained 14 introns. The sequences of the isoforms revealed that they were both bicupins that unexpectedly shared the greatest similarity to microbial bicupin oxalate decarboxylases rather than monocupin plant oxalate oxidases (also known as germins). We have shown that both fungal isoforms, one of which was heterologously expressed in Escherichia coli, are indeed oxalate oxidases that possess ≤0.2% oxalate decarboxylase activity and that the organism is capable of rapidly degrading exogenously supplied oxalate. They are therefore the first bicupin oxalate oxidases to have been described. Heterologous expression of active enzyme was dependent on the addition of manganese salts to the growth medium. Molecular modeling provides new and independent evidence for the identity of the catalytic site and the key amino acid involved in defining the reaction specificities of oxalate oxidases and oxalate decarboxylases.
Ceriporiopsis subvermispora is a white rot basidiomycete fungus that has potential in biomechanical pulping in the paper industry (32). This organism possesses two out of the three common extracellular ligninolytic enzymes: a copper-dependent laccase and a heme-dependent manganese peroxidase but no lignin peroxidase. While the laccase requires only dioxygen as a cosubstrate, manganese peroxidase requires hydrogen peroxide. Much of the hydrogen peroxide is thought to be produced in a series of well-characterized biochemical and chemical reactions that involve oxalate (6, 35), which is secreted by this organism (41). Manganese peroxidase catalyzes the conversion of Mn2+ and H2O2 to Mn3+ and H2O. The Mn3+ reacts with oxalate to form Mn2+, carbon dioxide, and a formyl radical, which produces carbon dioxide and a superoxide radical in the presence of dioxygen. The superoxide radical is then capable of reoxidizing Mn2+ to give hydrogen peroxide. The net result is that Mn3+ is produced as a diffusible and powerful oxidant capable of degrading many phenolic and possibly nonphenolic (13) components of lignin. At the same time, the concentration of hydrogen peroxide is amplified in the presence of oxalate, dioxygen, and protons to facilitate the further production of Mn3+.
In order for manganese peroxidase to commence the degradation of lignin, however, an initial source of hydrogen peroxide is required. An oxalate oxidase (EC 1.2.3.4) was identified in C. subvermispora that could be involved in a pathway leading to the production of hydrogen peroxide (1). This enzyme catalyzes the conversion of oxalate and dioxygen to carbon dioxide and hydrogen peroxide. The protein was shown to be a 400-kDa homohexamer of 65.5-kDa subunits (possibly including glycan) with a pI of 4.2 and a pH optimum of 3.5. Its Km for oxalate was 0.1 mM, and it had a kcat of 88 s−1. Histochemical studies showed the enzyme to be in membrane-bound vesicles (peroxisome-like structures and multivesicular bodies), some of which are in contact with the outer cell membrane and the periplasmic space, suggesting some kind of vesicular transport. The export of the enzyme to the periplasmic space and potentially extracellularly is consistent with its proposed role as a source for extracellular hydrogen peroxide. It must be noted that there has been a preliminary report of another fungal oxalate oxidase in the obligate wheat parasite Tilletia contraversa (42), although it is possible that this activity was of plant origin.
The best-characterized oxalate oxidase is from cereal plants such as barley and wheat (19). This plant cell-wall-associated enzyme is expressed in germinating seedling roots (hence its synonym, germin) and in mature leaves on infection by fungal pathogens. The enzyme was found to require a mononuclear manganese ion for catalysis (30), the resting states of the wild-type (30) and recombinant (43) enzymes being in the Mn2+ oxidation state.
The plant oxalate oxidases belong to the cupin (cupa, Latin for small barrel) protein superfamily (16). The members of this superfamily share a common β-barrel fold and two cupin motifs, defined as G(X)5HXH(X)3-4E(X)6G followed by G(X)5PXG(X)2H(X)3N, with typically 20 to 26 amino acids in the intermotif sequence. A crystal structure of the barley oxalate oxidase revealed a hexameric arrangement of subunits, each with the characteristic β-barrel fold (44). The mononuclear Mn2+ at the active site was coordinated by the one Glu as well as the three His of the cupin motifs as previously predicted (30). There are only three complete sequences that have been unequivocally verified as being associated with oxalate oxidase activity. The two sequences from barley (Hordeum vulgare, GenBank accession no. P45850 and CAA74595 [21, 46]) and one from wheat (GenBank accession no. P15290 [20]) share 96% sequence identity. These glycosylated proteins have a subunit molecular mass of about 23 kDa, of which 21 kDa is protein. Several other proteins that share very high sequence identity with the confirmed oxalate oxidases (>95% identity in pairwise comparisons) are likely to possess oxalate oxidase activity also (7). On the other hand, there are numerous germin-like proteins that share lower sequence identities and have no oxalate oxidase activity.
The cupin superfamily is subdivided into two broad groups: the monocupins, including oxalate oxidase and germin-like proteins, and the bicupins, which are roughly twice the size and contain a duplication of the cupin motifs (9, 16). It is thought that the bicupins arose by two distinct gene duplication events, one leading to the plant seed storage proteins and another leading to bacterial and fungal bicupins (36). The bacterial and fungal bicupins form a distinct monophyletic clade (16). Four members of this clade have been confirmed as having oxalate decarboxylase activity (EC 4.1.1.2): Flammulina velutipes (formerly known as Collybia velutipes) OxdC (GenBank accession no. AAF13275 [15]), Bacillus subtilis OxdC (formerly known as YvrK; GenBank accession no. O34714 [37]), B. subtilis OxdD (formerly known as YoaN; GenBank accession no. O34767 [37]), and an Aspergillus phoenices oxalate decarboxylase (GenBank accession no. AAE83943 [9]).
The C. subvermispora oxalate oxidase has a molecular mass (65.5 kDa, presumably including glycan) that resembles that of the fungal (55 kDa plus glycan = 64 kDa) and bacterial (43 kDa) oxalate decarboxylases more closely than that of the plant oxalate oxidases (21 kDa plus glycan = 23 kDa). This begs the question as to whether this enzyme belongs to the bicupin or some other protein family rather than the monocupin family to which the plant oxidases belong. Although one additional manganese-dependent oxalate oxidase has been reported from a Pseudomonas sp. that has a relatively large molecular mass of 38 kDa, no sequence information is available (17).
Oxalate-degrading enzymes have many established and potential uses, for example, in clinical assays for oxalate (a major component of kidney stones), human gene therapy, improved disease resistance in plants, reduced oxalate levels in food crops, the bioremediation of oxalate wastes, and the production of hydrogen peroxide (9). These uses, together with their potential role in lignin degradation and the desire to understand the novel chemistry that these enzymes catalyze, make them worthwhile subjects of study. The aim of this work was to obtain the gene sequence(s) of C. subvermispora oxalate oxidase, to establish whether it is a bicupin, and to gain insights into the structure-function relationships of oxalate-degrading enzymes.
MATERIALS AND METHODS
Materials.
