Abstract
Cryptosporidium is a significant cause of water-borne enteric disease throughout the world and represents a challenge to the water industry and a threat to public health. In this study we report the use of a cell culture-TaqMan PCR assay to measure oocyst inactivation rates in reagent-grade and environmental waters over a range of temperatures. While oocysts incubated at 4°C and 15°C remained infective over the 12-week holding period, we observed a 4 log10 reduction in infectivity for both 20 and 25°C incubation treatments at 12 and 8 weeks, respectively, for all water types examined, a faster rate of inactivation for oocysts than previously reported. This temperature-dependent inactivation was further investigated using a simple and rapid ATP assay described herein. Time course experiments performed in reagent-grade water at incubation temperatures of 4, 15, 20, 25, 30, and 37°C identified a close relationship between oocyst infectivity and oocyst ATP content, demonstrating that temperature inactivation at higher temperatures is a function of increased oocyst metabolic activity. While water quality did not affect oocyst inactivation, biological antagonism appears to be a key factor affecting oocyst removal from environmental waters. Both the cell culture-TaqMan PCR assay and the ATP assay provide a sensitive and quantitative method for the determination of environmental oocyst inactivation, providing an alternative to the more costly and time-consuming mouse infection assay. The findings presented here relating temperature to oocyst inactivation provide valuable information for determining the relative risks associated with Cryptosporidium oocysts in water.
Members of the genus Cryptosporidium are protozoan parasites that can cause the gastrointestinal disease cryptosporidiosis and represent an emerging and highly infectious risk (30). Cryptosporidia are transmitted as infective oocysts directly by the fecal-oral route or by ingestion of contaminated food or water (23). In immunocompetent individuals, infection causes self-limiting, watery diarrhea but can become persistent, leading to chronic diarrhea and wasting in immunocompromised individuals (4). The disease has been documented worldwide, with speculation that many undiagnosed cases of gastroenteritis may have been caused by this parasite (19, 42).
Oocysts of Cryptosporidium are excreted with the feces of the host and subjected to the rigors of the environment until rendered noninfectious or ingested by a susceptible host (6). The infectivity of sporozoites is maintained by the oocyst wall, an extremely robust multilayered structure (15). Upon ingestion, exposure to stomach acid and bile salts promotes destabilization of the oocyst wall, resulting in the release of sporozoites that can then invade host enterocytes (30).
Cryptosporidium oocysts are prevalent in surface waters (for reviews, see Rose et al. [32] and Smith and Rose [34]) and thick-walled oocysts are extremely resistant to chlorine and monochloramine at levels used in the disinfection of potable water (8, 25). In addition to being resistant to commonly used disinfectants, it has been thought that oocysts generally survive for several months in an aquatic environment (22, 26, 31). Cryptosporidia therefore represent a challenge to the water industry and a threat to public health. Accurate measurement of oocyst inactivation is critical for any risk assessment of cryptosporidia in water. However, little is known about the inactivation rates of cryptosporidia by natural processes in environmental waters, especially in regard to measuring infectivity.
A limited number of studies have examined the survival of oocysts in natural waters using indicators such as vital dye staining or in vitro excystation (3, 31), but such methods are only viability indicators and are known to overestimate infectivity in comparison to animal infectivity (2). Until recently, the only methods which appeared to be appropriate for evaluating infectivity were animal or cell culture models of infection (33). The development of a cell culture-TaqMan PCR assay (24), which has been demonstrated to be quantitative, sensitive, and reproducible for measurement of oocyst inactivation by disinfection, promises to dramatically improve our ability to quantify environmental inactivation of oocysts through biotic and abiotic mechanisms.
While water-borne Cryptosporidium oocysts appear to be resilient at a wide range of temperatures (3, 6, 10, 11, 21, 45), increased holding temperatures correspond to decreased oocyst infectivity (6). The ability of Cryptosporidium oocysts to initiate infection has been linked to finite carbohydrate energy reserves in the form of amylopectin, which are consumed in direct response to ambient environmental temperatures (6). Amylopectin has been identified as the carbohydrate storage in different life cycle stages of a number of apicomplexans (1, 27, 36, 37). It has been concluded that amylopectin constitutes the energy reserve needed for excystation and invasion of host cells by coccidian sporozoites and that when the content of amylopectin falls below a critical level, sporozoites lack sufficient energy to invade cells (43). However, assays measuring amylopectin content or levels of amyloglucosidase mRNA display only marginal agreement with mouse infection and cell culture infection (20).
We report the investigation of inactivation rates of Cryptosporidium parvum oocysts incubated in reagent water and an environmental water sample at temperatures representative of the conditions that water-borne oocysts may encounter in the environment. We also tested the hypotheses that oocyst infectivity is energy dependent and that prolonged exposure of oocysts to environmental temperatures results in the depletion of energy reserves by investigating the relationship between oocyst ATP content and cell culture infectivity at different temperatures over prolonged holding periods. Inactivation constants derived from the cell culture-TaqMan PCR and ATP assays over a wide range of environmental temperatures will provide a valuable framework for risk assessment and be particularly applicable for integration with hydrodynamic models to estimate the risk of infectious oocysts reaching the off-takes of reservoirs. This is critical for the risk assessment of cryptosporidia in water and other environmental matrices and in determining the effectiveness of residence in reservoirs as a barrier in the water treatment process.
MATERIALS AND METHODS
Source of oocysts.
Oocysts of C. parvum (cattle isolate Swiss cattle C26, type II genotype) were purchased from the Department of Veterinary and Biomedical Sciences, Murdoch University, Perth, Australia. Oocysts were passaged through mice as previously described (16, 28). Oocysts were stored in sterile phosphate-buffered saline supplemented with antibiotic solution (15 μl/ml) containing ampicillin (10 mg/ml) and lincomycin (4 mg/ml). On receipt, the infectivity of each oocyst batch was determined using a cell culture-TaqMan PCR assay (24). Standard curves of oocyst infectivity were constructed using serial dilutions of 105, 104, 103, 102, 101 and 100 oocysts. All oocyst standards were applied to cell culture in triplicate.
TaqMan cell culture infectivity assay.
