Abstract
The ability of Erwinia chrysanthemi to cope with environments of elevated osmolality is due in part to the transport and accumulation of osmoprotectants. In this study we have identified a high-affinity glycine betaine and choline transport system in E. chrysanthemi. By using a pool of Tn5-B21 ousA mutants, we isolated a mutant that could grow in the presence of a toxic analogue of glycine betaine (benzyl-glycine betaine) at high osmolalities. This mutant was impaired in its ability to transport all effective osmoprotectants in E. chrysanthemi. The DNA sequence of the regions flanking the transposon insertion site revealed three chromosomal genes (ousVWX) that encode components of an ABC-type transporter (OusB): OusV (ATPase), OusW (permease), and OusX (periplasmic binding protein). The OusB components showed a significant degree of sequence identity to components of ProU from Salmonella enterica serovar Typhimurium and Escherichia coli. OusB was found to restore the uptake of glycine betaine and choline through functional complementation of an E. coli mutant defective in both ProU and ProP osmoprotectant uptake systems. Competition experiments demonstrated that choline, dimethylsulfoniacetate, dimethylsulfoniopropionate, and ectoine were effective competitors for OusB-mediated betaine transport but that carnitine, pipecolate, and proline were not effective. In addition, the analysis of single and double mutants showed that OusA and OusB were the only osmoprotectant transporters operating in E. chrysanthemi.
The gram-negative bacterium Erwinia chrysanthemi is a plant pathogen, which is involved in a systemic soft rot disease in a wide range of plant species. E. chrysanthemi like other microbial pathogens faces frequent changes in the availability of water both inside and outside its hosts. Such sudden fluctuations in the osmolality of the environment profoundly influence the physiology of the bacterial cell (14, 39). Bacteria are required to maintain an intracellular osmotic pressure higher than that of the surrounding medium in order to generate cell turgor, which is thought to provide the mechanical force necessary for cell elongation and division (15, 70). Many bacterial species respond to increased osmotic pressure in the medium by accumulating high intracellular concentrations of low-molecular weight compounds called compatible solutes (14, 39). The most rapid response to a sudden osmotic upshock is a stimulation of potassium uptake, both in gram-negative and gram-positive bacteria (21, 69). In Escherichia coli glutamate is endogenously accumulated to counterbalance the positive charge of potassium (48), whereas in Bacillus subtilis the nature of the potassium counter-ion has not been elucidated since glutamate levels increase only slightly after osmotic upshock (39). The secondary response to osmotic stress consists of the synthesis and/or uptake of high amounts of compatible solutes that can be substituted for potassium glutamate (19). Important examples of compatible solutes are the iminoacid proline (13, 69), the disaccharide trehalose (19, 24), the trimethylammonium glycine betaine (GB) (15, 61, 70), and the tetrahydropyrimidine ectoine (23, 34, 35). Osmoprotectants are compatible solutes that are taken up from the culture medium and greatly stimulate bacterial growth under hyperosmotic conditions (14). In E. coli, the uptake of osmoprotectants occurs via two well-characterized transport systems, ProP and ProU. ProP, a secondary transporter, functions as an H+ symporter and is regulated mainly at the activity level (11). The ProU system is a binding-protein-dependent transport system, which belongs to the ABC superfamily (17, 29). E. coli, like many other bacteria, can also accumulate GB by synthesis from exogenously supplied choline (41, 43, 63). The synthesis of GB from choline is a function of the Bet system activities, including the choline high-affinity transport system (BetT), choline dehydrogenase (BetA), and betaine aldehyde dehydrogenase (BetB) (41, 43, 63).
To offset the deleterious effect of high osmolality, in the absence of supplied osmoprotectants, E. chrysanthemi synthesizes three endogenous osmolytes: glutamate, glutamine, and α-glucosylglycerate (25). Osmoprotectants such as GB, proline, ectoine, and pipecolic acid stimulate growth (26); these compounds have been shown to be accumulated within the cells and inhibit the synthesis of the endogenous osmolytes (25, 26). The accumulation of supplied osmoprotectants is achieved through osmoinducible systems (26) involving, OusA, a secondary transporter, homologue to ProP (28), and at least one other transport system (28). OusA is regulated at both expression and activity levels (28). To characterize the osmoprotectant uptake activity remaining in an ousA mutant of E. chrysanthemi, we have isolated an osmoprotectant transport mutant from a Tn5-B21 transposon insertional library, and we report here the nucleotide sequence of genes encoding an ABC-type transporter homologue to ProU from E. coli.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, chemicals, and growth conditions.