All materials and biochemicals, including F. velutipes oxalate decarboxylase, were of the highest grade available and, unless stated otherwise, were purchased from Sigma-Aldrich. Protein was determined using the Pierce Micro bicinchoninic assay. The PhastSystem (Amersham Pharmacia Biotech) with 8 to 25% PhastGels was used for native polyacrylamide gel electrophoresis. Horseradish peroxidase (HRP4C) was purchased from Biozyme Laboratories Ltd. (Blaenavon, Gwent, Wales, United Kingdom). Dithiothreitol was obtained from Melford Laboratories Ltd. (Suffolk, United Kingdom). Hydrogen peroxide and glass beads (5.0-mm diameter) were obtained from BDH Laboratory Supplies (Poole, United Kingdom). YM broth was obtained from DIFCO. AmpliTaq Gold DNA polymerase was obtained from Applied Biosystems. Oligo(dT)-cellulose columns were obtained from Amersham Pharmacia Biotech.
Strains and growth conditions.
Ceriporiopsis subvermispora ATCC 90466 was obtained from the American Type Culture Collection. The organism was maintained (1) and grown in liquid medium (32) with modifications (23). Homokaryotic strains of C. subvermispora CsA and CsB were obtained and grown as previously described (38).
Enzyme purification from C. subvermispora.
The oxalate oxidase from C. subvermispora was purified using anion exchange chromatography, pH precipitation of contaminant proteins, phosphocellulose chromatography, and gel filtration as described previously (1) but with minor modifications. Dithiothreitol was used instead of β-mercaptoethanol. DEAE-Sepharose and Superdex 200 were used instead of Q-Sepharose and Sephadex G-200, respectively. Quoted activities were obtained with samples after phosphocellulose chromatography.
Enzyme assays and activity stains.
One unit of enzyme activity was defined as the conversion of 1 μmol of substrate to product per min. Oxalate oxidase activity was determined using stopped assays, where the production of hydrogen peroxide was coupled to the oxidation of phenol red using horseradish peroxidase as described previously (1). It was important to subtract control reactions without peroxidase in order to distinguish between the production of hydrogen peroxide and the oxalate-dependent direct oxidation of the dye (37). Polyacrylamide gels were stained for oxalate oxidase activity using a similar coupled system with 4-chloro-1-naphthol as the dye as previously described (45). Oxalate decarboxylase activity was determined using a stopped assay, where the production of formate was coupled to the reduction of NAD by formate dehydrogenase as previously described (37). Superoxide dismutase activity was determined qualitatively in native polyacrylamide gels using a specific activity stain (5).
Quadrupole time-of-flight mass spectrometry.
The protein band of interest was excised from a polyacrylamide gel, reduced (10 mM dithiothreitol), alkylated (100 mM iodoacetamide; Sigma, Dorset, United Kingdom), and enzymically digested (sequencing-grade modified porcine trypsin; Promega, Southampton, United Kingdom) using an automated digest robot (Investigator ProGest; Genomic Solutions Ltd., Huntingdon, United Kingdom). Peptides generated from the tryptic digest were loaded at a high flow rate onto a reverse-phase trapping column (0.3-mm inside diameter by 1 mm, with 5-μm C18 100 Å PepMap packing; LC Packings, The Netherlands) and eluted through a reverse-phase capillary column (75-μm inside diameter by 50-mm column, with 5-μm C18 100 Å PepMap packing) directly into the nanoelectrospray ion source of a quadrupole time-of-flight mass spectrometer (Q-ToF2; Micromass UK Ltd., Manchester, United Kingdom). Sequences were interpreted from the resulting fragment ion spectra using the PepSeq de novo sequencing tool (Micromass).
Cloning.
Total RNA was extracted from cells grown and harvested as described above using a QIAGEN RNeasy Plant Mini kit, and mRNA was purified from this preparation using oligo(dT)-cellulose (Amersham Pharmacia Biotech). First-strand cDNA was prepared from the mRNA using Superscript II reverse transcriptase (Life Technologies) with a poly(A) tail-specific primer 5′-GACTCGACTCGACATCGATTTTTTTTTTTTTTTTT. Degenerate primers based on the first and forth peptide sequences obtained using quadrupole time-of-flight mass spectrometry were used to obtain an internal fragment of the oxalate oxidase cDNA from a total first-strand cDNA preparation, 5′-CCGGAGCGCGGYCAGCTCGGYGCBAAG and 5′-CTTGACGGTRCCRCCVGCGTACTGGGTVGG. The degree of degeneracy in these primers was guided by a codon usage table for C. subvermispora, where codons of a frequency of >6% only were represented. The rapid amplification of the 3′ cDNA end was achieved using the first-strand cDNA with the gene-specific primer 5′-CCCGTACTCATTCGAGTTCTCGAA and the poly(A) tail-specific primer used above but lacking its A(T)n 3′ end. This yielded a partial oxoX-C cDNA sequence. The use of rapid amplification of the 5′ cDNA end failed to provide the 5′ end of the sequence. A Lambda ZipLox (Life Technologies) cDNA library and a Lambda GEM-11 (Promega) genomic DNA library were prepared as previously described (24). A [32P]dC-labeled gene-specific probe was prepared from the double-stranded cDNA obtained with the degenerate primers described above using Rediprime II (Amersham Biosciences) and was purified using size exclusion chromatography. Lambda DNA was purified using a Wizard Lambda Preps DNA Purification System kit (Promega). Lambda clones containing cDNA and genomic DNA were digested with XbaI and XhoI/EcoRI, respectively, and the presence of DNA coding for the oxalate oxidase was confirmed using Southern blotting. cDNA from a Lambda ZipLox clone was recovered by excision into pZL1 (Life Technologies) or using PCR with the whole library using the gene-specific primer 5′-GGTGGCGCGACTCCCGGGAAGATG together with a Lambda ZipLox polylinker-specific primer, 5′-GATTACGCCAAGCTCTAATACG. This yielded more of the 5′ end of the oxoX-C cDNA sequence. Genomic DNA in a Lambda GEM-11 clone was sequenced directly, yielding the complete oxoX-G genomic sequence. Using total first-strand cDNA as a template and the gene-specific primers 5′-ATCTCAGACCATCTCCCCACTTGC and 5′-CGTCACATCAATGGACCTTGTCG based on the oxoX-G genomic sequence, the complete coding region of oxoX-G cDNA was obtained. The oxoX genes were amplified from genomic DNA isolated from the homokaryotic C. subvermispora strains CsA and CsB using the gene-specific primers 5′-AGAACGCAGACACGCTTG and 5′-AATGCCTTGCTCTTGTTCA. The PCR products from each strain were cloned into pGEM-T Easy (Promega) and analyzed by digestion with EcoRI. Sequencing confirmed the identity of both products. This yielded a partial genomic sequence of oxoX-C including all 14 expected introns. In order to obtain the 5′ coding sequence of oxoX-C, genomic DNA from CsB was digested with XmaI and ligated to produce circular DNA. This was used as a template in an “inverse” PCR with the gene-specific primers 5′-GGCTAGCACACACACCAAAGGACG and 5′-CCCGAGTCGGTCGTAGG. The PCR product was sequenced directly using the gene-specific primer 5′-CGTGTCTGCATTCTGGAGG, yielding the 5′ region of the oxoX-C genomic DNA. DNA was sequenced either by using the Applied Biosystems BigDye Terminator version 3.1 Cycle Sequencing kit or by MWG Biotech. At least two independent clones were sequenced in each case. Sequences were aligned using the Clustal W 1.81 algorithm (40), and phylogenetic trees were prepared using the neighbor-joining method (34).