Prior to cell culture, oocyst concentrations were determined for each sample by microscopy as described previously (24). Ten thousand oocysts were used in each cell culture infection experiment. In vitro culturing, cell culture infection of the HCT-8 cell line, and DNA extraction from the infected cell monolayer were as described by Keegan et al. (24). Real-time PCR and the preparation of DNA standards for the quantitation of the level of cell culture infection were conducted as described by Keegan et al. (24), except that an alternative TaqMan probe was used, EUK3 (5′-6-carboxyfluorescein (FAM)-AAGTCTGGTGCCAGCAGCCGC-BHQ1 3′), reaction volumes were decreased to 25 μl, and reactions were analyzed using a RotorGene 3000 and version 6 of the RotorGene analysis software.
Survival was calculated using the equation log10 survival = log10(treated sample cell culture TaqMan PCR result) − log10(control sample cell culture TaqMan PCR result). A result of >0 indicates that the treated sample has increased in infectivity relative to the control, whereas a result of <0 indicates that the treated sample has decreased in infectivity relative to the control. The 4°C time zero sample was used as the control to measure survival for the other 4°C samples. For samples incubated at other temperatures, the 4°C sample for that time point was used as the control. This approach was taken to minimize any interassay variation that could be attributed to changes in the cell line infectivity over time.
Water sources.
Reagent-grade MilliQ water (Milli-Q Plus ultrapure water system, Millipore) and Hope Valley reservoir water were used for oocyst incubation experiments. Hope Valley reservoir is located 10 km north of Adelaide, South Australia. Water samples for replicate experiments were collected during the months of May, June, and November 2003. A summary of water characteristics for Hope Valley raw water is presented in Table 1.
TABLE 1.
Characteristics of Hope Valley raw water samplesa
| Parameter | Range | Avg |
|---|---|---|
| pH | 7.9-8.7 | 8.3 |
| Alkalinity (mg of CaCo3 per liter) | 79.5-132 | 112 |
| Dissolved organic carbon (mg/liter) | 5.7-11.7 | 7.7 |
| Turbity (NTU) | 0.61-87 | 4 |
| Color (HU) | 8-95.9 | 33 |
Based on 12 months of routine monitoring. NTU, nephelometric turbidity units; HU, Hazen units.
Oocyst storage experiments for MilliQ, Hope Valley raw, and Hope Valley autoclaved waters.
Four incubators held at 4, 15, 20, and 25°C were utilized for oocyst incubation experiments. For each holding temperature and water type (MilliQ, Hope Valley raw, and Hope Valley autoclaved water), 5 × 105 oocysts were suspended in a 30-ml yellow-cap polystyrene container in a total volume of 10 ml of water. Oocysts were incubated at each temperature over a 12-week period; 1-ml sample aliquots were removed from the tubes after thorough vortexing at chosen time points for each temperature and oocyst concentrations were determined for all treatments at each sampling point so that 10,000 oocysts could be applied to cell monolayers for infection studies. Each sampling point was analyzed in triplicate using the cell culture infectivity assay.
ATP assay and ATP standard curve construction.
The ATPlite luminescence ATP detection assay system (PerkinElmer), based on the firefly luciferin-luciferase reaction (12), was used to determine ATP concentration according to the manufacturer's instructions. Samples were measured in a 96-well Isoplate (PerkinElmer) using a luminescence counter (Wallac 1420 multilabel counter, PerkinElmer) to measure light emission.
Serial dilutions were made from an ATP stock provided by the manufacturer for use as ATP standards. Lysis solution (50 μl) and enzyme-substrate solution (50 μl, luciferase/luciferin) was added to 1.5-ml centrifuge tubes containing ATP, and reactions were made to a final volume (200 μl) using MilliQ water. Standards were transferred to Isoplates and read using the luminescence counter. Each standard concentration was analyzed in triplicate for standard curve construction. The linearity of the relationship between bioluminescence and ATP concentration was initially tested over the range of 10−4 M to 10−9 M (R2 = 0.98). Appropriate standards were constructed for each experiment as required.
ATP extraction from oocysts.
The lysis solution (50 μl) provided by the manufacturer was added to 1.5-ml centrifuge tubes containing an oocyst suspension (100 μl) and snap frozen in liquid nitrogen. Centrifuge tubes were stored in the freezer at −20°C until use. Samples were taken through five cycles of freeze/thawing by immersing in liquid nitrogen for 30 seconds before transferring to a heating block at 100°C for 1 min. A final incubation for 2 min at 100°C was performed. Centrifuge tubes were spun at 10,000 × g for 5 min. Substrate-enzyme solution (50 μl) was added to centrifuge tubes in subdued light and mixed well before the total sample (200 μl) was transferred to an Isoplate and dark adapted for 10 min before measuring luminescence at 22°C using the plate counter.
Standard curve construction of oocyst number and extracted ATP concentration.
Oocysts (2 × 106) were sterilized with sodium hypochlorite (final concentration of 13 ppm of free available chlorine) which was neutralized after 2 h by 1% vol/vol 40 mM sodium thiosulfate. A standard curve of oocyst number versus ATP concentration was constructed using serial dilutions of 105, 5 × 104, 2.5 × 104, 104, 5 × 103, 103, 5 × 102, 102 and 101 oocysts. ATP oocyst extraction was performed as described above. ATP standard curves were determined over a concentration range of 1 × 10−7 M to 1 × 10−10 M.
Oocyst storage experiments comparing ATP concentration and cell culture infectivity.
Six incubators held at 4, 15, 20, 25, 30, and 37°C were utilized for oocyst incubation experiments. For each holding temperature, 5 × 105 oocysts were suspended in a yellow-cap polystyrene container in a total volume of 5 ml MilliQ water containing 13 ppm of chlorine, which was neutralized after 2 h by 1% vol/vol 40 mM sodium thiosulfate. Oocyst counts were performed for all treatments to confirm that oocyst concentrations were 10,000 oocysts/100 μl. To confirm the sterilization of oocyst-associated bacteria by sodium hypochlorite, 10 μl from each treatment was diluted in a total volume of 100 μl of MilliQ water and plated onto R2A (Oxoid) heterotrophic plate count medium and incubated at 20°C for 3 days. Sample aliquots (1 ml) for each treatment were transferred into 1.5-ml centrifuge tubes after thorough vortexing at each of the chosen time points. Aliquots were analyzed in triplicate by both ATP analysis and cell culture infectivity. ATP standard curves were determined over a concentration range of 2.5 × 10−9 M to 1 × 10−10 M.
