Abstract
DNA shuffling and saturation mutagenesis of positions F108, L190, I219, D235, and C248 were used to generate variants of the epoxide hydrolase of Agrobacterium radiobacter AD1 (EchA) with enhanced enantioselectivity and activity for styrene oxide and enhanced activity for 1,2-epoxyhexane and epoxypropane. EchA variant I219F has more than fivefold-enhanced enantioselectivity toward racemic styrene oxide, with the enantiomeric ratio value (E value) for the production of (R)-1-phenylethane-1,2-diol increased from 17 for the wild-type enzyme to 91, as well as twofold-improved activity for the production of (R)-1-phenylethane-1,2-diol (1.96 ± 0.09 versus 1.04 ± 0.07 μmol/min/mg for wild-type EchA). Computer modeling indicated that this mutation significantly alters (R)-styrene oxide binding in the active site. Another three variants from EchA active-site engineering, F108L/C248I, I219L/C248I, and F108L/I219L/C248I, also exhibited improved enantioselectivity toward racemic styrene oxide in favor of production of the corresponding diol in the (R) configuration (twofold enhancement in their E values). Variant F108L/I219L/C248I also demonstrated 10-fold- and 2-fold-increased activity on 5 mM epoxypropane (24 ± 2 versus 2.4 ± 0.3 μmol/min/mg for the wild-type enzyme) and 5 mM 1,2-epoxyhexane (5.2 ± 0.5 versus 2.6 ± 0.0 μmol/min/mg for the wild-type enzyme). Both variants L190F (isolated from a DNA shuffling library) and L190Y (created from subsequent saturation mutagenesis) showed significantly enhanced activity for racemic styrene oxide hydrolysis, with 4.8-fold (8.6 ± 0.3 versus 1.8 ± 0.2 μmol/min/mg for the wild-type enzyme) and 2.7-fold (4.8 ± 0.8 versus 1.8 ± 0.2 μmol/min/mg for the wild-type enzyme) improvements, respectively. L190Y also hydrolyzed 1,2-epoxyhexane 2.5 times faster than the wild-type enzyme.
Epoxide hydrolases (EHs; EC 3.3.2.3), ubiquitous enzymes found in mammals, insects, plants, and microorganisms (37), hydrolyze an epoxide to its corresponding vicinal diol with the addition of a water molecule (37). Whereas mammalian EHs have been extensively studied for their important role in the detoxification of epoxides derived from xenobiotics (1), their use as biocatalysts on a preparative scale has been impeded by the lack of availability of the enzymes (6). Recently, with the identification of more EHs from microbial sources which have excellent enantioselectivity (2), and given that they may be produced on an almost unlimited scale (6), interest has increased in using microbial EHs for preparing epoxides or vicinal diols at high enantiomeric purity (6, 33, 37).
Enantiomerically pure compounds are increasingly important to the chemical and pharmaceutical industries (12). Epoxides and vicinal diols in enantiomerically pure form are often prepared using chemical catalytic methods (24) such as the transition metal-based asymmetric epoxidation of allylic alcohols and unfunctionalized alkenes developed by Sharpless and Jacobsen (1), respectively, for the preparation of epoxides, and Sharpless osmium-catalyzed asymmetric dihydroxylation of olefins for the production of vicinal diols (1). Alternatively, catalysis using EHs with high enantioselectivity and activity enables the simple preparation of enantiomerically pure epoxides and vicinal diols starting from cheap racemic epoxides under mild conditions using aqueous solutions (Fig. 1).
FIG. 1.
Enantioselective hydrolysis of racemic styrene oxide by EchA to form (R)-1-phenylethane-1,2-diol and the unreacted epoxide enantiomer, (S)-styrene oxide.
Due to the potential importance of EHs in industrial applications, it is desirable to expand their current biosynthetic capabilities. This may be achieved either by finding new enzymes or by applying protein engineering to broaden the scope and enhance the performances of existing EHs. Recently, directed evolution was performed on the EH from Aspergillus niger (AnEH) (4, 20), yielding enzymes with either enhanced rates for 4-(p-nitrophenoxy)-1,2-epoxybutane (but the improved activity was due to an elevated expression level) (4) or more than doubled enantioselectivity in the hydrolytic kinetic resolution of glycidyl phenyl ether (20).
The 3-dimensional structure of the EH from Agrobacterium radiobacter AD1 (EchA; GenBank accession no. Y12804) has been solved (PDB accession code 1EHY) (21), which provides molecular-level insights into the reaction mechanism of this enzyme (17). EchA (294 amino acids) contains two domains: a core domain with typical α/β hydrolase fold topology formed by an eight-stranded β-sheet sandwiched by α-helices on both sides and an α-helical cap domain protruding from the core domain (17). The catalytic triads D107, D246, and H275 are located in a hydrophobic internal cavity between the two domains (17). The catalytic reaction follows a two-step mechanism involving an alkyl-enzyme ester intermediate, which is further hydrolyzed via the attack of a water molecule (17, 18). EchA purified from A. radiobacter AD1 hydrolyzed epichlorohydrin (the physiological substrate) at a rate of 32 μmol/min/mg (11). The kinetic mechanism of EchA was solved in detail for both enantiomers of styrene oxide (23), and wild-type EchA was determined to be moderately enantioselective toward (R)-styrene oxide (32). The EchA variant Y215F has also been found to have enhanced enantioselectivity toward styrene oxide (7, 31), but at the cost of decreased catalytic activity (24). Very recently, directed evolution of the same enzyme resulted in 13-fold-improved enantioselectivity toward p-nitrophenyl glycidyl ether (pNPGE) (35).
Previously, the EchA active-site residues F108, I219, and C248 were altered to enhance the degradation of cis-1,2-dichloroethylene epoxide (cis-DCE epoxide) (25). The variants created in that study (25) were evaluated here for altered enantioselectivity toward racemic styrene oxide, which has been used as a model substrate for studying the enantioselectivity of EchA (22, 31). Further, DNA shuffling was used to isolate EchA variants with altered enzymatic activity and enantioselectivity toward racemic styrene oxide, so that residues, especially those having a far-reaching effect on substrate binding that are not reliably predicted by rational design, may be identified; and saturation mutagenesis was subsequently used to evaluate all amino acids at the important sites identified from random mutagenesis in this study. Production of enantiomerically pure (R)-1-phenylethane-1,2-diol was emphasized here because it is a valuable chiral building block for the organic synthesis of important compounds (36) such as chiral catalysts (e.g., chiral ditertiary phosphine, used as a ligand in asymmetric homologous hydrogenation catalysts) (13) and pharmaceutically active compounds (e.g., optically pure isoproterenol analogues) (14). This is the first report of the importance of residue I219 (or its analogues) for enantioselectivity in epoxide hydrolases, and the importance of residue L190 for the catalysis rate is also disclosed.
MATERIALS AND METHODS
Chemicals, organisms, and growth conditions.