Degradation of oxalate in liquid cultures.
[13C]oxalic acid (Argo International Ltd., Basildon, Essex, United Kingdom), neutralized with NaOH, was added to cultures at the time when cells were usually harvested. Culture media including the cell pellets were divided equally (4.75 ml) into 25-ml sterile conical flasks before adding the [13C]oxalate to give a final concentration of 10 mM in a total volume of 5 ml. After 2, 4, or 6 h of incubation at 30°C with agitation at 200 rpm, the liquid medium was decanted from the cell pellets and frozen at −20°C. Controls without cells were performed in parallel. Thawed samples were, after the addition of 10% (vol/vol) D2O, analyzed by nuclear magnetic resonance (NMR) spectroscopy using a JEOL JNM LA400 spectrometer with sodium-2,2-dimethyl-2-silopentane-5-sulfonate as a reference compound. Some samples were subjected to ultrafiltration using an Amicon Microcon 10k immediately after thawing in order to remove any oxalate oxidase present in solution. Control samples of [13C]oxalate in 50 mM succinate buffers at pH 4.0 or 5.0 with or without authentic oxalate decarboxylase (37) or oxalate oxidase (Sigma) were similarly analyzed (oxalate, δC [100 MHz, H2O] 173.7 [s]; formate, δC [100 MHz, H2O] 171.7 [d, J 195.2 Hz]). The oxalate and formate signals were easily distinguished by obtaining decoupled and nondecoupled spectra. Signals associated with the glucose (56 mM at the start of the 2-week agitated growth phase) remaining in the growth medium were also observed between 61.5 and 96.8 ppm.
Heterologous expression.
The oxoX-G cDNA was amplified using the PCR with the primers 5′-TCTCTATCTATTTCATATGCGCCCCACCGGCAACG (inserts an NdeI site and a start codon to give a Met residue instead of the predicted signal peptide) and 5′-GGCTCGAGATCTGAGGCGACGACGAATGC (inserts an XhoI site in place of the stop codon). The PCR product was inserted into the NdeI and XhoI sites of pET32a (Novagen) to give pOxoxG. Escherichia coli BL21 Star (Invitrogen) was transformed with pOFX bad-KJ1, which harbors the genes dnaK and dnaJ (8), selected for kanamycin resistance, made competent using CaCl2, and transformed with pOxoxG. Cultures were grown at 37°C to an absorbance of 0.4 at 600 nm and induced with 0.4% (wt/vol) arabinose for 1 h to express DnaK and DnaJ. Expression of Oxox-G was then induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside after the addition of MnCl2 (5 mM). The cells were harvested after a further 4 h of incubation at 37°C with agitation and stored at −20°C. Thawed cells were resuspended in 50 mM phosphate buffer, pH 7.0, containing DNase and lysozyme and lysed with three passes though a French pressure cell. Cell debris was removed by centrifugation at 17,000 × g for 30 min at 4°C. The soluble fraction was dialyzed against 50 mM succinate buffer, pH 5.0, containing 200 mM NaCl. Precipitated protein was removed by centrifugation at 17,000 × g for 30 min at 4°C.
Molecular modeling.
The structures of the C. subvermispora oxalate oxidases were modeled on the basis of homology with oxalate decarboxylase OxdC (Protein Data Bank entry code 1UW8) (14) using Swiss-PDBViewer version 3.6b3 and energy minimized using Swiss-Model (11). A multiple sequence alignment between the two fungal oxidase isoforms and all of the known oxalate decarboxylases was used to help generate the models. Using the same software, the structure of barley oxalate oxidase (Protein Data Bank entry code 1FI2) (44) was independently superposed with the two domains of OxdC in order to derive a structure-based alignment of their sequences.
Nucleotide sequence accession numbers.
The following nucleotide sequences were deposited with EMBL: oxoX-C partial genomic DNA, AJ746414; oxoX-C partial cDNA, AJ563659; oxoX-G genomic DNA, AJ563660; oxoX-G cDNA, AJ746412.
RESULTS
Peptide sequencing.
Oxalate oxidase was purified from C. subvermispora to homogeneity using a previously published procedure (1). Amino acid sequencing of its N terminus was not possible due to it being blocked. However, quadrupole time-of-flight mass spectrometry of peptides obtained from the tryptic digestion of the enzyme gave five peptide sequences: PERGQ(L/I)GAK, (L/I)EAGA(L/I)R, GTTQ(L/I)TAVD(K/Q)NGR, V(K/Q)PT(K/Q)YAGGTVK, and SVAEVTVEPGAMR (where Leu and Ile are indistinguishable and some Lys and Gln were difficult to distinguish). Surprisingly, a BLAST search of these five peptide sequences in all available databases revealed maximum similarity with F. velutipes oxalate decarboxylase and a putative oxalate decarboxylase from Trametes versicolor (GenBank accession no. AAQ67425), rather than the known plant oxalate oxidases. The second and fifth peptides overlapped the start of the first and second set of cupin motifs of the oxalate decarboxylase sequences, strongly suggesting that the C. subvermispora oxalate oxidase was a bicupin as predicted (alignment not shown but see below). Degenerate primers based on the peptide sequences allowed the cloning of the gene as described below.
C-isoform gene.
A total of 1,544 nucleotides of the cDNA (henceforth called oxoX-C) coding for the C. subvermispora oxalate oxidase (henceforth called the C isoform) were cloned and sequenced. The 3′ end of oxoX-C included a stop codon together with a potential poly(A) signal sequence (AATTAA) 186 nucleotides downstream of the stop codon and 64 nucleotides upstream of a poly(A) tail. Although this is not the usual signal sequence (AATAAA), signal sequences are known to vary considerably in filamentous fungi and yeast (3, 12, 28, 31). The 5′ end was clearly incomplete with its start codon missing. Its cDNA in the Lambda library was therefore truncated at the 5′ end.
Two partial genomic DNA sequences (721 and 1,768 nucleotides covering 95% of the structural gene) that substantially overlapped with its cDNA sequence allowed the 5′ coding sequence and 14 introns to be identified. Only the last intron of oxoX-C did not comply with the GATG rule; it started with GC instead of GT. While some lariat sites conforming to the relatively less stringent fission yeast consensus sequence CTRAY (4) are obvious within the introns, most are not. Six out of seven nucleotides of the translation start site conformed to the Kozak consensus sequence [(A/G)NNATGG] (18). There was a potential TATA box at −69 in the 473-base-pair upstream region.
G-isoform gene.
A full-length genomic clone from a Lambda GEM-11 library gave a sequence with 3,752 nucleotides that was unexpectedly not identical to oxoX-C. This suggested that a distinct oxalate oxidase isoform (henceforth called the G isoform) exists in this organism. Its full-length cDNA of 1,418 nucleotides was then obtained. Sequence comparisons between oxoX-G and oxoX-C revealed the same number of introns that were located in the same positions relative to their aligned coding sequences. With two exceptions, the only insertions or deletions in the sequence alignment were within nine of the introns. At the nucleotide sequence level, the coding regions of oxoX-C and oxoX-G shared 79% identity, with insertions of 12 and 6 bases in the oxoX-G sequence near its 5′ and 3′ ends, respectively. All but one of the introns complied with the GT-AG rule; intron 5 starts with GC instead of GT. Many of the lariat sites conform to the consensus sequence CTRAY (4), but some do not.