RESULTS
Determination of oocyst removal and oocyst inactivation rates for three water types.
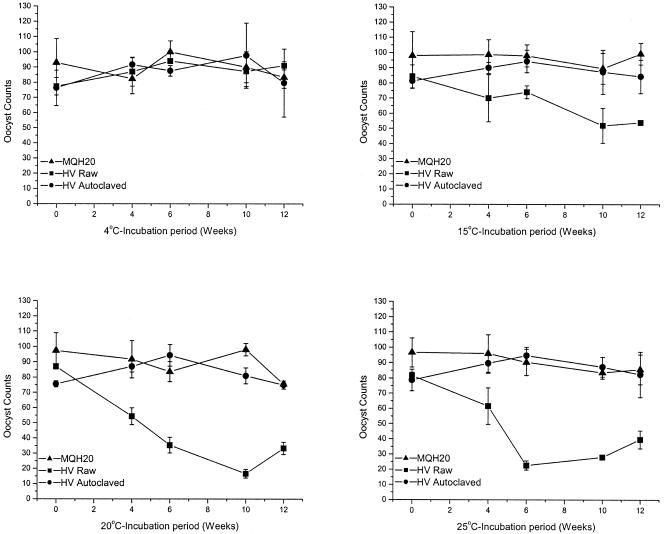
Three replicate in vitro experiments were performed in reagent-grade MilliQ water, Hope Valley raw water, and Hope Valley autoclaved water to assess oocyst removal rates and oocyst inactivation rates. Figure 1 illustrates oocyst removal for one of these experiments at four temperatures, 4, 15, 20, and 25°C. For MilliQ and autoclaved Hope Valley reservoir water treatments, oocyst counts remained constant throughout the incubation period for all four temperatures in all replicate experiments. Oocysts were not clumped when observed under fluorescence microscopy, were regular in shape, and showed consistent staining intensity with the fluorescein isothiocyanate (FITC) antibody. However, one replicate experiment conducted on Hope Valley raw water identified a gradual decrease in oocyst numbers at 15°C and more rapid decreases for both 20°C and 25°C over the 12-week incubation period (Fig. 1). Oocysts examined in these samples were often clumped together (as many as 18 in an individual clump, data not shown). Variation was also evident in FITC staining intensity between oocysts, especially within oocyst clumps (data not shown). In some instances there appeared to be deformation of the oocyst wall. Disappearance of oocysts in raw water varied between replicate experiments and was not consistent between replicates at the same temperature. For 20°C and 25°C samples where oocyst numbers decreased, the rate of decrease was similar to that presented in Fig. 1 (see 20°C and 25°C graphs for raw water).
FIG. 1.
Cryptosporidium oocyst counts for one replicate experiment in reagent-grade MilliQ water (MQH20), Hope Valley raw water (HV raw), and Hope Valley autoclaved water (HV autoclaved) at four temperatures: 4, 15, 20, and 25°C. Oocyst standard counts were determined by serial dilution and performed in triplicate. Error bars indicate standard deviations.
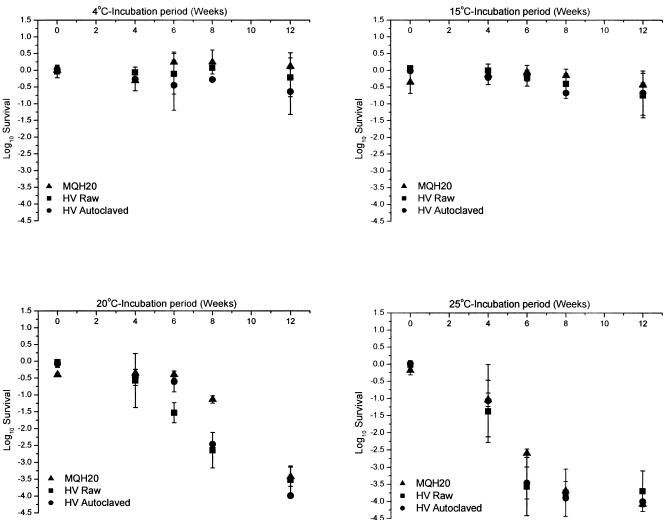
Figure 2 illustrates oocyst inactivation as determined by the cell culture-TaqMan PCR assay at four temperatures (4, 15, 20, and 25°C) in three different water types (MilliQ, Hope Valley raw, and Hope Valley autoclaved reservoir water). Microscopy counts of oocysts were performed prior to cell culture to ensure that 10,000 oocysts were assayed for each time point. Oocyst survival for 15, 20, and 25°C samples was calculated by using the 4°C samples as controls for each time point. The survival of the 4°C samples was calculated using the 4°C zero time point as a control. Following application of 10,000 fresh and infective oocysts to the HCT-8 cell line, the real-time PCR assay typically measured between 30,000 and 50,000 infectious bodies (sporozoite equivalents) in the monolayer after 48 h of incubation, allowing a 4 log10 reduction in oocyst infectivity to be measured.
FIG. 2.
Cryptosporidium oocyst inactivation rates for reagent-grade MilliQ water (MQH20), Hope Valley raw water (HV raw), and Hope Valley autoclaved water (HV autoclaved) at four temperatures: 4, 15, 20, and 25°C. Experiments were performed in triplicate for all three water types. Infectivity was calculated using the cell culture-TaqMan PCR assay. Oocysts were applied to cell monolayers in triplicate for each sampling point. Error bars indicate standard deviations.
Incubation temperatures of 20°C and 25°C showed the greatest reduction in oocyst infectivity as determined by cell culture-TaqMan PCR. However, there was no significant reduction in oocyst infectivity throughout the 12-week period for either the 4°C or 15°C incubation treatments. At 20°C, a 3 log10 reduction in infectivity was achieved after 8 weeks of incubation for all three water types. Inactivation at 25°C was more rapid, with a 3 log10 reduction in infectivity achieved as early as 6 weeks for both Hope Valley waters and at 8 weeks for MilliQ. A 4 log10 reduction, which represents complete inactivation as measured by the cell culture-TaqMan PCR assay, was achieved in all three water types by 12 weeks for 20°C and by 8 weeks for 25°C for all water types tested. The inactivation rate of cryptosporidia for all three water types appeared to be strongly temperature dependent, with no pronounced effect of water type on oocyst infectivity. This temperature-dependent inactivation led to the investigation of the relationship between oocyst ATP content, oocyst infectivity, and temperature over prolonged holding periods.