Racemic styrene oxide, pure (R)-styrene oxide, pure (S)-styrene oxide, and racemic epoxypropane were purchased from Sigma Chemical Co. (St. Louis, MO), and all the other chemicals were obtained from Fisher Scientific Company (Pittsburgh, PA). All materials were used without further purification. Escherichia coli strain TG1 (8) was utilized as the host for cloning and functional expression of genes. Recombinant strains were routinely cultivated at 37°C in Luria-Bertani (LB) broth (28) supplemented with kanamycin (Kan; 100 μg/ml) to maintain plasmid pBS(Kan)EH (25), which constitutively expresses EchA from a lac promoter. All experiments (unless otherwise stated) were conducted using exponential-phase cultures in LB-plus-Kan medium which were started from fresh colonies, induced fully with 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an optical density at 600 nm (OD600) of 0.2 to 0.3, and harvested after 2 h of IPTG induction with an OD600 of 1.5 to 2.0 (IPTG induction was used to ensure equal, high expression of the EchA isoforms).
Cell preparation for enzyme assays.
The exponentially grown cells were harvested by centrifugation (10,000 × g, 4°C) for 10 min and resuspended in TEM buffer (10 mM Tris-HCl, 1 mM EDTA, and 1 mM mercaptoethanol, pH 7.4) to a final OD600 of ∼1.0. The cell suspensions were disrupted with sonication by an F60 sonic dismembrator (Fisher Scientific, Pittsburgh, PA) performed 10 times, for 10 s each time, at 10 W (with a 1-min pause on ice). The resulting sonicated cells contained epoxide hydrolase concentrations of ∼50 μg/ml and were used for determining enzymatic activity (protein concentrations were estimated from sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE] using a known amount of bovine serum albumin as a standard and were approximately the same for all the variants unless otherwise stated).
Molecular techniques and protein analysis and purification.
Plasmid DNA was isolated using a Midi or Mini kit (QIAGEN, Inc., Chatsworth, CA), and PCR products were purified with a Wizard PCR Preps DNA purification system (Promega Corporation, Madison, WI). DNA fragments were isolated from agarose gels using a QIAquick gel extraction kit (QIAGEN, Inc.). Transformation of E. coli was carried out by electroporation using a Gene Pulser/Pulse Controller (Bio-Rad Laboratories, Hercules, CA) at 15 kV/cm, 25 μF, and 200 Ω. A dye terminator cycle sequencing protocol based on the dideoxy method of sequencing DNA developed by Sanger et al. (29) was used to sequence both strands of the wild-type and mutant echA. Expression of recombinant proteins was analyzed by standard Laemmli discontinuous SDS-PAGE (12%) (28). Purification of recombinant proteins was conducted using the method of Rink et al. (21) with the modifications described previously (25).
DNA shuffling of EchA.
DNA shuffling was performed using methods adapted from Stemmer (34) and Zhao and Arnold (38). To prepare genes to be shuffled, conventional PCR with Taq DNA polymerase (Promega) was used to amplify EchA from pBS(Kan)EH with primers EH Front (5′-AGCTATGACCATGATTACGCCAAGC-3′) and EH Rear (5′-CGTTGTAAAACGACGGCCAGTGA-3′), which were upstream and downstream of the natural KpnI and SacI restriction sites flanking the echA gene. The resulting 1.2-kb PCR product (∼2 μg) was mixed in the reaction buffer, containing 50 mM Tris-HCl (pH 7.4) and 10 mM MnCl2, to a final volume of 40 μl, equilibrated at 15°C for 5 min, digested with 0.15 U DNaseI (Promega) at 15°C for 6 min, and terminated by heating at 80°C for 10 min. The fragments (50 to 100 bp) were purified with Centri-Sep spin columns (Princeton Separations, Adelphia, NJ) and reassembled by PCR without primers using a 1:1 ratio of Taq and Pfu (Stratagene, La Jolla, CA) polymerases (2.5 U total in a 50-μl reaction volume). Three microliters of the reassembled products was amplified by a conventional PCR using Taq polymerase with primers EH Front and EH Rear. The shuffled fragment, containing the whole echA gene, was cloned into pBS(Kan)EH, replacing the part flanked by natural KpnI and SacI sites. This procedure mutated about 1 base pair over the 1-kb gene.
Saturation mutagenesis of EchA.
Overlap extension PCR (27) was used to perform saturation mutagenesis at positions L190, I219, and D235 of EchA in pBS(Kan)EH by replacing the target codon with NNN. Three pairs of degenerate primers, L190 Front (5′-ACTGGTCATACCGGGATGAGTTGNNNACTGAGGAAG-3′)-L190 Rear (5′-GAACCTCAAGTTCTTCCTCAGTNNNCAACTCATCC-3′), I219 Front (5′-CTTCAACTACTATCGTGCCAACNNNAGGCCCGATG-3′)-I219 Rear (5′-TGTCCACAGAGCGGCATCGGGCCTNNNGTTGGCAC-3′), and D235 Front (5′-GACAGACCTCGATCATACGATGAGCNNNCTTCCAG-3′)-D235 Rear (5′-CCCCATATCATTGTTACTGGAAGNNNGCTCATCG-3′) were designed to randomize codons L190, I219, and D235 in the nucleotide sequence, respectively (N = A/T/C/G in a 1:1:1:1 ratio), and primers EH Front and EH Rear (see above) were used for cloning. To minimize random point mutations, Pfu DNA polymerase was used in the PCR. Two degenerate PCR fragments were produced for each site, with 712 bp and 503 bp for site L190, 800 bp and 415 bp for site I219, and 848 bp and 370 bp for site D235. After purification from agarose gels, the two fragments for each site were combined at a 1:1 ratio as templates to obtain the full-length PCR product with the EH Front and EH Rear primers. The resulting randomized PCR product (1,167 bp), encoding all possible amino acids at position L190, I219, or D235, was cloned into pBS(Kan)EH after double digestion with KpnI and SacI, replacing the corresponding fragment in the original plasmid.
Screening of EchA mutagenesis libraries for enhanced styrene oxide activity and enantioselectivity.
Evolved EchA activity toward racemic styrene oxide was determined using the chromogenic reaction of styrene oxide with 4-nitrobenzylpyridine (21) in 96-well polypropylene plates. Whole TG1 cells harboring plasmid pBS(Kan)EH variants were grown in 150 μl LB-plus-Kan medium in 96-well plates at 37°C till exponential phase and were brought into contact with 5 mM racemic styrene oxide at 37°C for 30 min. Following the addition of 75 μl of 4-nitrobenzylpyridine (100 mM in 80 vol% ethylene glycol and 20 vol% acetone) to each well and incubation at 50°C for 50 min, 75 μl of triethylamine (1:1 in acetone) was added. The reaction of racemic styrene oxide with 4-nitrobenzylpyridine produced a blue color, which was proportional to the remaining styrene oxide and was measured at 620 nm with a Multiskan reader (Fisher Scientific). The wavelength of 620 nm rather than 600 nm was used in the plate reader due to reliance on a filter wheel; 600 nm was used for larger-scale reactions to characterize styrene oxide hydrolysis. Since the absorbance change was linear with the substrate concentration for both wavelengths and comparisons were made under identical conditions, variations due to slight changes in the wavelength may be excluded.
Enhanced styrene oxide enantioselectivity was screened by contacting the same exponentially growing cells harboring EH variants, which were split into two separate 96-well plates containing either 5 mM pure (R)-styrene oxide or 5 mM pure (S)-styrene oxide. After incubation with (R)-styrene oxide for 40 min and (S)-styrene oxide for 20 min at 37°C, the above spectrophotometric assay was used to determine the hydrolysis of both enantiomers of styrene oxide.