E. coli and E. chrysanthemi strains and plasmids used in the present study are listed in Table 1. Rich and minimal media for the growth of E. coli and E. chrysanthemi were previously described (66). The osmotic strength of the media was increased by the addition of NaCl at appropriate concentrations. The antibiotics ampicillin, kanamycin, and tetracycline were used in solid and liquid media for E. coli and E. chrysanthemi strains at final concentrations of 50, 50, and 10 μg/ml, respectively. The osmoprotectants used in the present study, which included GB, carnitine, pipecolate, proline, and choline, were purchased from Sigma-Aldrich (l'Isle d'Abeau Chesnes, France). Ectoine was extracted and purified from Brevibacterium linens as described previously (5, 33). Dimethylsulfonioacetate (DMSA) and dimethylsulfoniopropionate (DMSP) were synthesized as described previously (53). The osmoprotectant solutions were sterilized by passage through a 0.45-μm-pore-size sterile filter (Millipore), and these solutes were generally supplied at a concentration of 1 mM. The benzyl derivative of GB (BGB), which is chemically named carboxymethyl-dimethyl-(4-nitrobenzyl) ammonium chloride or N-benzyl-N,N-dimethylglycine betaine, was synthesized as described previously (12). The radiolabeled [methyl-14C]choline (2.07 GBq mmol−1), l-[methyl-14C]carnitine (1.86 GBq mmol−1), and [U-14C]proline (9.66 GBq mmol−1) were purchased from NEN-Dupont de Nemours. [methyl-14C]GB (2.07 GBq mmol−1) was prepared from [methyl-14C]choline as described earlier (34). [methyl-14C]DMSA (2.0 GBq mmol−1) and [1-14C]DMSP (37 MBq mmol−1) were synthesized and purified as previously described (53). [14C]ectoine (5.5 MBq mmol−1) was prepared biologically as described previously (34).
TABLE 1.
Bacterial strains, phage, and plasmids used in this study
| Strain, plasmid, or phage | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F′ endA1 hsdR17 glnV44 thi-1 recA1 gyrA96 relA1 Δ(lacIZYA-argF)U169 deoR φ80dlac Δ(lacZ)M15 | Bethesda Research Laboratories |
| SM10 | leu RP4-2TC::Mu-Kmr | 58 |
| MKH13 | Δ(betTIBA) U169 Δ(putPA)101 Δ(proP)2 andΔ(proU)608 | 31 |
| E. chrysanthemi | ||
| 3937 | Wild-type strain isolated from Saintpaulia ionanthia | 66 |
| 5512 | 3937 ousA::uidA-Kmr | 28 |
| W245 | 3937 ousB::Tn5-B21 | This work |
| W246 | 3937 ousA::uidA-Kmr, ousB::Tn5-B21 | This work |
| A350 | 3937 lmrTc lacZ2 | 49 |
| A3779 | A350 ousV::uidA-Cmr | 49 |
| Plasmids | ||
| pBluescript | ori colE1 AprlacZα | Stratagene |
| pSUP102::Tn5-B21 | Cmr Tcr ′lacZ | 59 |
| pSU8 | ori p15A CmrlacZα | 46 |
| pSU9 | ori p15A CmrlacZα | 46 |
| pB6855 | pBluescript carrying W245 right junction | This work |
| pB6864 | pBluescript carrying W245 left junction | This work |
| pB7056 | pB6855 ΔXhoI | This work |
| pB7265 | 1.5-kb KpnI fragment from pB6855 cloned into pBluescript | This work |
| pB7156 | 5.0-kb XhoI-EcoRI fragment from pB7056 cloned into pSU9 | This work |
| Phage ΦEC2 | E. chrysanthemi generalized transducing phage | 54 |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance.
DNA manipulations and analysis.
Chromosomal and plasmids DNA isolation and the construction of recombinant plasmids were carried out according to standard procedures (56). Searches for homologies were performed at the NCBI and EMBL databases using the BLAST and CLUSTAL W programs (1, 65).
Insertional mutagenesis.