Development of an assay for measuring ATP concentration from Cryptosporidium oocysts.
The reliability of ATP measurement depends upon an extraction procedure that completely transfers ATP into solution and does not destroy ATP (18). Standard methods used for direct ATP measurement from most microorganisms are unlikely to be suitable for oocysts due to the robustness of the oocyst wall (35). To overcome this, a freeze-thaw process was investigated as a possible method for quick and reliable extraction of ATP from Cryptosporidium oocysts.
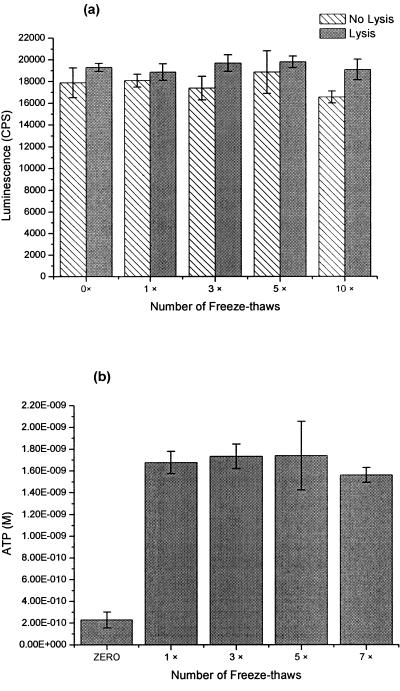
Experiments were initially conducted to determine if freeze-thawing affected ATP stability. A solution of 10−7 M ATP in either the presence or absence of lysis buffer was subjected to a series of freeze-thaws (1, 3, 5, or 10 times). No significant degradation of ATP was observed during the freeze-thaw treatments in comparison to the control that had no freeze-thawing of the ATP (Fig. 3a). With the exception of 10 cycles of freeze-thawing, addition of lysis buffer before the freeze-thaw process did not result in significantly greater luminescence than addition of lysis buffer after processing (Fig. 3a). However, a consistent trend towards higher luminescence for freeze-thawing in the presence of lysis buffer was evident. Lysis buffer addition before processing was used in our methodology thereafter and was considered essential for inactivation of endogenous ATPases that may be present within samples and capable of ATP degradation after oocysts have been freeze-thawed.
FIG. 3.
a. Effect of freeze-thaws on ATP as quantified by bioluminescence using the luciferin-luciferase reaction (n = 3). Error bars indicate standard deviations. b. Effect of freeze-thaws on ATP extracted from 10,000 Cryptosporidium oocysts (n = 3). Error bars indicate standard deviations.
Cryptosporidium oocysts were suspended in 50 μl of lysis buffer and taken through a series of multiple freeze-thaw treatments to determine optimal conditions for ATP extraction (Fig. 3b). The absence of any freeze-thawing resulted in significantly less (P > 0.01) ATP extracted in comparison to treatments utilizing freeze-thawing. The inclusion of only one cycle of freeze-thawing appeared to be sufficient to extract all ATP from oocysts, and increasing the number of freeze-thaw cycles did not result in any further improvement.
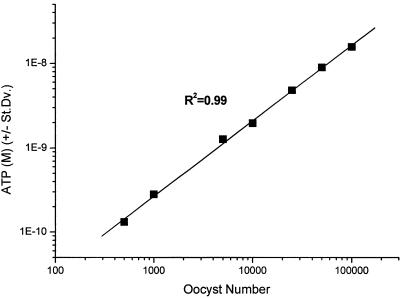
A standard curve to examine the relationship between oocyst number and ATP extracted by freeze-thawing was constructed (Fig. 4). A linear relationship was observed between ATP extracted and oocyst number over the range of 500 to 100,000 oocysts. The lower level of sensitivity for this test was determined to be 500 oocysts.
FIG. 4.
Standard curve of Cryptosporidium oocyst number and ATP extracted using freeze-thaw methodology (n = 3). Error bars which indicate standard deviations are smaller than data points plotted.
Relationship between oocyst infectivity and oocyst ATP concentration.
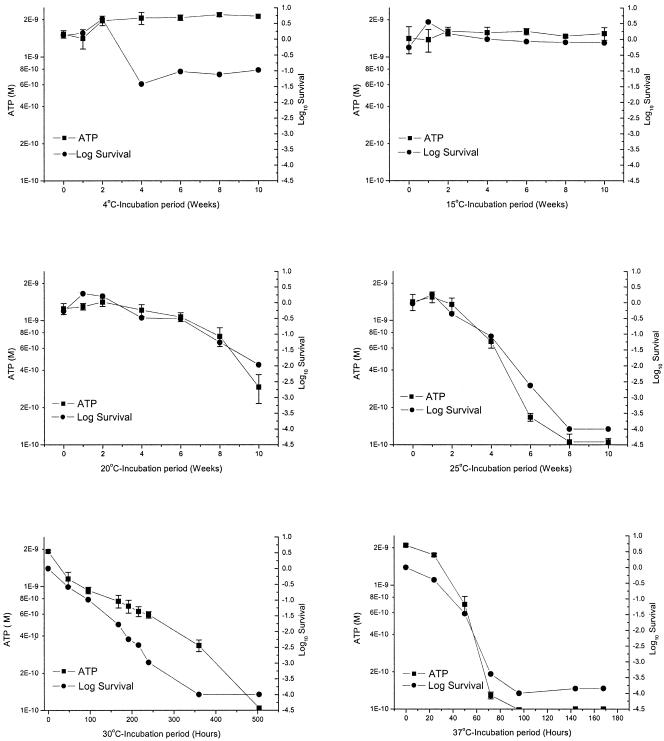
Time course experiments were performed in MilliQ water at incubation temperatures of 4, 15, 20, 25, 30, and 37°C to investigate the relationship between oocyst ATP content and oocyst infectivity as a function of temperature (Fig. 5). Ten thousand oocysts were analyzed by each method for each time point because this number of oocysts was within the linear range of the ATP assay and also the upper limit for the number of oocysts that can be analyzed by the infectivity assay. For 4°C, oocyst ATP concentration was constant over the 10-week incubation period. At week 4 there was greater than a 1 log10 reduction in infectivity for oocysts, after which oocyst infectivity remained constant (Fig. 5). A similar reduction was apparent in the raw infectivity values (data not shown) for the same time periods for all other incubation temperatures for this time point. This reduction in infectivity is likely to be an artifact of the assay due to a decrease in the sensitivity of the HCT-8 cell line to infection, which can occur during continuous passage. Variation in infectivity caused by the cell line was normalized for temperatures other than 4°C by using the 4°C treatment as the control for the measurement of inactivation at each time point.