EchA variants identified by the 96-well plate screening as having enhanced racemic styrene oxide hydrolysis activity or improved enantioselectivity were further verified by a whole-cell, larger-volume assay. Exponentially growing cells (2.0 ml) sealed in 15-ml glass serum vials were incubated with 5 mM racemic, (R)-, or (S)-styrene oxide at 37°C and 250 rpm for 30 min, 40 min, or 20 min, respectively, followed by the sequential addition of 100 mM 4-nitrobenzylpyridine (1.0 ml) and triethylamine (1.0 ml). The mixture was then measured spectrophotometrically at 600 nm to determine the hydrolysis of racemic, (R)-, or (S)-styrene oxide. TG1/pBS(Kan) cells were used as the negative control.
Separation and identification of products from kinetic resolution of styrene oxide by wild-type and evolved EchA.
Racemic styrene oxide, (R)-styrene oxide, or (S)-styrene oxide at a final concentration of 5 mM was added to 4.0 ml of sonicated cells sealed in 15-ml glass serum vials and brought into contact with cells by shaking at 37°C. In the case of purified enzymes, 80 μg wild-type EchA or 60 μg F108/I219L/C248I was contacted with 5 mM racemic styrene oxide in 4 ml TEM buffer sealed in a 15-ml glass vial. The reaction was monitored by periodically (at a 5- or 10-min interval within a 0- to 50-min period) withdrawing 500-μl samples from the reaction mixture, which were immediately quenched, extracted with 800 μl diethyl ether, and analyzed by chiral high-performance liquid chromatography (HPLC). The HPLC system (Waters Corp., Milford, MA) includes 515 HPLC pumps, a 996 photodiode array detector, and Millenium32 Chromatography Manager software. The enantiomers of 1-phenylethane-1,2-diol formed from hydrolyzing styrene oxide were resolved with a Chiralcel OB (4.6 by 250 mm) column from Chiral Technologies, Inc. (Exton, PA) using hexane-2-propanol (76:24, vol/vol) as the eluent with a flow rate of 0.5 ml/min. Under such conditions, the (R)- and (S)-1-phenylethane-1,2-diol eluted with retention times of 10.2 min and 11.2 min, respectively; and styrene oxide had a retention time of 14.5 min with (R) and (S) enantiomers partially separated. UV-visible spectra were acquired online using a diode array detector (scanning from 200 to 600 nm) to characterize and quantify the products and remaining substrate. Products were identified by comparing the retention times and UV-visible spectra with authentic standards and were confirmed by coelution with the standards. Sonicated TG1/pBS(Kan) cells prepared the same way were used as the negative control and to make calibration curves under the same HPLC conditions to determine the concentration of each compound.
The values for the enantiomeric ratio (E values) were calculated using the equation E = ln{1 − c · [1 + ee(P)]}/ln{1 − c · [1 − ee(P)]} (5), where ee(P) is the enantiomeric excess of the product fraction (R)-1-phenylethane-1,2-diol and c is the extent of conversion of racemic styrene oxide. The E value of each variant was calculated when about 50% of the racemic styrene oxide was converted (c = 0.5).
EchA activity toward epoxypropane and 1,2-epoxyhexane.
The specific activities of wild-type EchA and variants toward epoxypropane (5 mM) and 1,2-epoxyhexane (5 mM) were assayed at 37°C with 2.5 ml of sonicated cells in a 15-ml serum vial sealed with a Teflon-coated septum and aluminum crimp (cells were diluted with TEM buffer two or five times as necessary). The specific activities of wild-type EchA and variant F108L/I219L/C248I toward 1,2-expoxyhexane were also determined with purified enzymes by using 150 μg enzyme for each isoform contacting 5 mM 1,2-expoxyhexane in 2.5 ml Tris-HCl buffer (50 mM, pH 7.4) in a sealed 15-ml glass vial. The hydrolysis rate of each substrate was determined by headspace concentrations and was monitored using gas chromatography (GC). The GC (Agilent 6890N series) was equipped with a 0.10% AT-1000 packed column (Alltech; length, 1.829 m; inner diameter, 3.175 mm; film thickness, 2.159 mm) and a flame ionization detector (FID). The FID was supplied with hydrogen (30 ml/min) and air (300 ml/min), and nitrogen was used as the carrier gas. Headspace samples were injected into the GC periodically and analyzed isothermally. For epoxypropane, the carrier gas was supplied at 10 ml/min and the temperature was kept at 100°C; for 1,2-epoxyhexane, the N2 carrier flow rate was maintained at 20 ml/min and samples were analyzed at 210°C. Under these conditions, the retention times for epoxypropane and 1,2-epoxyhexane were 1.48 min and 1.75 min, respectively.
Homology structural modeling and substrate docking.
Substrate docking simulations were performed using the Lamarckian genetic algorithm (16) of AutoDock 3.0 (9). Multiple docking runs (50) were performed for each substrate and each variant; the conformations with the overall lowest energies were considered for further analysis. The 3-dimensional coordinates of the EchA variants based on an EchA structure model (17) were generated with the SWISS-MODEL server (10, 19, 30) as described previously (25). Polar hydrogen atoms were added to the protein, and charges were assigned to each atom by using AutoDockTools. The structure of the substrate molecule (R)-styrene oxide was obtained from the Protein Data Bank with the accession code 1PWZ, while the 3-dimensional coordinates of (S)-styrene oxide were generated by reversing the stereo-bond with Chem3D (CambridgeSoft Inc., Cambridge, MA) and minimizing the energy by MOPAC using the PM3 method (CAChe Group, Beaverton, OR). Prior to automatic docking using AutoDock, the ligand coordinates were manually modified so that they were near the active-site center of EchA. The conformations of the substrates in the context of macromolecules were analyzed via Swiss-PdbViewer (10, 19, 30).
RESULTS
DNA shuffling, screening, sequence analysis, and expression level.
The vector constructed previously, pBS(Kan)EH (25), enabled stable and constitutive expression of EchA, generation of large libraries via electroporation, and reliable screening, because the segregational instability and feeder colony artifacts associated with ampicillin resistance vectors were avoided. A random library of 10,000 colonies expressing EchA mutants was created, and more than 4,000 colonies were screened using whole cells in 96-well plates for enhanced racemic styrene oxide hydrolysis. All potentially positive variants were initially tested in triplicate in 96-well plates and were further examined in 15-ml vials for racemic styrene oxide hydrolysis activity. Two variants, L190F and D235H, exhibited activities toward 5 mM racemic styrene oxide that were about fourfold improved over that of the wild-type enzyme. Hence, in both cases, single amino acid substitutions were responsible for the improved activity.