The start and stop codons were readily identified. As with oxoX-C, six out of seven nucleotides of the translation start site conformed to the Kozak consensus sequence (18). An analysis of the 850-base-pair sequence, upstream of the protein coding region, revealed few obvious features except for a potential TATA box at position −83 and a possible metal responsive element (inverted TGCRCNC [39]) at position −142. It is interesting that there is a potential metal responsive element because the oxidase is likely to require manganese ions, in common with the plant oxalate oxidase and the bacterial oxalate decarboxylase (30, 37). The 3′ region contains a potential poly(A) signal (AAATAA) that is 207 nucleotides downstream of the stop codon.
The C and G isoforms are allelic.
C. subvermispora is a dikaryotic organism. In order to establish whether the C and G isoforms were allelic or different genes, the oxoX genes were amplified using the PCR with genomic DNA isolated from two C. subvermispora homokaryotic strains (38) together with two gene-specific primers that were complementary to highly conserved sequences in both of their coding regions. The nucleotide sequences of the isoforms showed that only the C-isoform DNA should be cleaved by EcoRI. Therefore, if the genes were not allelic, both PCRs would amplify both genes and both samples would be partially cleaved. On the other hand, if the genes were allelic, only one of the samples would be cleaved. The experiment gave the latter result, showing that the two oxoX genes were two alleles that resided on separate chromosomes. The oxoX-C and oxoX-G genes were associated with the strains CsB and CsA, respectively. Sequencing of each PCR product confirmed this result.
C-isoform protein sequence.
The protein sequence predicted from the overlapping partial oxoX-C sequences is identical to the five above-described peptide sequences obtained (Fig. 1). The protein sequence is 455 amino acids long and is predicted to have a molecular mass of 49.8 kDa. It has a putative extracellular secretion signal of 20 amino acids at its N-terminal end (10, 27), consistent with the known secretion of the enzyme activity (1). The mature protein is therefore predicted to have a molecular mass of 47.7 kDa and a pI of 4.8, close to that of 4.2 determined experimentally (1). Importantly, the duplication of the cupin motif showed that the enzyme is indeed the first bicupin with oxalate oxidase activity to be described. The sequence shows three potential N-glycosylation sites with the NX(S/T) motif. Intriguingly, a BLAST search of this protein sequence against the protein databases confirmed that maximum similarity was shared with the putative T. versicolor oxalate decarboxylase (e−148; 64% identity and 78% similarity) and F. velutipes oxalate decarboxylase (e−139; 61% identity and 75% similarity). However, a Glu residue thought to be required for decarboxylase activity (14) is not conserved in the oxidase sequence (see Discussion). The next best matches were with a putative Bacillus cereus oxalate decarboxylase (e−105; GenBank accession no. NP_830817) and then the B. subtilis oxalate decarboxylases OxdC and OxdD (e−103 and e−101, respectively) followed by several other hypothetical bicupins (2e−94 ≤ E value ≤ 6e−61) but with no other significant matches (E values, ≥7e−19). Importantly, no plant oxalate oxidases gave a significant score (E value cutoff of 0.72).
FIG. 1.
Protein sequences predicted from nucleic acid sequences of C. subvermispora oxalate oxidase allelic isoforms OxoX-C and OxoX-G. Peptide sequences of the purified oxalate oxidase obtained using quadrupole time-of-flight mass spectrometry are aligned above the protein sequences. Conserved, highly conservatively substituted, and moderately conservatively substituted amino acids are highlighted with asterisks, colons, and stops, respectively. Putative signal peptide sequences are underlined. The duplicated cupin motifs are boxed and consecutively labeled 1a, 1b, 2a, and 2b in italics. Putative N-glycosylation sites are highlighted in boldface type.
G-isoform protein sequence.
The protein sequence of the G isoform predicted from its DNA sequence is 461 amino acids long (Fig. 1). It is predicted to have a molecular mass of 50.0 kDa. Like the C isoform, it has a putative extracellular secretion signal of 20 amino acids at its N-terminal end (10, 27). The mature protein is therefore predicted to have a molecular mass of 48.0 kDa and a pI of 4.5. The G isoform has three potential N-glycosylation sites, the second and third of these being conserved in the C isoform. Given the predicted masses of the complete G-isoform protein (50.0 kDa) and that of the C isoform (49.8 kDa), it is likely that the enzyme is glycosylated in vivo because its mass has been determined to be 65.5 kDa (1). The C-isoform protein sequence shares high identity with that of the G isoform (85% identity and 96% similarity) with an insertion of 4 amino acids near the G-isoform N-terminal end and an extra 2 amino acids at the C-terminal end. The five peptide sequences described above share complete identity with the C isoform only. Two of the four elements of the duplicated cupin motifs are completely conserved between both isoforms, while the other two elements have only one highly conservative substitution each. Most of the sequence variation between the C isoform and the G isoform occurs at their N-terminal ends. A BLAST search of the G isoform gave similar result to those with the C isoform as described above. The similarity of the G-isoform protein sequence to the F. velutipes oxalate decarboxylase was also high (61% identity and 74% similarity). As with the C isoform, a Glu residue thought to be required for decarboxylase activity (14) is not conserved (see Discussion).
Degradation of exogenous oxalate during growth.
The degradation of exogenous oxalate by the organism at the time that cells were usually harvested was assessed by adding 10 mM [13C]oxalate to the growth medium. After just 2 h of continued incubation, NMR spectroscopy of the growth medium revealed that the oxalate had been completely degraded. This result is consistent with studies described previously (41) and implies that oxalate is rapidly degraded by a combination of the reactions described above together with the oxalate oxidase reaction. The only other peaks that were discernible in the spectrum were associated with glucose and a trace of formate. Control samples without added [13C]oxalate showed that the glucose was not derived from the added oxalate but was simply a component of the growth medium that contained the natural abundance of 13C. The trace amount of formate produced was presumably formed by the oxalate decarboxylase side reaction catalyzed by the C isoform.
Enzyme activity of the C isoform.
Given the high identity between the C. subvermispora oxalate oxidase isoform protein sequences and those of the fungal and bacterial oxalate decarboxylases, it was important to confirm that the enzyme was an oxalate oxidase and to establish whether it had any decarboxylase activity. The specific oxalate oxidase activity of the enzyme purified from C. subvermispora (now known to be predominantly the C isoform) was typically 9.2 U mg−1, which compares well with the value of 10.4 U mg−1 reported previously (1). The relative specific oxalate decarboxylase activity was determined to be only 0.2%, confirming its main activity to be oxalate oxidation. Furthermore, the relative oxalate decarboxylase activity was less than 0.6% in the crude extracts and in each fraction throughout the purification procedure, showing that the organism did not express oxalate decarboxylase activity in the conditions in which it was grown. In order to ensure that the oxalate decarboxylase from F. velutipes is indeed primarily a decarboxylase, the rate of oxalate oxidation relative to decarboxylation was determined to be 3.7%.