FIG. 5.
Relationship between oocyst infectivity and oocyst ATP for 10,000 Cryptosporidium oocysts incubated in reagent-grade MilliQ water at incubation temperatures of 4, 15, 20, 25, 30, and 37°C. For cell culture results, survival at 4°C was calculated using the 4°C time zero sample as a control. For other temperatures, the corresponding 4°C sample was used as a control for each time point. Error bars indicate the standard deviations for ATP measurements.
Both ATP concentration and oocyst survival remained constant throughout the 15°C incubation period. However, significant decreases in ATP concentration and oocyst infectivity were evident for both the 20 and 25°C treatments. At week 8 for 20°C, significant decreases occurred for both ATP concentration and oocyst infectivity, with greater than a 1 log10 reduction in oocyst infectivity. The rate of decrease in ATP concentration and oocyst infectivity was more rapid at 25°C. By week 8 at 25°C, there was no detectable ATP or infectivity for any of the oocysts sampled. Decreases in oocyst ATP concentration and infectivity for 30 and 37°C were so rapid that experiments were sampled over a time scale of hours. For 30°C, significant decreases in ATP concentration and oocyst infectivity were evident after 48 h incubation, followed by a gradual decrease over a 2-week period, with complete inactivation and no detectable ATP observed by 3 weeks. At 37°C, a rapid decrease in ATP and infectivity occurred by 50 h, with complete inactivation and no detectable ATP observed after 72 h. For all six temperatures examined, the ATP concentration closely paralleled oocyst infectivity as measured by the cell culture-TaqMan assay.
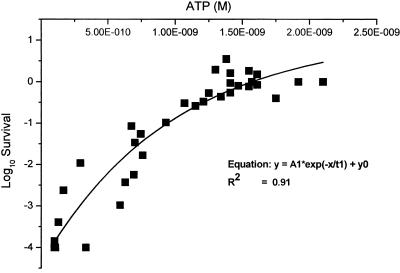
ATP concentrations and infectivity data derived from five temperature trials (excluding the 4°C trial) were plotted to determine the relationship between these two parameters (Fig. 6). A gradual decrease in oocyst infectivity was accompanied by a gradual decrease in ATP until the amount fell below 10−9 M. Beyond this point, the decrease in infectivity was much more rapid. An exponential decay curve calculated using Origin-7 graphing software provided a good fit for the data (R2 = 0.91).
FIG. 6.
Single plot defining the relationship between oocyst infectivity and ATP for 10,000 Cryptosporidium oocysts after incubation at 15, 20, 25, 30, and 37°C.
DISCUSSION
One of the main goals of this work was to evaluate the survival of cryptosporidia in three water types incubated under common environmental temperatures and quantify changes in oocyst infectivity using a cell culture-TaqMan assay. Data from Chauret et al. (3) identified that total oocyst counts must be accounted for to estimate relative oocyst viability. We employed fluorescence microscopy so that consistent oocyst numbers were applied to cell culture for inactivation studies and to determine oocyst removal rates in the three water types examined. It was noted in some replicate experiments for raw Hope Valley reservoir water that oocyst numbers decreased, but this was not consistent between replicate experiments.
Chauret et al. (3) proposed that natural biological antagonism has a pronounced influence in determining the environmental stability of cryptosporidia. They hypothesized that the observed biological antagonism may be due to microbial degradation, environmental modification by microbial metabolism, or microbial predation by larger organisms. Heterotrophic bacterial plate counts for Hope Valley raw waters were consistent between replicate raw water experiments (data not shown), suggesting that variation in oocyst removal is more likely to result from predation by larger organisms and not via bacterial degradation. This conclusion is strengthened by the results of a limited study of Hope Valley raw water with the addition of an antibiotic cocktail (penicillin, streptomycin, gentamicin, and lincomycin), which found removal of oocysts to occur at a similar rate to that described herein (A. R. Keegan, unpublished data). The variation in oocyst removal between replicate experiments is most likely explained as a result of sampling variation, spatial variation within the reservoir, or seasonal variation of predator densities.
For Hope Valley raw water experiments where oocyst numbers decreased, other effects, such as clumping of oocysts, differential FITC staining with the monoclonal antibody, and in some instances deformation of the oocyst wall, were apparent. Clumping of oocysts in raw waters has also been observed by Chauret et al. (3). These results suggest that predation may play an important role in the removal of oocysts in natural waters. Preliminary examination of raw Hope Valley reservoir water from the same location used in the experiments described herein has identified a number of organisms, including rotifers, ciliates, and amoebae (K. Harvey, unpublished data). Such organisms have previously been reported as being capable of ingesting oocysts (7, 38, 39), and it is known that rotifers excrete oocysts in boluses (clumps) (7). This behavior suggests that the clumped oocysts observed in this study are due to oocyst predation by rotifers.
The observed variation in oocyst FITC staining may be the result of partial digestion of oocysts by predatory organisms, resulting in degradation of the epitope to which the Cryptosporidium antibody is targeted. This change in staining intensity might provide a useful marker for separating ingested oocysts from noningested oocysts, by techniques such as flow cytometry, to determine if such treatment alters long-term survival or susceptibility of oocysts to disinfectants such as chlorine and monochloramine. Minimal work has been directed towards characterizing the organisms in the water column that are capable of ingesting Cryptosporidium oocysts (7, 38, 39). An understanding of their ecology and predation kinetics will be invaluable in determining their contribution to oocyst removal from reservoirs and whether they provide a viable means of biological control of oocysts in environmental waters.