Tn5-B21 mutagenesis was performed by mating E. coli SM10 (58) bearing a mobilizable and nonreplicative plasmid in E. chrysanthemi [pSUP102::Tn5-B21 (Tcr)] with E. chrysanthemi strain 5512 carrying a ousA::uidA-Kmr fusion as described previously (59). E. chrysanthemi transconjugants bearing a Tn5-B21 insertion were selected as tetracycline-resistant (Tcr) clones on LB medium. Mutants resistant to BGB were identified by replica plating on M63 medium with 0.4 M NaCl and 1 mM BGB. The corresponding mutations were transduced into the wild-type 3937 and 5512 (ousA) strains with the generalized transducing phage ΦEC2 as described previously (54), producing the strains W245 and W246, respectively. The mutant strains 5512, W245, and W246 are derivatives of the wild-type strain 3937. A Tcr colony that grew on M63 supplemented with 0.4 M NaCl plus 1 mM BGB was retained. This clone was named W246; the presence of a single copy of Tn5-B21 in its chromosome was confirmed by Southern blotting with pSUP102::Tn5-B21 DNA as the probe.
Cloning and sequencing of the chromosomal fragments carrying the transposon ends.
To isolate the chromosomal DNA flanking the inserted transposon in W246, we took advantage of the presence of unique restriction sites in Tn5-B21, either upstream (HindIII) or downstream (EcoRI) from its Tcr gene. These features enable the isolation of junctions as Tcr fragments. The right and left Tn5-B21 flanking regions were cloned into pBluescript (Stratagene) as a 5.0-kb EcoRI fragment in pB6855 and as a 3-kb HindIII fragment in pB6864, respectively. The E. chrysanthemi DNA contained in the recombinant plasmids was sequenced by Genaxis (Nîmes, France) by using a primer-walking strategy on both strands.
GUS assays.
The β-Glucuronidase (GUS) assays were carried out as described previously (28). Specific GUS were expressed as micromoles of para-nitrophenol liberated per minute per milligram of protein. The protein concentration was determined by the method of Lowry (44) with bovine serum albumin as the standard.
Transport assays and fate of radioactive choline.
Cells grown in M63 with or without NaCl were centrifuged (5,000 × g for 10 min), washed twice with an isotonic medium, and resuspended to an optical density at 570 nm of 0.5 to 1. For the kinetic studies, the GB or choline concentration in the uptake assay was varied from 1 to 100 μM. Transport assays were carried out as described previously (34) except that cells were resuspended into an isotonic medium. To study the fate of choline, cells were cultured in M63 containing 0.3 M NaCl and 100 μM [methyl-14C]choline. Cells were collected by centrifugation (5,000 × g, 10 min) and extracted twice with 80% ethanol, and the ethanol-soluble fraction was analyzed by paper electrophoresis (34). The spots of the radioactive molecules (choline and GB) were visualized and quantified by using a Packard Instant Imager.
Binding assays.
Cells were grown to mid-exponential phase in M63 with 0.3 M NaCl, harvested by centrifugation, washed, and resuspended in 10 mM Tris-HCl buffer with 0.3 M NaCl (pH 7.5). Periplasmic proteins were released by a cold osmotic shock procedure as described previously (34). Periplasmic fractions were filtered through a 0.22-μm-pore-size membrane filter (Millipore), centrifuged at 100,000 × g, dialyzed against 10 mM Tris-HCl (pH 7.5), freeze-dried, and dissolved in the same buffer. Protein concentration was estimated by the Lowry method (44). The binding activity of shock fluid was detected by the ammonium sulfate precipitation technique (55) in the presence of 5 μM [methyl-14C]GB (2.07 GBq mmol−1) or other radiolabeled compounds. The protein concentration was 2 mg ml−1. Samples of shock fluid (50 μl) in 10 mM Tris-HCl (pH 7.5) were mixed with the 14C-labeled substrate in the presence or absence of unlabeled competitors and then incubated at room temperature for 5 min. Proteins were precipitated by adding 950 μl of ice-cold saturated ammonium sulfate solution and, after incubation for 15 min on ice, the precipitated proteins were collected by filtration onto GF/F glass microfiber filters (Whatman). Each filter was then washed twice with 2 ml of an ice-cold ammonium sulfate solution, and the radioactivity on the filters was determined by scintillation counting.
Nucleotide sequence accession number.
The nucleotide sequence of ousV, ousW, and ousX and the flanking sequences have been deposited in GenBank under accession number AF494101.