It was possible to detect the oxalate oxidase activity of the C. subvermispora enzyme in polyacrylamide gels by incubation with oxalate, 4-chloro-1-naphthol, and horseradish peroxidase (not shown). However it was essential to run the gel under native conditions because enzyme activity was lost in the presence of sodium dodecyl sulfate (SDS). This is in sharp contrast to the plant oxalate oxidases, which are stable in the presence of SDS, provided the samples are not boiled or exposed to reductants such as dithiothreitol (21).
No superoxide dismutase activity of the purified enzyme was detected. However, the purified enzyme did exhibit another side reaction: the oxalate-dependent oxidation of a dye. The relative rate of direct phenol red oxidation was determined to be 9% compared with normal oxalate oxidation. These results show similarities with the B. subtilis oxalate decarboxylase OxdC in that this enzyme exhibits oxalate oxidase activity and oxalate-dependent but hydrogen peroxide-independent dye oxidation activity at 0.2 and 0.5% of the rate of oxalate decarboxylation (75 U mg−1) (37). Overall, these results show that the control of reaction specificity by the oxidase and decarboxylase enzymes is tight but not absolute.
Heterologous expression and activity of the G isoform.
The oxoX-G isoform without its putative signal peptide was expressed in E. coli. Soluble and active protein was obtained only when the enzyme was coexpressed with the chaperones DnaK and DnaJ and when a manganese salt was added to the growth medium. The enzyme was partially purified to about 40% purity according to SDS-polyacrylamide gel electrophoresis by precipitating contaminating proteins by dialysis at pH 5.0. Its 56-kDa size was close to that expected (51 kDa). The enzyme did not have any detectable oxalate decarboxylase or oxalate-dependent dye oxidation activities but had oxalate oxidase activity with a specific activity of about 20 U mg−1, a higher specific activity than that of OxoX-C purified from the fungus.
Molecular modeling.
Homology models of the two fungal oxidase isoforms were developed, based on the structures of the bacterial decarboxylase (2, 14), to help identify amino acids responsible for defining reaction specificity. The only amino acid substitutions in the N-terminal domain metal ion binding sites (site 1) of the oxidases were in an active-site lid covering the substrate binding pocket (E162S in OxoX-C and E162A in OxoX-G and T165Q in both isoforms [decarboxylase numbering]) together with a conservative change at the back of the pocket (I142L for both). There were no other changes within 6 Å of the metal ion binding site of this domain. There were two amino acid substitutions within 6 Å of the metal ion binding sites of the C-terminal domains (site 2) of the oxidases: a moderate change (Q282T in both isoforms) and a conservative change (V313I in both). Both of these changes in site 2 were separated in space from the metal ion binding site by residues that coordinated the metal ion; i.e., they did not line a potential substrate binding pocket. Incidentally, the potential N-glycosylation sites of the fungal oxidase isoforms are all solvent accessible in the model.
DISCUSSION
General considerations.
This paper describes the cloning and sequencing of the C-isoform cDNA and genomic DNA (oxoX-C) of oxalate oxidase from C. subvermispora, whose predicted protein sequence matches the peptide sequences obtained from the purified enzyme. It is highly likely that the original description of the enzyme from this organism was also of the C isoform because the methods employed to grow the organism and purify the enzyme were essentially identical (1). This is supported by preliminary data that suggested that the mRNA of this isoform, which is digested by EcoRI as described above, is dominant in the dikaryotic strain under normal growth conditions. The genomic DNA and cDNA of a distinct G isoform was also cloned and sequenced (oxoX-G). The heterologous expression of the oxoX-G cDNA in E. coli gave a protein with oxalate oxidase activity. It is noteworthy that its specific activity was twice that of the C isoform purified from the fungus despite it being only ∼40% pure. This implies a fivefold-higher specific activity when pure, which could be due to differences in intrinsic activity or other factors such as the absence of glycosylation when expressed in E. coli.
The protein sequences of both oxalate oxidase isoforms show them to contain a duplication of the cupin motifs as predicted. Surprisingly, they share most sequence identity with fungal and bacterial oxalate decarboxylases, which are also bicupins. In order to rule out the possibility that the C. subvermispora enzymes were in fact decarboxylases, it was established that both the enzyme purified from C. subvermispora (predominantly the C isoform) and the heterologously expressed G isoform exhibited at least 500-fold more oxalate oxidase than decarboxylase activity. By contrast, the F. velutipes bicupin oxalate decarboxylase was shown to exhibit 27-fold less oxidase than decarboxylase activity. Therefore, a key conclusion from this work is that the C. subvermispora oxalate oxidases are the first to be identified as bicupins, while all previously sequenced examples have been monocupins that were from plants (also known as germins). It is noteworthy that the fungal oxidase was SDS sensitive, unlike the plant oxidase (21). The lack of robustness of the fungal oxidase could have practical biotechnological advantages over the plant enzymes. This is because the cupin superfamily contains members that are strongly allergenic (26), and commercial products for human or animal use that contain a cupin protein could present fewer problems if the cupin can be rapidly degraded in vitro or in vivo.
OxoX-C did not possess any detectable superoxide dismutase activity. There is one report of barley oxalate oxidase having superoxide dismutase activity (44), but this has yet to be confirmed by other groups (7, 43). The active site of barley oxalate oxidase does resemble that of the otherwise unrelated mitochondrial manganese superoxide dismutase, albeit with some geometric differences (30, 44). However, it is important to appreciate that the resting state of barley oxalate oxidase is in the Mn2+ oxidation state, while that of the mitochondrial superoxide dismutase is Mn3+, presumably due to differences in their reduction potentials.
Relationship between the two isoforms.
The identification of two protein isoforms from this organism is not unusual, as multiple isoforms of other enzymes involved in lignin degradation have been reported previously (23). However, C. subvermispora is dikaryotic, allowing the possibility that the isoforms are allelic. The 79% nucleotide identity between the two oxoX genes is lower than that of >95% which is usually associated with alleles in basidiomycetes (22, 38). Nevertheless, using homokaryotic strains, it was possible to demonstrate that the C and G isoforms do indeed represent two alleles of the oxalate oxidase rather than different genes. Although the peptide sequencing and preliminary reverse transcriptase PCR experiments indicate that the C isoform is the dominant isoform to be transcribed and translated, the fact that a cDNA sequence was obtained for the G isoform shows that oxoX-G is also a structural gene that is expressed under these growth conditions. Why the allele that codes for the enzyme with an apparently lower specific activity is more highly expressed is not quite clear. It is important that the ligninolytic enzymes of C. subvermispora appear to be constitutively expressed throughout growth under the conditions employed, unlike the corresponding enzymes of Phanerochaete chrysosporium, which are expressed during nutrient limitation (33).
Gene organization.