The initial screening was performed with whole cells in the absence of IPTG induction. Comparison of the total cellular protein expression profiles via SDS-PAGE revealed fourfold-elevated expression of EchA by variant D235H compared to that of the wild-type enzyme. Because DNA sequencing showed that the lac promoter and the ribosome-binding regions of EchA variant D235H are unaltered, the higher protein concentration of variant D235H was possibly due to altered lability of the protein or mRNA due to modifications in the DNA sequence. Further, with 1 mM IPTG induction, D235H expression was equivalent to that of wild-type EchA and the specific activity of D235H toward racemic styrene oxide was almost the same as that of the wild-type enzyme. Therefore, the elevated expression level of D235H in the absence of IPTG was responsible for the fourfold increase in activity toward racemic styrene oxide. For L190F, the enhanced activity was not due to a change in expression level, and there was no alteration in either its lac promoter region or its ribosome-binding region based on DNA sequencing.
Saturation mutagenesis and screening.
Random mutagenesis identified two sites in the amino acid sequence, L190 and D235, as contributing to enhanced enzymatic activity, and saturation mutagenesis was conducted individually at these two hot spots to seek further improvements in EH activity for racemic styrene oxide hydrolysis. By cloning DNA fragments from saturation mutagenesis back into the corresponding position of pBS(Kan)EH, all possible amino acids were introduced at the two sites, respectively. About 300 of those colonies were screened in 96-well plates for racemic styrene oxide hydrolysis, since 292 independent clones from saturation mutagenesis at one site need to be screened for a 99% probability that each possible codon has been tested (26).
One new variant from L190 saturation mutagenesis, L190Y, was identified from the 96-well plate assay and further confirmed with 15-ml vials for enhanced racemic styrene oxide hydrolysis using the spectrophotometric assay. Compared to 20% hydrolysis of 5 mM racemic styrene oxide by wild-type EchA over a 30-min period, L190Y hydrolyzed 50% of racemic styrene oxide under the same conditions. But L190Y was not superior to L190F in terms of racemic styrene oxide hydrolysis activity. There was no further beneficial amino acid substitution identified from saturation mutagenesis at site D235, probably because the only beneficial mutation, D235H, simply elevated the expression level, and because it is located at the surface of the enzyme (far from the active site).
Enhanced enantioselectivity toward styrene oxide by engineered EchA.
The cis-DCE epoxide EchA variants, F108L, I219F, C248I, F108L/I219L, F108L/C248I, and F108L/I219L/C248I (25), the two variants identified from DNA shuffling, D235H and L190F, and the additional variant from saturation mutagenesis, L190Y, were tested for possible enhanced enantioselective production of (R)-1-phenylethane-1,2-diol from racemic styrene oxide. Variant D235H showed specific activity and enantioselectivity toward styrene oxide equivalent to those of the wild-type enzyme when expressed equally and was not characterized fully. All the experiments were conducted in sealed containers, since the substrate is volatile. Because linear calibration curves of the substrate [racemic, (R)-, and (S)-styrene oxides] were obtained for both the spectrophotometric assay and HPLC analysis (data not shown), evaporation of the substrate during sampling due to volatility should not be a concern.
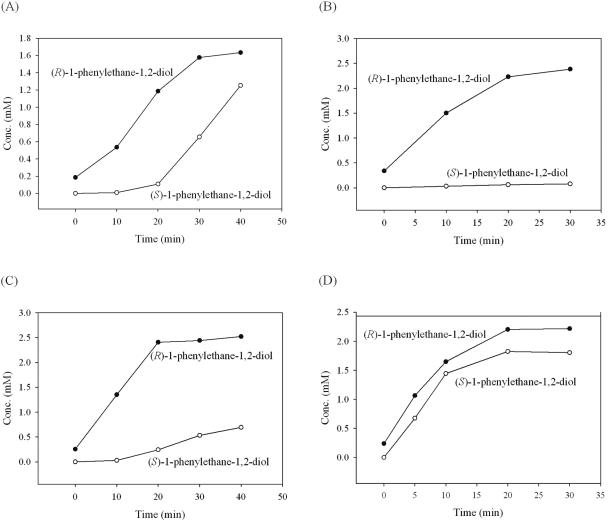
The enantioselective hydrolysis of racemic styrene oxide catalyzed by the wild-type and mutant enzymes is summarized in Table 1, and the reactions of the EchA variants with pure (R)- and (S)-styrene oxide are summarized in Table 2. The rates of Table 2 basically corroborate the data of the racemic styrene oxide reactions (Table 1), except that the absolute values of (S)-diol formation rates are sometimes different due to competitive substrate inhibition in the racemic styrene oxide reaction system. Representative reaction progress curves of the formation of (R)- and (S)-1-phenylethane-1,2-diol from racemic styrene oxide catalyzed by wild-type EchA and variants I219F, F108L/I219L/C248I, and L190F are shown in Fig. 2. No nonenzymatic hydrolysis of the substrate was observed, as confirmed by the negative control (sonicated cells of TG1/pBSKan).
TABLE 1.
Determination of the E value for the enantioselective production of (R)-1-phenylethane-1,2-diol from racemic styrene oxide catalyzed by wild-type EchA and variants
| Enzymea | Formation of:
|
Hydrolysis of racemic styrene oxideb
|
Enantioselectivityd
|
|||||
|---|---|---|---|---|---|---|---|---|
| (R)-1-phenylethane-1,2-diolb
|
(S)-1-phenylethane-1,2-diolc
|
|||||||
| Rate (μmol/min/mg) | Fold increase relative to WT | Rate (μmol/min/mg) | Fold increase relative to WT | Rate (μmol/min/mg) | Fold increase relative to WT | E value | Fold increase relative to WT | |
| Wild-type EchA | 1.04 ± 0.07 | 1.0 | 1.00 ± 0.18 | 1.0 | 1.77 ± 0.18 | 1.0 | 17 ± 0.5 | 1.0 |
| F108L | 2.23 ± 0.01 | 2.1 | 1.41 ± 0.05 | 1.4 | 2.80 ± 0.03 | 1.6 | 9 ± 1.0 | 0.5 |
| 1219F | 1.96 ± 0.09 | 1.9 | 0.06 ± 0.003 | 0.06 | 2.26 ± 0.28 | 1.3 | 91 ± 12 | 5.3 |
| C2481 | 1.60 ± 0.13 | 1.5 | 0.61 ± 0.14 | 0.6 | 2.02 ± 0.04 | 1.1 | 10 ± 0 | 0.6 |
| F108L/C2481 | 1.85 ± 0.08 | 1.8 | 0.29 ± 0.03 | 0.3 | 2.57 ± 0.23 | 1.4 | 46 ± 11 | 2.7 |
| 1219L/C2481 | 1.35 ± 0.07 | 1.3 | 0.40 ± 0.26 | 0.4 | 1.87 ± 0.13 | 1.0 | 35 ± 11 | 2.0 |
| F108L/I219L/C248I | 2.16 ± 0.01 | 2.1 | 0.43 ± 0.04 | 0.4 | 3.01 ± 0.36 | 1.7 | 31 ± 0.1 | 1.8 |
| L190F | 2.60 ± 0.32 | 2.5 | 2.46 ± 0.60 | 2.5 | 8.55 ± 0.26 | 4.8 | 2 ± 0.5 | 0.1 |
| L190Y | 1.94 ± 0.14 | 1.9 | 1.61 ± 0.70 | 1.6 | 4.82 ± 0.82 | 2.7 | 14 ± 0.6 | 0.8 |
Crude enzymes were prepared by cell sonication, and the EchA concentration from each strain was 50 μg/ml as estimated from SDS-PAGE.