RESULTS
Isolation and analysis of an osmoprotectant transport-deficient mutant.
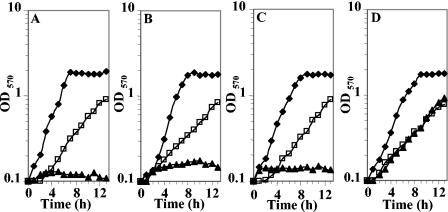
BGB, a structural analogue of GB, has been shown to be toxic to E. coli strains in which either ProP or ProU was functional (12). We used this characteristic to isolate a mutant, which was resistant to BGB, from a pool of Tn5-B21 ousA::uidA-Kmr E. chrysanthemi mutants. That mutant W246 was able to grow on elevated-osmolality minimal plates (M63 with 0.4 M NaCl) in the presence of 1 mM BGB, whereas the wild-type strain 3937 and the ousA strain 5512 failed to grow on the same medium. The Tn5-B21 mutation was transduced into 3937 and 5512 strains by using the generalized transducing phage ΦEC2 (54), resulting in strains W245 and W246, respectively. The wild-type strain 3937 and the mutants 5512, W245, and W246 were cultivated in M63 without or with 0.4 M NaCl in the presence or absence of 1 mM BGB. The growth patterns of the four strains are presented in Fig. 1. All of the strains had similar growth rates in medium without NaCl, and they all grew slowly in the presence of NaCl (Fig. 1). As expected, the double mutant W246 strain was insensitive to BGB (Fig. 1D), whereas the single mutants 5512 (Fig. 1B) and W245 (Fig. 1C) and the wild-type strain 3937 (Fig. 1A) were highly sensitive to this compound. These observations are similar to those obtained with E. coli strains impaired in ProP and/or ProU uptake activity (12). They suggest strongly that the Tn5-B21 insertion had occurred in gene(s) encoding an osmoprotectant transport system other than the OusA transporter.
FIG. 1.
Effect of benzylglycine betaine on growth of E. chrysanthemi 3937 and derivatives strains in M63 medium with 0.4 M NaCl. Strains 3937 (wild type) (A), 5512 (ousA) (B), W245 (ousB) (C), and W246 (ousA ousB) (D) were grown in M63 medium containing 0 M NaCl (⧫), 0.4 M NaCl (□), or 0.4 M NaCl and 1 mM BGB (▴). Results are the means of triplicate determinations; the standard error did not exceed 10%.
Cloning and nucleotide sequence of ousB locus.
Transposon-flanking chromosomal DNA was cloned and sequenced. The sequence analysis revealed that Tn5-B21 was inserted 52 bp upstream of the first open reading frame of a gene cluster composed of three genes. The open reading frames are oriented in the same direction and constitute the ousB locus. They are 1,202 (ousV), 1,220 (ousW), and 998 (ousX) bp in length. The entire sequence of these genes is carried by the plasmid pB6855. ATG start codons of the three genes are all preceded by a ribosome-binding site. The intergenic distance between ousW and ousX is 121 bp, and the end of ousV overlaps the beginning of ousW by 8 bp. The tight physical organization of these genes suggests strongly that they are organized in an operon. Consistent with this suggestion is the fact that a region upstream from ousV is homologous to the promoter sequences of the opuA operon from B. subtilis (38) and the proU operon from E. coli (47). The sequence analysis suggested that the Tn5-B21 insertion had occurred between the ousB promoter region and the start codon of ousV gene, which is sufficient to prevent the ousB expression. The inactivation of ousV by insertional mutagenesis in an ousA background also abolished the uptake of GB (data not shown), which confirms that ousB was involved in GB uptake. The deduced amino acid sequences of ousV, ousW, and ousX genes exhibit features characteristic of binding-protein-dependent transport systems (18, 32). A comparison of the amino acid sequences of the three open reading frames showed striking homologies to ProU transporter from E. coli and Salmonella enterica serovar Typhimurium (10, 29).
The ousV gene encodes a hydrophilic protein of 400 amino acids (Mr = 44,338), which showed strong sequence similarities to many prokaryotic ATPases involved in ABC transport systems. The analysis of the OusV amino acid sequence revealed 74 and 73% identities to ProV from the ProU system from serovar Typhimurium and E. coli, respectively (29, 62). However, lower homologies (57 and 49%, respectively) were obtained with HisV from S. meliloti and OpuAA from B. subtilis (6, 40). Moreover, the OusV sequence contained the Walker A and B motifs, and the linker peptide, which are the ATP binding sites, and the signature of the ABC transporter family, respectively (18).