Water-borne Cryptosporidium oocysts have been described in the literature as being resilient to a wide range of environmental temperatures (3, 6, 10, 11, 21, 45). One of the main goals of this work was to evaluate the survival of cryptosporidia in different water types incubated under common environmental temperatures using a quantitative cell culture-TaqMan PCR assay. Real-time PCR has previously been used for the quantification of Cryptosporidium numbers in environmental water samples and sewage samples (9, 13), but until this study has not been used in conjunction with cell culture for the determination of oocyst inactivation rates in environmental waters. The data clearly demonstrate that temperature is a major factor in the inactivation of oocysts in water. The inactivation rate was not affected by the water quality, with oocysts in MilliQ and Hope Valley reservoir raw water showing the same inactivation rate. Similarly, no difference in oocyst inactivation rates between autoclaved and raw Hope Valley water samples indicates that biological antagonism due to bacterial degradation was not present within the water samples examined.
Our results are in accordance with other observations previously described in the literature for the survival of oocysts at 4°C and 15°C (6). The current data suggest that oocysts can maintain infectivity for at least 3 months when stored at between 4°C and 15°C. This is in agreement with the results of Fayer et al. (6), who used a mouse infectivity model to demonstrate that oocysts stored in deionized water at 5°C or 15°C maintained infectivity for at least 24 weeks. In comparison, inactivation measured using cell culture-TaqMan PCR was much more rapid at 20°C and 25°C, leading to complete (4 log10-fold) inactivation after 12 weeks and 8 weeks, respectively. These results contrast with those of Fayer et al. (6), who found that oocysts held at 20°C for 24 weeks or at 25°C for 12 weeks were still infectious to mice. However, the doses used in the Fayer et al. (6) experiments were 10-fold higher than those used herein and the infections were only low level, as determined by histology, with only 1/10 mice becoming infected. In addition, oocysts held at 20°C for 20 weeks were not infectious to mice, suggesting that the number of infectious oocysts present was near the limit of detection for the assay.
The recent observation of the ability of cryptosporidia to complete their life cycle in a cell-free system (17) provides a possible explanation of these differences in inactivation rates. In our experimental design, oocysts used in incubation experiments were stored at a concentration of 50,000 oocysts/ml, 2 orders of magnitudes less than in the experiments conducted by Fayer et al. (6), where oocysts were stored at 3 × 106/ml. Hijjawi et al. (17) identified that high densities of cryptosporidia are able to complete their life cycle in RPMI 1640 maintenance medium. It is therefore feasible that in high densities of Cryptosporidium oocysts stored in deionized water, some excysted sporozoites may be able to complete their life cycle using the available energy sources of amylopectin, lipid, and protein from broken and excysted oocysts, resulting in the production of new infective oocysts and increasing the apparent length of time that oocysts retain their infectivity. This hypothesis is further strengthened by the recent report of the cell-free growth of cryptosporidia in rain water (Boxell et al., presented at the 4th International Giardia Conference and first combined Giardia/Cryptosporidium meeting, Amsterdam, The Netherlands, 20 to 24 September 2004). This questions the use of high densities of oocysts in temperature storage experiments for determination of temperature-dependent inactivation.
Following the demonstration that temperature plays an important role in oocyst inactivation, an additional objective of this study was to determine the mechanism of this inactivation. One hypothesis was that elevated temperatures result in the exhaustion of oocyst energy reserves, resulting in the inability of sporozoites to initiate infection. This hypothesis was tested by measuring the levels of ATP in oocysts exposed to different temperatures and comparing this with cell culture-TaqMan PCR. Reductions in oocyst ATP content were found to closely parallel decreases in oocyst infectivity for all temperatures investigated. Importantly, the rate of decrease in ATP content and oocyst infectivity became more rapid as incubation temperatures approached 37°C, indicating that temperature-dependent inactivation of oocysts is indeed a function of oocyst metabolic activity.
The ability of Cryptosporidium oocysts to initiate infection has previously been linked to finite carbohydrate energy reserves in the form of amylopectin, which are consumed in direct response to ambient environmental temperatures (6). However, mixed results had been reported for its use as a surrogate marker (20). The use of amylopectin or assays targeting a particular amyloglucosidase activity (21) as a surrogate marker for oocyst infectivity may be problematic. Total amylopectin includes the amylopectin content of broken oocysts and oocysts already inactivated. Additionally, assays targeting a particular mRNA or activity of an amyloglucosidase may only represent part of the spectrum of glucosidase activity within the oocyst (21, 40, 44). Finally, measurement of the substrate amylopectin or enzymatic processes targeting amylopectin as a surrogate marker for oocyst infectivity makes the assumption that amylopectin is the principal energy source utilized by sporozoites.
Within the oocyst residuum, a large lipid body and crystalline protein body reside alongside amylopectin granules (14). It is possible that these structures contribute as a source of metabolites for energy production during long storage periods. Glycolysis is thought to be the principal source of energy production in coccidia (5), so the breakdown of lipid could enter this pathway via the conversion of glycerol to dihydroxyacetone phosphate. Quantification of amylopectin or targeting enzymatic reactions acting upon this substrate may therefore underestimate energy reserves available for Cryptosporidium oocysts during long periods of storage.
The energy-rich triphosphate moiety ATP is the free-energy donor in most energy-requiring processes in biological systems (41). We propose that measuring oocyst ATP provides a more accurate measurement of total metabolic energy and therefore the potential sporozoite infectivity than measurement of amylopectin content or quantification of specific metabolites or enzymatic processes within the oocyst. Measurement of ATP using the luciferase reaction has been described for the evaluation of the growth of parasitic protozoa, including Entamoeba histolytica, Trichomonas vaginalis, Giardia spp., and Leishmania spp. (29). Recently, the measurement of oocyst ATP content has been identified as a suitable indicator of viability for cryptosporidia in regards to ozone disinfection (35). Here we present data demonstrating that quantification of oocyst ATP levels can provide a simple and rapid alternative to estimating oocyst viability and determination of temperature dependent oocyst inactivation rates.
In conclusion, we demonstrate that the cell culture-TaqMan PCR assay is able to provide a sensitive and quantitative method for the determination of environmental oocyst inactivation, providing an alternative to the more costly, time-consuming, and ethically questionable mouse infection assay. This work also identifies oocyst predation as an important biotic factor responsible for oocyst removal. However, we emphasize the need for further attention to this process in order to understand the fate of Cryptosporidium oocysts in the environment. An increased rate of inactivation for oocysts incubated at temperatures greater than 15°C, higher than that previously reported, was also identified, suggesting that temperature inactivation is a key abiotic factor affecting oocyst survival and infectivity in the environment. Finally, we have demonstrated that temperature inactivation at higher temperatures is a function of increased metabolic activity of oocysts and describe a rapid and simple ATP assay which can be used as an alternative for determination of oocyst inactivation rates.