The formation rate of (R)-1-phenylethane-1,2-diol and the hydrolysis rate of racemic styrene oxide were determined via chiral HPLC over a linear range beginning at time zero. WT, wild type.
The formation rate of (S)-1-phenylethane-1,2-diol was determined over a linear range via chiral HPLC from the time when competitive substrate inhibition ceased (different for each strain).
The E values for the formation of (R)-1-phenylethane-1,2-diol were calculated using the equation E = ln{1 − c · [1 + ee(P)]}/ln{1 − c · [1 − ee(P)]} (5) at c = 0.5.
TABLE 2.
Specific activities of wild-type EchA and variants toward pure (R)-styrene oxide and (S)-styrene oxide
| Enzymea | Formationb of:
|
(R)-diol rate vs (S)-diol rate
|
||||
|---|---|---|---|---|---|---|
| (R)-1-phenylethane-1,2-diol
|
(S)-1-phenylethane-1,2-diol
|
|||||
| Rate (μmol/min/mg) | Fold difference from WT rate | Rate (μmol/min/mg) | Fold difference from WT rate | Ratio | Fold difference from WT rate | |
| Wild-type EchA | 1.00 ± 0.12 | 1.0 | 3.21 ± 0.30 | 1.0 | 0.31 | 1.0 |
| F108L | 1.89 | 1.9 | 4.51 ± 0.43 | 1.4 | 0.42 | 1.3 |
| I219Fc | 1.43 ± 0.02 | 1.4 | 0.93 ± 0.09 (0-10 min) | 0.29 | 1.54 | 4.9 |
| 0.27 ± 0.03 (10-30 min) | 0.08 | 5.3 | 1.7 | |||
| C248I | 1.58 ± 0.36 | 1.6 | 1.99 ± 0.10 | 0.55 | 0.79 | 2.5 |
| F108L/C248I | 1.04 ± 0.06 | 1.0 | 1.27 ± 0.10 | 0.40 | 0.82 | 2.6 |
| I219L/C248I | 0.93 ± 0.13 | 0.9 | 1.18 ± 0.11 | 0.37 | 0.78 | 2.5 |
| F108L/I219L/C2481 | 2.94 ± 0.27 | 2.9 | 1.78 ± 0.23 | 0.55 | 1.65 | 5.3 |
| L190F | 2.76 ± 0.71 | 2.8 | 8.76 ± 0.62 | 2.7 | 0.32 | 1.0 |
| L190Y | 2.18 ± 0.17 | 2.2 | 7.39 ± 0.22 | 2.3 | 0.29 | 0.95 |
Crude enzymes were prepared by cell sonication; the EchA concentration from each strain was 50 μg/ml as estimated from SDS-PAGE.
The formation rates of (R)- and (S)-1-phenylethane-1,2-diol were determined via chiral HPLC and were from a linear range.
I219F exhibited distinctive two-stage (S)-1-phenylethane-1,2-diol formation, and the rates for these two stages (0 to 10 min and 10 to 30 min) were determined separately.
FIG. 2.
Representative reactions for the formation of (R)- and (S)-1-phenylethane-1,2-diol from racemic styrene oxide catalyzed by sonicated cells expressing (A) wild-type EchA, (B) variant I219F, (C) variant F108L/I219L/C248I, and (D) variant L190F. Because of the time elapsed between substrate addition and reaction quenching, some (R)-diol was already formed at 0 min.
Wild-type EchA (in crude extracts) hydrolyzed racemic styrene oxide at 1.77 μmol/min/mg protein, which is comparable to the specific activity measured for purified EchA from A. radiobacter AD1 (2.56 μmol/min/mg protein) (11). The E value we measured for wild-type EchA was 17, in favor of the production of 1,2-diol in the (R) configuration, and it agrees well with the reported E value of 16 (32). After the start of the reaction, the (R) enantiomer of styrene oxide was hydrolyzed to (R)-1-phenylethane-1,2-diol, with the retention of the stereochemical configuration, indicating that the attack was preferentially at the least-hindered carbon atom (1). The (R) enantiomer was almost completely hydrolyzed before the (S) enantiomer reacted, but the (S) enantiomer was subsequently converted at a higher rate (Fig. 2A). Such kinetic behavior could be due to the higher affinity of the (R) enantiomer for the active site and its subsequent inhibition of binding by the (S) enantiomer. The threefold-higher activity of the (S) enantiomer was confirmed by investigating the conversion of both enantiomers separately (Table 2).
EchA variants I219F, F108L/C248I, I219L/C248I, and F108L/I219L/C248I showed significant increases in enantioselectivity (Table 1). That of I219F was most distinct, with the E value increasing from 17 for the wild-type enzyme to 91, a >5-fold enhancement. During the first 50% conversion of racemic styrene oxide, the enantiomeric excess (ee) (the same as optical purity; represents how much more of one enantiomer than of the other is present) of (R)-1-phenylethane-1,2-diol was 75% with the wild-type enzyme and 94% with the I219F variant. In addition, I219F also appeared to be quite active, producing (R)-1,2-diol from racemic styrene oxide at a rate 1.9-fold higher than that of the wild-type enzyme (Table 1). The significant improvement in enantiomeric purity and the enhanced catalytic rate make I219F an EchA variant of considerable interest for preparing enantiomerically pure diols. The other three variants, F108L/C248I, I219L/C248I, and F108L/I219L/C248I, showed modest improvements of about twofold in E values (E values, 30 to 45 [Table 1]). Interestingly, all three mutants have higher activity for (R)-1-phenylethane-1,2-diol production (Table 1), especially F108L/I219L/C248I (2.1-fold). The enhanced enantioselectivity of all the above variants could be explained by the fact that the rate of hydrolysis of (S)-styrene oxide decreased significantly, in addition to the improved catalytic activity toward (R)-styrene oxide compared to that of the wild-type enzyme.
Enantioselectivity was most pronounced with the I219F variant: it hydrolyzed the second enantiomer, (S)-styrene oxide, more than 30 times more slowly than the (R) enantiomer during the sequential hydrolysis of racemic styrene oxide (Table 1). One peculiar characteristic of the I219F variant was observed for the conversion of pure (S)-styrene oxide: (S)-1-phenylethane-1,2-diol was consistently formed in two stages, and the first reaction stage was 3.6 times faster than the second stage (Table 2). This suggested that the product (S)-diol inhibits conversion of (S)-styrene oxide and may further explain the remarkably improved enantioselectivity of this variant (with racemic mixtures). To further test the possible inhibition by the (S)-diol, it was added at concentrations of 0.5 mM and 2 mM to the reaction system along with 5 mM pure (S)-styrene oxide. The reaction rates under these two conditions were very similar to the second-stage rate found in the absence of additional (S)-diol; hence, the (S)-diol saturated the active site at concentrations less than 0.5 mM. The I219F substitution was found to be the most significant mutation for improving enantioselectivity based on screening of the I219 saturation mutagenesis library.