The similarity between both isoforms of the C. subvermispora oxalate oxidase and the F. velutipes oxalate decarboxylase sequences extends to the positions of their introns (not shown). The oxidases have fewer introns, with 14 as opposed to 17 with introns 5, 7, and 10 of the decarboxylase not being present in the oxidases. Although many of the introns of each sequence harbor insertions and deletions, only intron 11 of the oxidases is in a nonequivalent position relative to the aligned exon sequence of the decarboxylase. There has therefore been considerable sequence conservation at the level of gene organization as well as at the protein level.
Phylogeny.
A phylogenetic analysis of protein sequences reveals the relationship of the two fungal oxalate oxidase isoforms to the four confirmed oxalate decarboxylases, 19 other bicupins that were identified in the BLAST search, and barley oxalate oxidase (not shown). (Many additional bicupin sequences have been identified in preliminary genome sequences but have not been included in the analyses.) The two fungal oxidase isoforms cluster together. In turn, these isoforms belong to a group that includes the F. velutipes oxalate decarboxylase and the putative T. versicolor oxalate decarboxylase. There are several other groupings that include the remaining three confirmed oxalate decarboxylases and the bicupins. The barley oxalate oxidase is the most distantly related and aligns best with the C-terminal domains of these bicupins. Together with the BLAST search, this analysis emphasizes the closer relationship of the fungal oxalate oxidase to the oxalate decarboxylases than to the plant oxalate oxidases. It is not clear from the phylogenetic analysis whether the bicupin oxalate oxidase arose by gene duplication of the monocupin oxalate oxidase and only subsequently evolved to give the bicupin oxalate decarboxylases. However, the lack of a known monocupin oxalate decarboxylase makes this sequence of events still likely.
Conservation of metal ion binding sites.
Fig. 2 shows an alignment of the protein sequences of the two fungal oxalate oxidase isoforms with the four confirmed oxalate decarboxylases from fungi and bacteria together with the oxalate oxidase from barley. The duplication of the cupin motifs is obvious with considerable conservation of residues within these regions in all bicupin sequences. The degree of sequence conservation is clearly less in the barley oxidase. The Glu and three His of each pair of motifs known to ligate a manganese ion in each of the two B. subtilis OxdC decarboxylase domains (2) are completely conserved in all of these protein sequences (Fig. 2, boldface type). The same set of residues binds the manganese ion (30) in the plant oxalate oxidase (44). These residues are suitably placed to bind a metal ion in both sites of both homology models of the fungal oxidases. Just as with B. subtilis OxdC (37), there is a requirement for the addition of manganese salts to the growth medium in order to express soluble and active OxoX-G. This strongly suggests that OxoX-G and, by close association, OxoX-C also bind manganese ions in the same way as the other oxalate-degrading enzymes. Incidentally, preliminary reverse transcriptase PCR experiments indicated that the level of expression of oxoX-C in wild-type C. subvermispora was not dependent on manganese salt concentration in the growth medium, arguing against a manganese-sensitive metal response element associated with the oxoX-C gene. It must also be noted that there was no indication of such a regulatory element in the upstream region of oxoX-G.
FIG. 2.
Protein motif sequence alignments of the C. subvermispora oxalate oxidase allelic isoforms with oxalate-degrading enzymes. The OxoX-C and OxoX-G allelic isoforms of C. subvermispora (C. sub) oxalate oxidase are aligned with the oxalate decarboxylases from F. velutipes (F. vel) (GenBank accession no. AAF13275), A. phoenices (A. pho) (GenBank accession no. AAE83943), and B. subtilis (B. sub) (OxdD GenBank accession no. O34767; OxdC GenBank accession no. O34714). The sequence of barley (H. vulgare [H. vul]) oxalate oxidase (GenBank accession no. P45850) is also shown for comparison, but this was structurally aligned with the B. subtilis OxdC C-terminal and N-terminal domains. Only the alignment of the duplicated cupin motifs (labeled in italics) together with regions downstream are shown. The first amino acid of each cupin motif is numbered, and the number of any intervening amino acids is indicated with underlining. The residues known to ligate mononuclear manganese ions in B. subtilis OxdC and barley OxoX are highlighted in boldface type. Conserved amino acids are highlighted with a grey background. Arg92 (B. subtilis OxdC sequence and numbering) is indicated by #, Glu162 is indicated by @, and Glu333 is indicated by +.
Implications for reaction specificity.
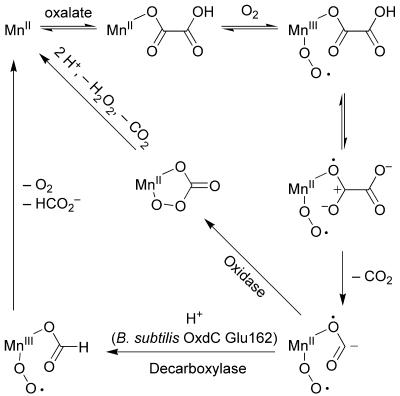
We have proposed divergent mechanisms for oxalate oxidation and oxalate decarboxylation that involve common free-radical intermediates based on the presence of Mn2+ in the resting state of both enzymes and the unique requirement of dioxygen for the decarboxylase reaction (30, 37). The detection of formyl radicals produced by the recombinant plant oxidase in the presence of oxalate and the crystal structures of the plant oxidase and the bacterial decarboxylase have provided additional support for such mechanisms (2, 14, 43, 44). Recently, a detailed kinetic isotope effect study has provided the first evidence for a free-radical mechanism in the oxalate decarboxylase reaction (29). The unexpectedly high identity between the fungal oxalate oxidases and the microbial oxalate decarboxylases reported here provides an opportunity to better understand what controls the reaction specificity of these enzymes.
Figure 3 summarizes our current understanding of the divergent free-radical mechanisms of oxalate oxidases and decarboxylases. After oxalate and dioxygen bind to the Mn2+ ion, electron transfer from the oxalate to the manganic-superoxo species facilitates decarboxylation to give a formyl radical. The lack of an amino acid proton donor in the active site of the plant oxidase suggests that percarbonate is an intermediate with this enzyme. By contrast, the decarboxylase requires the specific protonation of the carbon atom of the formyl radical intermediate. We had therefore predicted that it is primarily the presence or absence of a proton donor at the active site of these enzymes that dictates the outcome of their catalytic cycles (37).
FIG. 3.
Current model of divergent free-radical mechanisms for oxalate oxidation and decarboxylation (2, 14, 29, 30, 37, 43).
More recently, we have described evidence from crystallographic and mutagenic studies with B. subtilis OxdC that Glu162 (Fig. 2) is the proton donor and that site 1 is the sole or dominant catalytic site (14). The Glu162 residue is found in a specific position in a loop that forms a lid over site 1 that is anchored by flanking Phe residues (160 and 166). If Glu162 were primarily responsible for defining an oxalate decarboxylase, it would be expected that the fungal oxalate oxidases would not have an acidic residue at this exact position. This is indeed the case with Ser and Ala residues being present in the C and G isoforms, respectively. It is important that the flanking Phe residues are conserved in both fungal oxidase sequences and that the molecular models show how they would anchor the fungal oxalate oxidase lid. These constraints dictate that the positions of the Ser and Ala residues of the oxidase isoforms are identical to that of Glu162 in the decarboxylase. The high identity and similarity between the fungal oxidases and the decarboxylases give confidence in the molecular modeling in general.