The data described in this paper should provide valuable information when combined with hydrodynamic models in determining guides for risk analysis and determining the effectiveness of residence in reservoirs as a barrier. Assumptions can also be made regarding oocyst survival in waterways with regard to prevailing ambient temperatures.
Acknowledgments
We acknowledge the financial support received from the Co-operative Research Centre for Water Quality and Treatment, Australian Water Quality Centre and South Australian Water Cooperation.
We thank David Daminato and Stella Fanok for excellent technical support. We also thank Andrew Humpage for use of the luminescence counter and fruitful discussions.
REFERENCES
- 1.Alvarez-Pellitero, P., O. Palenzuela, and A. Sitja-Bobadilla. 1997. Ultrastructure and cytochemistry study of Eimeria sparis (Protozoa:Apicomplexa) stages from the intestine of gilthead sea bream Sparus aurata (Pisces: teleostei). Parasitol. Res. 83:126-136. [DOI] [PubMed] [Google Scholar]
- 2.Black, E. K., G. R. Finch, R. Taghi-Kilani, and M. Belosevic. 1996. Comparison of assays for Cryptosporidium parvum oocysts viability after chemical disinfection. FEMS Microbiol. Lett. 135:187-189. [DOI] [PubMed] [Google Scholar]
- 3.Chauret, C., K. Nolan, P. Chen, S. Springthorpe, and S. Sattar. 1998. Aging of Cryptosporidium parvum oocysts in river water and their susceptibility to disinfection by chlorine and monochloramine. Can. J. Microbiol. 44:1154-1160. [DOI] [PubMed] [Google Scholar]
- 4.Current, W. L., N. C. Reese, J. V. Ernst, W. S. Bailey, M. B. Heyman, and W. M. Weinstein. 1983. Human cryptosporidiosis in immunocompetent and immunodeficient persons. Studies of an outbreak and experimental transmission. N. Engl. J. Med. 308:1252-1257. [DOI] [PubMed] [Google Scholar]
- 5.Entrala, E., and C. Mascaro. 1997. Glycolytic enzyme activities in Cryptosporidium parvum oocysts. FEMS Microbiol. Lett. 151:51-57. [DOI] [PubMed] [Google Scholar]
- 6.Fayer, R., J. M. Trout, and M. C. Jenkins. 1998. Infectivity of Cryptosporidium parvum oocysts stored in water at environmental temperatures. J. Parasitol. 84:1165-1169. [PubMed] [Google Scholar]
- 7.Fayer, R., J. M. Trout, E. Walsh, and R. Cole. Rotifers ingest oocysts of Cryptosporidium parvum. J. Eukaryot. Microbiol. 47:161-163, 2000. [DOI] [PubMed]
- 8.Finch, G. R., E. K. Black, L. Gyurek, and M. Belosevic. 1993. Ozone inactivation of Cryptosporidium parvum in demand-free phosphate buffer determined by in vitro excystation and animal infectivity. Appl. Environ. Microbiol. 59:4203-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fontaine, M., and E. Guillot. 2003. An immunomagnetic separation-real-time PCR method for quantification of Cryptosporidium parvum in water samples. J. Microbiol. Methods 54:29-36. [DOI] [PubMed] [Google Scholar]
- 10.Freire-Santos, F., A. M. Oteiza-López, C. A. Vergara-Castiblanco, and E. Ares-Mazás. 2000. Study of the combined influence of environmental factors on viability of Cryptosporidium parvum oocysts in water evaluated by fluorogenic vital dyes and excystation techniques. Vet. Parasitol. 89:253-259. [DOI] [PubMed] [Google Scholar]
- 11.Freire-Santos, F., A. M. Oteiza-López, C. A. Vergara-Castiblanco, and M. E. Ares-Mazás. 1999. Effect of salinity, temperature and storage time on mouse experimental infection by Cryptosporidium parvum. Vet. Parasitol. 87:1-7. [DOI] [PubMed] [Google Scholar]
- 12.Green, A., and W. D. McElroy. 1956. Function of adenosine triphosphate in the activation of luciferin. Arch. Biochem. Biophys. 64:257-271. [DOI] [PubMed] [Google Scholar]
- 13.Guy, R. A., P. Payment, U. J. Krull, and P. A. Horgen. 2003. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl. Environ. Microbiol. 69:5178-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris, J. R., M. Adrian, and F. Petry. 2004. Amylopectin: a major component of the residual body in Cryptosporidium parvum oocysts. Parasitology 128:269-282. [DOI] [PubMed] [Google Scholar]
- 15.Harris, J. R., and F. Petry. 1999. Cryptosporidium parvum: structural components of the oocyst wall. J. Parasitol. 85:839-849. [PubMed] [Google Scholar]
- 16.Hijjawi, N. S., B. P. Meloni, U. M. Morgan, and R. C. Thompson. 2001. Complete development and long-term maintenance of Cryptosporidium parvum human and cattle genotypes in cell culture. Int. J. Parasitol. 31:1048-1055. [DOI] [PubMed] [Google Scholar]
- 17.Hijjawi, N. S., B. P. Meloni, M. Ng'anzo, U. M. Ryan, M. E. Olson, P. T. Cox, P. T. Monis, and R. C. Thompson. 2004. Complete development of Cryptosporidium parvum in host cell-free culture. Int. J. Parasitol. 34:769-777. [DOI] [PubMed] [Google Scholar]
- 18.Holm-Hansen, O., and C. R. Booth. 1966. The measurement of adenosine triphosphate in the ocean and its ecological significance. Limnol. Oceanogr. 11:510-519. [Google Scholar]
- 19.Hunter, P., Q. Syed, and E. N. Naumova. 2001. Possible undetected outbreaks of cryptosporidiosis in areas of the north west of England supplied by an unfiltered surface water source. Commun. Dis. Public Health 4:136-138. [PubMed] [Google Scholar]