F108L, C248I, and L190Y have E values similar to or lower than that of wild-type EchA, but all have improved (R)-1-phenylethane-1,2-diol formation rates. In addition, L190Y hydrolyzed racemic styrene oxide at a rate of 4.8 ± 0.8 μmol/min/mg, 2.7-fold faster than the wild-type enzyme (Table 1), which was consistent with the result of the previous spectrophotometric assay. C248I had a lower rate of formation of (S)-1-phenylethane-1,2-diol and thus was expected to have a higher E value than the wild-type enzyme, but the (S) enantiomer reaction was not inhibited as much as with the wild-type enzyme (data not shown), resulting in the decreased E value.
Variant L190F had an exceptionally low E value (E ≈ 2), so this enzyme is not useful for resolution of racemic mixtures; however, its enhanced rate and much-reduced enantioselectivity may be of interest in exploring the mechanism of this mutant. L190F seemed to be the only variant for which the binding of (S)-styrene oxide was not competitively inhibited by the binding of (R)-styrene oxide, and the conversion of the two enantiomers proceeded almost simultaneously (which explains its low E value [Fig. 2D]). In addition, this variant hydrolyzed racemic styrene oxide 4.8 times faster than the wild-type enzyme (confirming the previous spectrophotometric assay result) and enhanced the formation rates of both enantiomers of the products 2.5- to 3.3-fold (Table 1).
These results were corroborated using purified wild-type EchA enzyme and variant F108L/I219L/C248I, as similar relative differences were measured for racemic styrene oxide hydrolysis. With purified enzymes, (R)-1-phenylethane-1,2-diol formation rates of 4.0 and 7.1 μmol/min/mg, (S)-1-phenylethane-1,2-diol formation rates of 4.5 and 2.8 μmol/min/mg, and racemic styrene oxide hydrolysis rates of 5.2 and 9.1 μmol/min/mg were measured for the wild-type enzyme and F108L/I219L/C248I, respectively. In addition, the enantioselectivity for (R)-1-phenylethane-1,2-diol production with the two purified enzymes (E values were 10 for the wild-type enzyme and 24 for variant F108L/I219L/C248I) agreed with the results reported above using crude enzymes prepared by cell sonication (E values were 17 and 31 for sonicated cells expressing the wild-type and F108L/I219L/C248I enzymes, respectively). Hence, the crude enzyme results were robust for determining changes in enzymatic activity and enantioselectivity.
Improved catalytic activity toward aliphatic epoxides by the evolved EchA.
To further characterize the enzymatic activity of the variants, two additional aliphatic epoxides, epoxypropane and 1,2-epoxyhexane, were used as substrates to test the catalytic efficiency of the evolved EchA enzymes. The specific activities were determined using GC by periodically monitoring substrate depletion of the headspace samples (5 mM initial concentration of each substrate). Variant F108L/I219L/C248IT had a 10-fold increase in activity for epoxypropane over that of the wild-type enzyme (24 ± 2 versus 2.4 ± 0.3 μmol/min/mg protein). Variants F108L/C248I, I219L/C248I, F108L, I219F, C248I, and L190F resulted in 3.9-, 3.8-, 3.4-, 2.2-, 2.2-, and 1.9-fold enhancements. In addition, the beneficial mutations seemed cumulative, as a similar trend in the enhancement of activity toward cis-DCE epoxide was seen (25).
For the hydrolysis of 1,2-epoxyhexane using crude enzymes (sonicated cells), variant L190Y showed 2.5-fold enhancement in enzymatic activity relative to that of wild-type EchA (6.3 ± 0.2 versus 2.6 ± 0 μmol/min/mg protein), while the F108L, C248I, F108L/I219L/C248I, and L190F variants showed approximately 2-fold improvement in 1,2-epoxyhexane hydrolysis. In addition, the results for the wild-type EchA enzyme and variant F108L/I219L/C248I were corroborated using purified enzymes; there was a 2.3-fold enhancement in the hydrolysis of 1,2-epoxyhexane by F108L/I219L/C248I relative to that by the wild-type enzyme (16 ± 1 versus 7 ± 1 μmol/min/mg protein). As with styrene oxide (above), this indicates that the crude extracts were robust for characterizing the relative activities of the EchA variants.
No enantioselectivity toward epoxypropane and 1,2-epoxyhexane was characterized, as these two substrates were not the focus of the present study. These substrates were investigated to show that the enzymes produced here are robust for more than just styrene oxide.
Homology structural modeling and substrate docking.
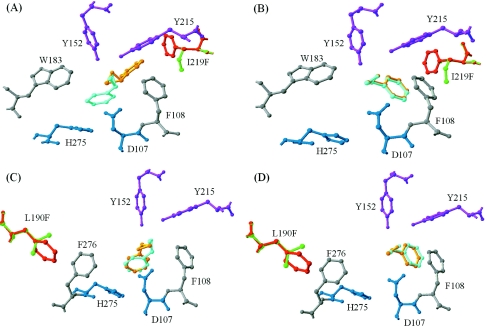
The 3-dimensional coordinates of the two EchA variants with the most significant changes in styrene oxide hydrolysis, I219F and L190F, were constructed, and the possible effects of the mutation on the interaction with substrates were further explored by computer docking. A plausible picture of the binding model of both enantiomers of styrene oxide into the active site of both the wild-type EchA enzyme and variants is shown in Fig. 3. The substrate styrene oxide is positioned between the nucleophile D107 and the proton donor residues Y152 and Y215 in a way that should facilitate nucleophilic attack by D107 and hydrogen bond formation between the substrate and the tyrosine pair.
FIG. 3.
Part of active-site cavity of wild-type EchA, variant I219F, and variant L190F with (R)- or (S)-styrene oxide docked. Catalytic residues D107 and H275 are dark blue, the proton donor residues Y152 and Y215 are purple, and the other active-site residues, F108 and W183, are grey. The amino acid substitutions F219 and F190 (red) are superimposed on the wild-type enzyme residues I219 and L190 (green). The conformation of (R)- or (S)-styrene oxide docked in wild-type EchA is shown in light blue, while the docked conformation in the variants is shown in orange. (A) Wild-type EchA and variant I219F docked with (R)-styrene oxide; (B) wild-type EchA and variant I219F docked with (S)-styrene oxide; (C) wild-type EchA and variant L190F docked with (R)-styrene oxide; (D) wild-type EchA and variant L190F docked with (S)-styrene oxide.
In the I219F variant, F219 has a T-shaped interaction (15) with the Y215 side chain (Fig. 3A), which might help stabilize the phenolate anion intermediate formed as Y215 donates a proton to facilitate substrate ring opening during the nucleophilic attack initiated by D107. The substrate (R)-styrene oxide is docked with a very different orientation from that in the wild-type enzyme, so that its aromatic ring can form both T-shaped interactions with F219 and F108 and parallel displaced π-stacking (15) with the side chain of Y215 (Fig. 3A and B). The combination of favorable aromatic interactions between the three amino acids that gives rise to the significantly altered position of the substrate (R)-styrene oxide may explain the accelerated hydrolysis of (R)-styrene oxide versus (S)-styrene oxide by the I219F variant. In contrast, despite the substantial decrease in the (S)-styrene oxide hydrolysis rate by variant I219F compared to the wild-type enzyme, docking calculations of the enzyme isoforms with (S)-styrene oxide showed very similar binding modes (Fig. 3B).