There are only two other changes in site 1 of the models. A conservative change at the back of the substrate binding pocket is unlikely to have any effect on the reaction specificity (I142L). Other than Glu162, there is only one other amino acid side chain of the lid that forms part of the substrate binding pocket: Thr165. This residue is completely conserved in all known and putative oxalate decarboxylases. It is substituted by a Gln residue in both oxidase isoforms, and it could help stabilize negatively charged catalytic intermediates through hydrogen bonding. While the substitution of Glu162 by Ala causes a loss of decarboxylase activity in bacterial oxalate decarboxylase, this change alone does not give rise to enhanced oxalate oxidase activity (14). The role of other amino acids to affect such a conversion, such as those discussed above, is the subject of current study.
As described previously (14), the plant oxidase does not have a loop that can form a lid because of a deletion of two amino acids in this part of the sequence. Of note is the observation that all of the bicupins from the BLAST search had an acidic residue at the key position, with a single exception: an Aspergillus nidulans hypothetical protein (GenBank accession no. EAA59771) had a Lys residue at this position together with an amino acid insertion in the sequence of the loop. This suggests that this enzyme is not an oxalate decarboxylase but possibly has oxalate oxidase or some other enzyme activity. The preliminary genome sequence of the ligninolytic organism P. chrysosporium (25) also revealed three bicupins (Pc137.3.1, Pc137.17.1, and Pc45.42.1) with high identity with the OxoX isoforms, but all had an acidic residue at the key position, suggesting that they are all decarboxylases.
Mutagenesis and modeling studies have shown that Arg92 of B. subtilis OxdC site 1 (Fig. 2) is probably involved in the binding of oxalate and the stabilization of the oxalate radical intermediate by interacting with the negative charge on the proximal carboxyl group. This residue is conserved in the fungal oxidase sequences and is situated in the appropriate position in the molecular models, as would be expected. It is noteworthy that at this position, there is an Asn residue in the plant oxalate oxidase, which aligns best with the C-terminal domain as described previously (9).
Anand et al. had previously suggested that B. subtilis OxdC Glu333 (Fig. 2) was the proton donor in site 2. However, recent mutagenesis studies are more consistent with this manganese binding site having a structural role and, at most, a minor catalytic role (14). The complete conservation of this acidic residue in the fungal oxalate oxidase isoforms makes such a structural role extremely likely because the enzymes exhibit very low oxalate decarboxylase activity (≤0.2%). Furthermore, the homology models of the oxidases indicate that there are no amino acid changes in a potential site 2 substrate binding site and that the nearest changes could not obviously define the reaction specificity. This result therefore provides strong independent evidence against site 2 having any catalytic activity in either the fungal oxidases or the bacterial decarboxylases. The subtle differences in sequence between the bicupin oxalate oxidases and decarboxylases revealed by this work will help in the correct prediction and annotation of the function of new bicupin sequences.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council with Competitive Strategic Grant and Joint Research Equipment Initiative proteomics facility grants (JE412631 and JE412701) awarded to the John Innes Centre. Marta R. Escutia was a recipient of a John Innes Foundation studentship.
We thank Cathie Martin for helpful discussions, Shirley A. Fairhurst for the NMR spectrometry, Olivier Fayet for providing pOFX bad-KJ1, and Carol Gormal for peptide sequencing.
REFERENCES
- 1.Aguilar, C., U. Urzúa, C. Koenig, and R. Vicuña. 1999. Oxalate oxidase from Ceriporiopsis subvermispora: biochemical and cytochemical studies. Arch. Biochem. Biophys. 366:275-282. [DOI] [PubMed] [Google Scholar]
- 2.Anand, R., P. C. Dorrestein, C. Kinsland, T. P. Begley, and S. E. Ealick. 2002. Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 Å resolution. Biochemistry 41:7659-7669. [DOI] [PubMed] [Google Scholar]
- 3.Ballance, D. J. 1986. Sequences important for gene-expression in filamentous fungi. Yeast 2:229-236. [DOI] [PubMed] [Google Scholar]
- 4.Barker, D. G., J. H. M. White, and L. H. Johnston. 1987. Molecular characterization of the DNA-ligase gene, Cdc17, from the fission yeast Schizosaccharomyces pombe. Eur. J. Biochem. 162:659-667. [DOI] [PubMed] [Google Scholar]
- 5.Beauchamp, C., and L. Fridovich. 1971. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44:276-287. [DOI] [PubMed] [Google Scholar]
- 6.Benson, D. 1976. Mechanisms of oxidation by metal ions. Elsevier Scientific Publishing Company, Amsterdam, The Netherlands.
- 7.Bernier, F., and A. Berna. 2001. Germins and germin-like proteins: plant do-all proteins. But what do they do exactly? Plant Physiol. Biochem. 39:545-554. [Google Scholar]
- 8.Castanie, H. P., H. Berges, J. Oreglia, M. F. Prere, and O. Fayet. 1997. A set of pBR322-compatible plasmids allowing the testing of chaperone-assisted folding of proteins overexpressed in Escherichia coli. Anal. Biochem. 254:150-152. [DOI] [PubMed] [Google Scholar]
- 9.Dunwell, J. M., S. Khuri, and P. J. Gane. 2000. Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 64:153-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emanuelsson, O., H. Nielsen, S. Brunak, and G. von Heijne. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300:1005-1016. [DOI] [PubMed] [Google Scholar]
- 11.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 12.Heidmann, S., C. Schindewolf, G. Stumpf, and H. Domdey. 1994. Flexibility and interchangeability of polyadenylation signals in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:4633-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen, K. A., W. L. Bao, S. Kawai, E. Srebotnik, and K. E. Hammel. 1996. Manganese-dependent cleavage of nonphenolic lignin structures by Ceriporiopsis subvermispora in the absence of lignin peroxidase. Appl. Environ. Microbiol. 62:3679-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Just, V. J., C. E. M. Stevenson, L. Bowater, A. Tanner, D. M. Lawson, and S. Bornemann. 2004. A closed conformation of Bacillus subtilis oxalate decarboxylase OxdC provides evidence for the true identity of the active site. J. Biol. Chem. 279:19867-19874. [DOI] [PubMed] [Google Scholar]
- 15.Kesarwani, M., M. Azam, K. Natarajan, A. Mehta, and A. Datta. 2000. Oxalate decarboxylase from Collybia velutipes—molecular cloning and its overexpression to confer resistance to fungal infection in transgenic tobacco and tomato. J. Biol. Chem. 275:7230-7238. [DOI] [PubMed] [Google Scholar]