- 20.Jenkins, M., J. M. Trout, J. Higgins, M. Dorsch, D. Veal, and R. Fayer. 2003. Comparison of tests for viable and infectious Cryptosporidium parvum oocysts. Parasitol. Res. 89:1-5. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins, M. C., J. Trout, M. S. Abrahamsen, C. A. Lancto, J. Higgins, and R. Fayer. 2000. Estimating viability of Cryptosporidium parvum oocysts using reverse transcriptase-polymerase chain reaction (RT-PCR) directed at mRNA encoding amyloglucosidase. J. Microbiol. Methods 43:97-106. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, D. C., C. E. Enriquez, I. L. pepper, C. P. Gerba, and J. B. Roe. 1997. Survival of Giardia, Cryptosporidium, Poliovirus and Salmonella in marine waters. Water Sci. Technol. 35:261-268. [Google Scholar]
- 23.Juranek, D. D. 1997. Cryptosporidium and water: a public health handbook-1997. Clin. Lab. Sci. 10:272. [PubMed] [Google Scholar]
- 24.Keegan, A. R., S. Fanok, P. T. Monis, and C. P. Saint. 2003. Cell culture-TaqMan PCR assay for evaluation of Cryptosporidium parvum disinfection. Appl. Environ. Microbiol. 69:2505-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korich, D. G., J. R. Mead, M. S. Madore, N. A. Sinclair, and C. R. Sterling. 1990. Effects of ozone, chlorine dioxide, chlorine, and monochloramine on Cryptosporidium parvum oocyst viability. Appl. Environ. Microbiol. 56:1423-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medema, G. J., M. Bahar, and F. M. Schets. 1997. Survival of Cryptosporidium parvum, Escherichia coli, faecal enterococci and Clostridium perfringens in river water: influence of temperature and autochthonous microorganisms. Water Sci. Technol. 35:249-252. [Google Scholar]
- 27.Medina, H., J. M. Barboza, H. Urdaneta, M. Rondon, and N. V. Joshi. 2001. Morphological investigation of Toxoplasma gondii in vivo by a multiple beam interference microscope. Mem. Inst. Oswaldo Cruz 96:983-986. [DOI] [PubMed] [Google Scholar]
- 28.Meloni, B. P., and R. C. Thompson. 1996. Simplified methods for obtaining purified oocysts from mice and for growing Cryptosporidium parvum in vitro. J. Parasitol. 82:757-762. [PubMed] [Google Scholar]
- 29.Miyahira, Y., and T. Takeuchi. 1991. Application of ATP measurement to evaluation of the growth of parasitic protozoa in vitro with a special reference to Pneumocystis carinii. Comp. Biochem. Physiol. A. 100:1031-1034. [DOI] [PubMed] [Google Scholar]
- 30.O'Donoghue, P. J. 1995. Cryptosporidium and cryptosporidiosis in man and animals. Int. J. Parasitol. 25:139-195. [DOI] [PubMed] [Google Scholar]
- 31.Robertson, L. J., A. T. Campbell, and H. V. Smith. 1992. Survival of Cryptosporidium parvum oocysts under various environmental pressures. Appl. Environ. Microbiol. 58:3494-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose, J. B., J. T. Lisle, and M. LeChevallier. 1997. Waterborne cryptosporidiosis: incidence, outbreaks and treatment strategies, p. 93-109. In R. Fayer (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Boca Raton, Fla.
- 33.Slifko, T. R., D. Friedman, J. B. Rose, and W. Jakubowski. 1997. An in vitro method for detecting infectious Cryptosporidium oocysts with cell culture. Appl. Environ. Microbiol. 63:3669-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, H. V., and J. B. Rose. 1990. Waterborne cryptosporidiosis. Parasitol. Today 6:8-12. [DOI] [PubMed] [Google Scholar]
- 35.Somiya, I., S. Fujii, N. Kishimoto, and R. H. Kim. 2000. Development of ATP assay as a surrogate indicator of viability of Cryptosporidium parvum oocysts. Water Sci. Technol. 41:181-188.11381990 [Google Scholar]
- 36.Speer, C. A., S. Clark, and J. P. Dubey. 1998. Ultrastructure of the oocysts, sporocysts, and sporozoites of Toxoplasma gondii. J. Parasitol. 84:505-512. [PubMed] [Google Scholar]
- 37.Speer, C. A., J. P. Dubey, M. M. McAllister, and J. A. Blixt. 1999. Comparative ultrastructure of tachyzoites, bradyzoites, and tissue cysts of Neospora caninum and Toxoplasma gondii. Int. J. Parasitol. 29:1509-1519. [DOI] [PubMed] [Google Scholar]
- 38.Stott, R., E. May, E. Matsushita, and A. Warren. 2001. Protozoan predation as a mechanism for the removal of Cryptosporidium oocysts from wastewaters in constructed wetlands. Water Sci. Technol. 44:191-198. [PubMed] [Google Scholar]
- 39.Stott, R., E. May, E. Ramirez, and A. Warren. 2003. Predation of Cryptosporidium oocysts by protozoa and rotifers: implications for water quality and public health. Water Sci. Technol. 47:77-83. [PubMed] [Google Scholar]
- 40.Strong, W. B., and R. G. Nelson. 2000. Preliminary profile of the Cryptospo-ridium parvum genome: an expressed sequence tag and genome survey sequence analysis. Mol. Biochem. Parasitol. 107:1-32. [DOI] [PubMed] [Google Scholar]
- 41.Stryer, L. 1981. Metabolism: basic concepts and design, p. 235-254. In L. Stryer (ed.), Biochemistry, 2nd ed. W. H. Freeman and Company, New York, NY.
- 42.Tillett, H. E., J. de Louvois, and P. G. Wall. 1998. Surveillance of outbreaks of water-borne infectious disease: categorizing levels of evidence. Epidemiol. Infect. 120:37-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vetterling, J. M., and D. J. Doran. 1969. Storagen polysaccharide in coccidial sporozites after excystation and penetration of cells. J. Protozool. 16:772-775. [DOI] [PubMed] [Google Scholar]
- 44.Wang, C. C., R. M. Weppelman, and B. Lopez-Ramos. 1975. Isolation of amylopectin granules and identification of amylopectin phosphorylase in the oocysts of Eimeria tenella. J. Protozool. 22:560-564. [DOI] [PubMed] [Google Scholar]
- 45.Widmer, G., E. A. Orbacz, and S. Tzipori. 1999. β-Tubulin mRNA as a marker of Cryptosporidium parvum oocyst viability. Appl. Environ. Microbiol. 65:1584-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]