The ousW gene encoded a hydrophobic protein of 406 amino acids (Mr = 43,125) that shares high homology (71% identity) to the integral inner membrane protein ProW of both the serovar Typhimurium and E. coli ProU transport systems. Much less homology was obtained with HisW from S. meliloti (45%) and OpuAB (40%) from B. subtilis, the transmembrane protein components of their respective transporters.
The last gene in the operon, ousX, encoded a 332-amino-acid hydrophilic protein (Mr = 36,357), which is presumed to be the substrate-binding protein. This protein had significantly greater homology to ProX from serovar Typhimurium and E. coli (75% identity) than to HisX from S. meliloti (24% identity) and OpuAC from B. subtilis (21% identity). It is noteworthy that the first 20 amino acids of OusX contain a positively charged N-terminal end, followed by a high proportion of hydrophobic residues, which is a characteristic signature of signal peptides. Furthermore, OusX contained the consensus sequence recognized by signal peptidase I. Also, the three tryptophan residues, forming a rectangular aromatic box, which are the key determinants for the binding of GB and proline betaine by E. coli ProX (57) were conserved in OusX.
Regulation of ousB expression.
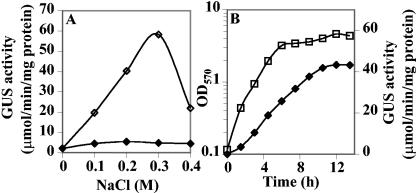
To study the expression of the ousB operon, ousV was fused to the uidA reporter gene in the mutant strain A3779 (49), and the induction of ousB was monitored by measuring the level of GUS activity. The transcription of ousB was at basal level when cells were grown at low osmolality (Fig. 2A), and the expression was proportionally correlated with increasing medium osmolality up to 0.3 M NaCl; at higher salinities, the expression of ousB decreased (Fig. 2A). The addition of 1 mM GB strongly repressed the ousB induction regardless of medium salinity (Fig. 2A). When ousB expression was monitored during the growth cycle of cells cultivated in M63 with 0.3 M NaCl, we observed that GUS activity increased immediately after osmotic upshift, reaching a maximal level in the early exponential growth phase. Then, it remained constant until the cells entered into the stationary phase (Fig. 2B).
FIG. 2.
Osmotic regulation of ousB expression. (A) Effect of GB and high osmolality on the expression of ousB. An ousV-uidA fusion strain was grown overnight in M63 minimal medium whose osmolality had been increased by the indicated concentration of NaCl. The cultures were grown in the absence (⋄) or in the presence (⧫) of 1 mM GB. (B) Influence of medium osmolality on ousB expression during the growth cycle of E. chrysanthemi A3779. The strain was grown in M63 medium with 0.3 M NaCl. The GUS specific activity (□) and the optical density at 570 nm (⧫) were determined at various times during the growth cycle. The results are averages of at least three independent experiments and standard error did not exceed 20%.
Functional complementation of E. coli MKH13.