- 16.Khuri, S., F. T. Bakker, and J. M. Dunwell. 2001. Phylogeny, function, and evolution of the cupins, a structurally conserved, functionally diverse superfamily of proteins. Mol. Biol. Evol. 18:593-605. [DOI] [PubMed] [Google Scholar]
- 17.Koyama, H. 1988. Purification and characterization of oxalate oxidase from Pseudomonas sp. OX-53. Agric. Biol. Chem. 52:743-748. [Google Scholar]
- 18.Kozak, M. 1981. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 9:5233-5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lane, B. G. 2002. Oxalate, germins, and higher-plant pathogens. IUBMB Life 53:67-75. [DOI] [PubMed] [Google Scholar]
- 20.Lane, B. G., F. Bernier, E. Dratewka-Kos, R. Shafai, T. D. Kennedy, C. Pyne, J. R. Munro, T. Vaughan, D. Walters, and F. Altomare. 1991. Homologies between members of the germin gene family in hexaploid wheat and similarities between these wheat germins and certain physarium spherulins. J. Biol. Chem. 266:10461-10469. [PubMed] [Google Scholar]
- 21.Lane, B. G., J. M. Dunwell, J. A. Ray, M. R. Schmitt, and A. C. Cuming. 1993. Germin, a protein marker of early plant development, is an oxalate oxidase. J. Biol. Chem. 268:12239-12242. [PubMed] [Google Scholar]
- 22.Larrondo, L. F., B. González, D. Cullen, and R. Vicuña. 2004. Characterization of a multicopper oxidase gene cluster in Phanerochaete chrysosporium and evidence of altered splicing of the mco transcripts. Microbiology 150:2775-2783. [DOI] [PubMed] [Google Scholar]
- 23.Lobos, S., J. Larraín, L. Salas, D. Cullen, and R. Vicuña. 1994. Isoenzymes of manganese-dependent peroxidase and lactase produced by the lignin-degrading basidiomycete Ceriporiopsis subvermispora. Microbiology 140:2691-2698. [DOI] [PubMed] [Google Scholar]
- 24.Lobos, S., L. Larrondo, L. Salas, E. Karahanian, and R. Vicuña. 1998. Cloning and molecular analysis of a cDNA and the Cs-mnp1 gene encoding a manganese peroxidase isoenzyme from the lignin-degrading basidiomycete Ceriporiopsis subvermispora. Gene 206:185-193. [DOI] [PubMed] [Google Scholar]
- 25.Martinez, D., L. F. Larrondo, N. Putnam, M. D. Gelpke, K. Huang, J. Chapman, K. G. Helfenbein, P. Ramaiya, J. C. Detter, F. Larimer, P. M. Coutinho, B. Henrissat, R. Berka, D. Cullen, and D. Rokhsar. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 22:695-700. [DOI] [PubMed] [Google Scholar]
- 26.Mills, E. N. C., J. Jenkins, N. Marigheto, P. S. Belton, A. P. Gunning, and V. J. Morris. 2002. Allergens of the cupin superfamily. Biochem. Soc. Trans. 30:925-929. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 28.Peterson, J. A., and A. M. Myers. 1993. Functional-analysis of messenger-RNA 3′ end formation signals in the convergent and overlapping transcription units of the Saccharomyces cerevisiae genes Rho1 and Mrp2. Nucleic Acids Res. 21:5500-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinhardt, L. A., D. Svedruzic, C. H. Chang, W. W. Cleland, and N. G. J. Richards. 2003. Heavy atom isotope effects on the reaction catalyzed by the oxalate decarboxylase from Bacillus Subtilis. J. Am. Chem. Soc. 125:1244-1252. [DOI] [PubMed] [Google Scholar]
- 30.Requena, L., and S. Bornemann. 1999. Barley (Hordeum vulgare) oxalate oxidase is a manganese-containing enzyme. Biochem. J. 343:185-190. [PMC free article] [PubMed] [Google Scholar]
- 31.Russo, P., W. Z. Li, D. M. Hampsey, K. S. Zaret, and F. Sherman. 1991. Distinct cis-acting signals enhance 3′ end-point formation of Cyc1 messenger-RNA in the yeast Saccharomyces cerevisiae. EMBO J. 10:563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rüttimann, C., E. Schwember, L. Salas, D. Cullen, and R. Vicuña. 1992. Ligninolytic enzymes of the white rot basidiomycetes Phlebia brevispora and Ceriporiopsis subvermispora. Biotechnol. Appl. Biochem. 16:64-76. [Google Scholar]
- 33.Rüttimann-Johnson, C., L. Salas, R. Vicuña, and T. K. Kirk. 1993. Extracellular enzyme production and synthetic lignin mineralization by Ceriporiopsis subvermispora. Appl. Environ. Microbiol. 59:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitou, N., and M. Nei. 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 35.Shimada, M., Y. Akamtsu, T. Tokimatsu, K. Mii, and T. Hattori. 1997. Possible biochemical roles of oxalic acid as a low molecular weight compound involved in brown-rot and white-rot wood decays. J. Biotechnol. 53:103-113. [Google Scholar]
- 36.Shutov, A. D., F. R. Blattner, and H. Baumlein. 1999. Evolution of a conserved protein module from Archaea to plants. Trends Genet. 15:348-349. [DOI] [PubMed] [Google Scholar]
- 37.Tanner, A., L. Bowater, S. A. Fairhurst, and S. Bornemann. 2001. Oxalate decarboxylase requires manganese and dioxygen for activity—overexpression and characterization of Bacillus subtilis YvrK and YoaN. J. Biol. Chem. 276:43627-43634. [DOI] [PubMed] [Google Scholar]
- 38.Tello, M., D. Seelenfreund, S. Lobos, J. Gaskell, D. Cullen, and R. Vicuna. 2001. Isolation and characterization of homokaryotic strains from the ligninolytic basidiomycete Ceriporiopsis subvermispora. FEMS Microbiol. Lett. 199:91-96. [DOI] [PubMed] [Google Scholar]
- 39.Thiele, D. J. 1992. Metal-regulated transcription in eukaryotes. Nucleic Acids Res. 20:1183-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Urzúa, U., P. J. Kersten, and R. Vicuña. 1998. Manganese peroxidase-dependent oxidation of glyoxylic and oxalic acids synthesized by Ceriporiopsis subvermispora produces extracellular hydrogen peroxide. Appl. Environ. Microbiol. 64:68-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaisey, E. B., V. H. Cheldelin, and R. W. Newburgh. 1961. Oxalate oxidation by an obligately parasitic fungus Tilletia contraversa. Arch. Biochem. Biophys. 95:66-69. [DOI] [PubMed] [Google Scholar]
- 43.Whittaker, M. M., and J. W. Whittaker. 2002. Characterization of recombinant barley oxalate oxidase expressed by Pichia pastoris. J. Biol. Inorg. Chem. 7:136-145. [DOI] [PubMed] [Google Scholar]
- 44.Woo, E. J., J. M. Dunwell, P. W. Goodenough, A. C. Marvier, and R. W. Pickersgill. 2000. Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Biol. 7:1036-1040. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Z. G., J. Yang, D. B. Collinge, and H. Thordalchristensen. 1996. Ethanol increases sensitivity of oxalate oxidase assays and facilitates direct activity staining in SDS gels. Plant Mol. Biol. Rep. 14:266-272. [Google Scholar]
- 46.Zhou, F. S., Z. G. Zhang, P. L. Gregersen, J. D. Mikkelsen, E. de Neergaard, D. B. Collinge, and H. Thordal-Christensen. 1998. Molecular characterization of the oxalate oxidase involved in the response of barley to the powdery mildew fungus. Plant Physiol. 117:33-41. [DOI] [PMC free article] [PubMed] [Google Scholar]