The docking conformation of (R)-styrene oxide in the L190F variant relative to that in the wild-type enzyme (Fig. 3C) indicates a slight rotation of the epoxide side chain that may be responsible for the threefold increase in the rate of reaction for this variant. There are no significantly different binding modes between the wild-type enzyme and the variant for the (S)-styrene oxide enantiomer. Considering that F190 is relatively far away from substrate binding cleft and seems not to be in the appropriate location for a catalytic role, its long-distance effect on catalysis could be due to its interaction with the aromatic side chain of H275 relayed by F276 (Fig. 3C and D). Such a series of interactions might affect the chemical properties of H275 in a manner that is important in catalysis and might in turn affect the hydrolysis reaction and enhance the conversion rates of both enantiomers, possibly without discrimination.
DISCUSSION
It is shown clearly in this paper that rational evolution via active-site engineering may be used to develop epoxide hydrolases with enhanced enantioselectivity. This may create more applications for asymmetric catalysis to produce enantiomerically pure epoxides and vicinal diols. In addition, the reaction rates of the epoxide hydrolase from A. radiobacter AD1 toward various epoxides can be improved by both rational and directed evolution.
The most pronounced enhancement in enantioselectivity toward racemic styrene oxide was achieved with the I219F substitution. Because of their close proximity (within 5 Å), introducing the phenylalanine residue at this position allows extensive van der Waals aromatic-aromatic interactions between F219, F108, Y215, and the substrate (R)-styrene oxide that shifts the position of the substrate (R)-styrene oxide (Fig. 3A); Y215 functions as the proton donor during catalysis (23), and substitution at this residue (Y215F or Y215H) results in enhanced enantioselectivity toward styrene oxide and its derivatives (31) as well as pNPGE (35). This may explain the enhanced catalytic activity toward (R)-styrene oxide versus (S)-styrene oxide. However, the 17-fold-impaired activity of variant I219F toward the (S) enantiomer (Table 2) cannot be inferred from the docking of the substrate. Since (S)-1-phenylethane-1,2-diol, formed as the hydrolysis product or added prior to substrate transformation, substantially decreased activity toward (S)-styrene oxide, the diminished activity must be the result of a change in the active site that leads to tighter binding of the product (S)-1-phenylethane-1,2-diol, which subsequently inhibits further binding of the substrate (S)-styrene oxide.
The individual mutations at F108 and C248 did not bring appreciable improvement in enantioselectivity for the hydrolysis of racemic styrene oxide, whereas the combination of the two mutations (F108L and C248I) together with the amino acid substitution I219L resulted in an enhanced E value, all accompanied by an impaired (S)-styrene oxide transformation rate and improved (R)-styrene oxide conversion. It was postulated that the enantioselectivity of EchA for racemic styrene oxide (E value) was determined primarily by the difference in the alkylation rates between the two enantiomers (22), while the alkylation rate was dictated largely by stabilization of the transition state of the alkyl-enzyme intermediate and/or the Michaelis complex formed by substrate binding (24). Thus, the concerted effects from the substitutions of the two or three residues may change the size, shape, and aromatic character of the active site to facilitate differentiated binding of the two enantiomers of styrene oxide and/or stabilization of the transitional-state intermediates.
(R)-1-Phenylethane-1,2-diol is synthesized chemically either via selective reduction of phenylglyoxalates derived from bile acids, followed by reductive cleavage (3), or by using a commercially available enzyme (Candida antarctica lipase B) as the catalyst (36). However, for both methods, multiple reaction steps are involved and organic solvents are used. With an enantiomeric excess of about 95%, the I219F variant may hold commercial promise for synthesis of (R)-1-phenylethane-1,2-diol via kinetic resolution of cheap racemic styrene oxide in one easy step. Further, the downstream separation and recovery of (R)-1-phenylethane-1,2-diol should be facilitated by the physical-property differences between the diol and the remaining unreacted styrene oxide, mainly in the (S) form.
Epoxide hydrolases from microbial sources have been increasingly recognized as highly versatile biocatalysts for the preparation of enantiopure epoxides and vicinal diols (6, 33, 37). Despite the excellent enantioselectivity found with certain substrates, the enantioselectivity of microbial epoxide hydrolases is in general often low (6). Furthermore, in most reported cases, bacterial epoxide hydrolases are useful in the resolution of disubstituted epoxides, and specific activities are often not high (37). Our effort in transforming one moderately enantioselective bacterial epoxide hydrolase to one with excellent enantioselectivity in the hydrolysis of styrene oxide and with enhanced activity toward various epoxides increases the options for producing enantiomerically pure epoxides and vicinal diols. Such efforts can easily be extended to other substrates and new enzymes, and they emphasize that protein engineering is a valuable approach for improving the versatility of epoxide hydrolases.
Acknowledgments
This research was supported by the National Science Foundation (BES-9911469 and BES-0331416).
We are grateful for the gifts of plasmid pEH20 from Dick Janssen and the EchA structure model from Bauke Dijkstra (both of the University of Groningen).
REFERENCES
- 1.Archelas, A., and R. Furstoss. 1998. Epoxide hydrolases: new tools for the synthesis of fine organic chemicals. Trends Biotechnol. 16:108-116. [DOI] [PubMed] [Google Scholar]
- 2.Archer, I. V. J. 1997. Epoxide hydrolase as asymmetric catalysts. Tetrahedron 53:15617-15662. [Google Scholar]
- 3.Bandyopadhyaya, A. K., N. M. Sangeetha, A. Radha, and U. Maitra. 2000. Synthesis of both enantiomers of 1-phenylethane-1,2-diol via chirality transfer from bile acid derivatives. Tetrahedron Asymmetry 11:3463-3466. [Google Scholar]
- 4.Cedrone, F., S. Niel, S. Roca, T. Bhatnagar, N. Ait-Abdelkader, C. Torre, H. Krumm, A. Maichele, M. T. Reetz, and J. C. Baratti. 2003. Directed evolution of the epoxide hydrolase from Aspergillus niger. Biocatalysis Biotransformation 21:357-364. [Google Scholar]
- 5.Chen, C.-S., Y. Fujimoto, G. Girdaukas, and C. J. Sih. 1982. Quantitative analyses of biochemical kinetic resolutions of enantiomers. J. Am. Chem. Soc. 104:7294-7299. [Google Scholar]
- 6.Faber, K., M. Mischitz, and W. Kroutil. 1996. Microbial epoxide hydrolases. Acta Chem. Scand. 50:249-258. [Google Scholar]
- 7.Genzel, Y., A. Archelas, J. H. L. Spelberg, D. B. Janssen, and R. Furstoss. 2001. Microbiological transformations. Part 48. Enantioselective biohydrolysis of 2-, 3-, and 4-pyridyloxirane at high substrate concentration using the Agrobacterium radiobacter AD1 epoxide hydrolase and its Tyr215Phe mutant. Tetrahedron 57:2775-2779. [Google Scholar]
- 8.Gibson, T. J. 1984. Studies on the Epstein-Barr virus genome. Ph.D. thesis. Cambridge University, Cambridge, England.