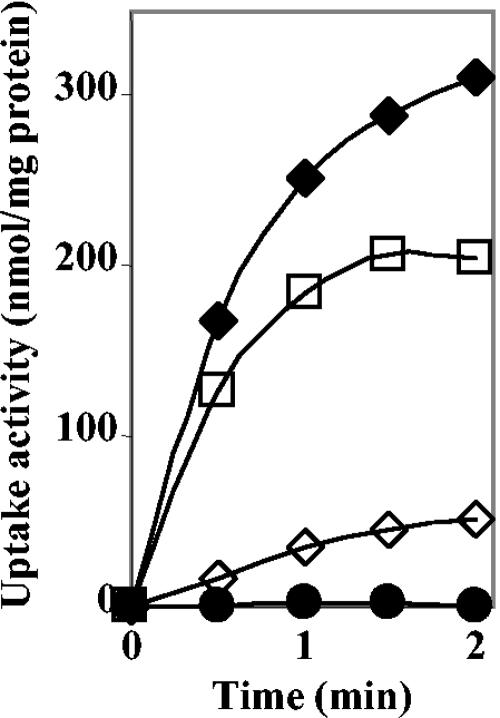
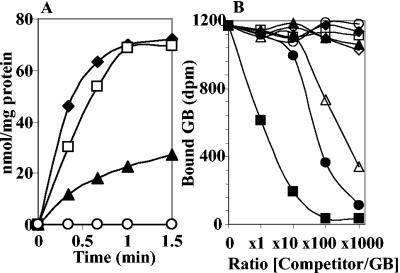
The E. coli strain MKH13 lacks both the ProU and ProP osmoprotectant uptake systems, as well as the bet genes involved in the uptake and oxidation of choline into GB (43, 63). Hence, supplied osmoprotectants cannot improve the growth of this strain at an elevated osmolality (27, 31). Plasmid pB7156, which harbors the ousB genes cloned into pSU9 (46), was transferred into MKH13. The expression of ousB genes carried by pB7156 plasmid was under the control of plac promoter of pSU9. Osmotically stressed transformants containing pB7156 but not pSU9 showed their growth restored on high-osmolality (0.5 M NaCl) minimal-medium plates supplemented with 1 mM GB. This phenotype was also observed with the plasmid pB7056 that contained the entire ousB operon but not with pB7265, which lacked the ousX gene. These observations were confirmed by measuring the initial rate of [14C]GB uptake in cultures of MKH13 (pSU9) and MKH13(pB7156). [14C]GB transport at high affinity (5.5 μM) was not detected in MKH13(pSU9), but GB uptake was detected in MKH13(pB7156) grown in minimal medium without or with 0.3 M NaCl (Fig. 3). Thus, the functional complementation observed in MKH13(pB7156) clearly demonstrates that ousB encodes a GB transporter. Since the ProU transporter, which is highly homologous to OusB, is involved in the transport of various osmoprotectants, we have analyzed the uptake of various osmoprotectants by using strain MKH13(pB7156) grown in the presence of 0.3 M NaCl. The results showed that OusB was also a transporter of choline (Fig. 3) and several other osmoprotectants such as ectoine, DMSA, DMSP, proline, and carnitine (data not shown).
FIG. 3.
OusB-mediated glycine betaine and choline uptake in E. coli MKH13. Cells of MKH13(pB7156)(OusB+) were grown to mid-log phase in M63 medium without or with 0.3 M NaCl and then assayed for the uptake of [14C]GB at low (⋄) or high (⧫) osmolality and for [14C]choline uptake at high osmolality (□). The final concentrations of GB and choline were 5.5 and 4.5 μM, respectively. Uptake of [14C]GB and [14C]choline in MKH13(pSU9) (•) was used as a control. The results are means of triplicate determinations; the standard deviation did not exceed 5%.
Osmoprotection is mediated by OusA and OusB osmoporters in E. chrysanthemi.
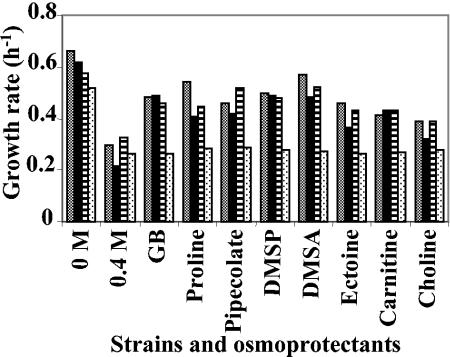
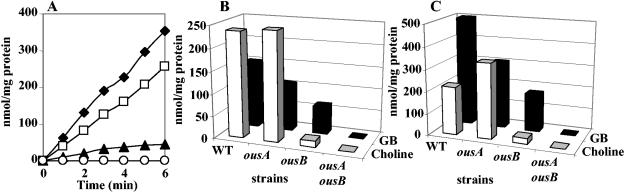
To assess the contribution of OusA and OusB to osmoprotection by various osmoprotectants (26, 28), we grew the wild-type strain and mutants impaired in one or two osmoprotectant transport systems under elevated osmolality conditions. As shown in Fig. 1, the growth of all strains was impaired when cells were grown in M63 medium supplemented with 0.4 M NaCl. The growth rates ranged from 0.5 to 0.65 h−1 at low osmolalilty, and they decreased to 0.2 to 0.3 h−1 at high osmolality (Fig. 4). All of the assayed compounds showed a stimulatory effect on the growth of the wild-type strain and that of the mutants synthesizing a single osmoprotectant uptake system (Fig. 4). Growth of the double mutant strain W246 (ousA ousB) was not protected from the detrimental effects of elevated osmolality by anyone osmoprotectant. Thus, the ability of these compounds to restore growth of E. chrysanthemi in media of high osmolality was exclusively dependent on functional OusA and OusB osmoporters.