- 9.Goodsell, D. S., and A. J. Olson. 1990. Automated docking of substrates to proteins by simulated annealing. Proteins 8:195-202. [DOI] [PubMed] [Google Scholar]
- 10.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs, M. H., A. J. Van den Wijngaard, M. Pentenga, and D. B. Janssen. 1991. Characterization of the epoxide hydrolase from an epichlorohydrin-degrading Pseudomonas sp. Eur. J. Biochem. 202:1217-1222. [DOI] [PubMed] [Google Scholar]
- 12.Jaeger, K. E., T. Eggert, A. Eipper, and M. T. Reetz. 2001. Directed evolution and the creation of enantioselective biocatalysts. Appl. Microbiol. Biotechnol. 55:519-530. [DOI] [PubMed] [Google Scholar]
- 13.King, R. B., J. Bakos, C. D. Hoff, and L. Marko. 1979. 1,2-Bis(diphenylphosphino)-1-phenylethane: a chiral ditertiary phosphine derived from mandelic acid used as a ligand in asymmetric homogenous hydrogenation catalysts. J. Org. Chem. 44:1729-1731. [Google Scholar]
- 14.Marki, H. P., Y. Crameri, R. Eigenmann, A. Krasso, H. Ramuz, K. Bernauer, M. Goodman, and K. L. Melmon. 1988. Optically pure isoproterenol analogues with side chains containing an amide bond: synthesis and biological properties. Helv. Chim. Acta 71:320-336. [Google Scholar]
- 15.McGaughey, G. B., M. Gagne, and A. K. Rappe. 1998. π-Stacking interactions. Alive and well in proteins. J. Biol. Chem. 273:15458-15463. [DOI] [PubMed] [Google Scholar]
- 16.Morris, G. M., D. S. Goodsell, R. S. Halliday, R. Huey, M. E. Hart, R. K. Belew, and A. J. Olson. 1998. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19:1639-1662. [Google Scholar]
- 17.Nardini, M., I. S. Ridder, H. J. Rozeboom, K. H. Kalk, R. Rink, D. B. Janssen, and B. W. Dijkstra. 1999. The X-ray structure of epoxide hydrolase from Agrobacterium radiobacter AD1. An enzyme to detoxify harmful epoxides. J. Biol. Chem. 274:14579-14586. [PubMed] [Google Scholar]
- 18.Nardini, M., R. Rink, D. B. Janssen, and B. W. Dijkstra. 2001. Structure and mechanism of the epoxide hydrolase from Agrobacterium radiobacter AD1. J. Mol. Catal. B 11:1035-1042. [Google Scholar]
- 19.Peitsch, M. C. 1993. Protein modeling by E-mail. Biotechnology 13:658-660. [Google Scholar]
- 20.Reetz, M. T., C. Torre, A. Eipper, R. Lohmer, M. Hermes, B. Brunner, A. Maichele, M. Bocola, M. Arand, A. Cronin, Y. Genzel, A. Archelas, and R. Furstoss. 2004. Enhancing the enantioselectivity of an epoxide hydrolase by directed evolution. Org. Lett. 6:177-180. [DOI] [PubMed] [Google Scholar]
- 21.Rink, R., M. Fennema, M. Smids, U. Dehmel, and D. B. Janssen. 1997. Primary structure and catalytic mechanism of the epoxide hydrolase from Agrobacterium radiobacter AD1. J. Biol. Chem. 272:14650-14657. [DOI] [PubMed] [Google Scholar]
- 22.Rink, R., and D. B. Janssen. 1998. Kinetic mechanism of the enantioselective conversion of styrene oxide by epoxide hydrolase from Agrobacterium radiobacter AD1. Biochemistry 37:18119-18127. [DOI] [PubMed] [Google Scholar]
- 23.Rink, R., J. Kingma, J. H. Lutje Spelberg, and D. B. Janssen. 2000. Tyrosine residues serve as proton donor in the catalytic mechanism of epoxide hydrolase from Agrobacterium radiobacter. Biochemistry 39:5600-5613. [DOI] [PubMed] [Google Scholar]
- 24.Rink, R., J. H. L. Spelberg, R. J. Pieters, J. Kingma, M. Nardini, R. M. Kellogg, B. W. Dijkstra, and D. B. Janssen. 1999. Mutation of tyrosine residues involved in the alkylation half reaction of epoxide hydrolase from Agrobacterium radiobacter AD1 results in improved enantioselectivity. J. Am. Chem. Soc. 121:7417-7418. [Google Scholar]
- 25.Rui, L., L. Cao, W. Chen, K. F. Reardon, and T. K. Wood. 2004. Active site engineering of the epoxide hydrolase from Agrobacterium radiobacter AD1 to enhance aerobic mineralization of cis-1,2-dichloroethylene in cells expressing an evolved toluene ortho-monooxygenase. J. Biol. Chem. 279:46810-46817. [DOI] [PubMed] [Google Scholar]
- 26.Rui, L., Y. M. Kwon, A. Fishman, K. F. Reardon, and T. K. Wood. 2004. Saturation mutagenesis of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chloroform degradation. Appl. Environ. Microbiol. 70:3246-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakamoto, T., J. M. Joern, A. Arisawa, and F. H. Arnold. 2001. Laboratory evolution of toluene dioxygenase to accept 4-picoline as a substrate. Appl. Environ. Microbiol. 67:3882-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spelberg, J. H. L., R. Rink, A. Archelas, R. Furstoss, and D. B. Janssen. 2002. Biocatalytic potential of the epoxide hydrolase from Agrobacterium radiobacter AD1 and a mutant with enhanced enantioselectivity. Adv. Synth. Catal. 344:980-985. [Google Scholar]
- 32.Spelberg, J. H. L., R. Rink, R. M. Kellogg, and D. B. Janssen. 1998. Enantioselectivity of a recombinant epoxide hydrolase from Agrobacterium radiobacter. Tetrahedron Asymmetry 9:459-466. [Google Scholar]
- 33.Steinreiber, A., and K. Faber. 2001. Microbial epoxide hydroxylases for preparative biotransformations. Curr. Opin. Biotechnol. 12:552-558. [DOI] [PubMed] [Google Scholar]
- 34.Stemmer, W. P. C. 1994. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. USA 91:10747-10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Loo, B., J. H. L. Spelberg, J. Kingma, T. Sonke, M. G. Wubbolts, and D. B. Janssen. 2004. Directed evolution of epoxide hydrolase from A. radiobacter toward higher enantioselectivity by error-prone PCR and DNA shuffling. Chem. Biol. 11:981-990. [DOI] [PubMed] [Google Scholar]
- 36.Virsu, P., A. Liljeblad, A. Kanerva, and L. T. Kanerva. 2001. Preparation of the enantiomers of 1-phenylethane-1,2-diol. Regio- and enantioselectivity of acylase I and Candida antarctica lipases A and B. Tetrahedron Asymmetry 12:2447-2455. [Google Scholar]
- 37.Weijers, C. A. G. M., and J. A. M. de Bont. 1999. Epoxide hydrolases from yeasts and other sources: versatile tools in biocatalysis. J. Mol. Catal. B 6:199-214. [Google Scholar]
- 38.Zhao, H., and F. H. Arnold. 1997. Optimization of DNA shuffling for high fidelity recombination. Nucleic Acids Res. 25:1307-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]