FIG. 4.
Influence of medium osmolality and osmoprotectants on the growth rate of strain 3937 and derivatives. Cells were grown in minimal M63 medium without or with 0.4 M NaCl. Osmoprotectants were added at 1 mM. Columns in each group illustrate findings for strains 3937 (▩), 5512 (▪), W245 (▤), and W246 ( ) (from left to right). The results are the means of triplicate determinations; the standard error did not exceed 10%.
) (from left to right). The results are the means of triplicate determinations; the standard error did not exceed 10%.
Role of OusB in osmoprotectant uptake in E. chrysanthemi.
OusA is involved in the uptake of various osmoprotectants (28), and an ousA mutant strain still retained an uncharacterized osmoprotectant uptake activity (28). To assess whether OusB is responsible for this activity, we measured [14C]GB uptake activity in the wild type and in single (ousA or ousB) and double (ousA ousB) mutants. The preliminary [14C]GB uptake experiments showed that the transport activity of this solute, via OusA or OusB, was very low when cells were cultured in M63 without NaCl (data not shown), and it increased to reach a maximal level when cells were grown in M63 with 0.3 M NaCl. At higher salinities, the GB uptake activity decreased (data not shown). Thus, the salinity of 0.3 M was retained for all transport assays.
At high affinity (5.5 μM), transport of GB was moderately affected in 5512 (ousA) strain, whereas in the W245 (ousB) strain transport was strongly affected (Fig. 5A). In the double-mutant strain W246, GB uptake activity was completely abolished (Fig. 5A), indicating that no GB transport systems other than OusA and OusB operated in E. chrysanthemi. To evaluate the distinctive contributions of the OusA and OusB transporters to the overall GB transport activity and their kinetic parameters, mutant strains that synthesize only one of these transporters were used. The Michaelis-Menten parameters of GB uptake by OusA and OusB were determined when cells of strains 5512 (ousA) and W245 (ousB) were grown in M63 with 0.3 M NaCl. The apparent Km values of GB uptake were 1.6 and 50 μM, with Vmax values of 92 and 270 nmol/min/mg of protein for OusB and OusA, respectively. Thus, OusB is a high-affinity GB transport system, whereas OusA is a relatively low-affinity transport system.
FIG. 5.
Uptake and binding activities of GB in E. chrysanthemi. (A) Strains 3937 (OusA+ OusB+) (⧫), 5512 (OusA− OusB+) (□), W245 (OusA+ OusB−) (▴), and W246 (OusA− OusB−) (○) were grown to mid-log phase in M63 medium with 0.3 M NaCl and then assayed for the uptake of [14C]GB at a final substrate concentration of 5.5 μM. (B) Effect of various osmoprotectants on [14C]GB binding to shock fluid proteins. Shock fluid periplasmic proteins (100 μg) obtained from cells of 3937 strain grown in M63 with 0.3 M NaCl were incubated with 250 pmol of [14C]GB (2.07 GBq mmol−1) in 10 mM Tris-HCl (pH 7.5). The assay volume was 50 μl. The competitors were added in 1-, 10-, 100-, and 1,000-fold excesses. Symbols: ⧫, control; ▪, GB; ▴, ectoine; ⋄, choline; ○, carnitine; •, DMSA; ▵, DMSP; □, proline. After 5 min, the proteins were precipitated by adding 950 μl of ice-cold saturated ammonium sulfate solution. Values are means of triplicate determinations; the standard error did not exceed 15%.
To analyze the substrate specificity of OusB, competition assays were undertaken with strain 5512 (ousA) grown in M63 with 0.3 M NaCl. The uptake of 5.5 μM [14C]GB (Table 2) was greatly inhibited by the addition of unlabeled GB and DMSA, with 100 and 95% inhibition, respectively, by a 100-fold excess of competitor (Table 2). A moderate inhibition was observed in the presence of unlabeled DMSP, choline, and ectoine with 68, 64, and 47% inhibition, respectively, using a 100-fold excess of competitor (Table 2). In contrast, pipecolate, carnitine, and proline had no effect on GB uptake activity via OusB under similar experimental conditions. These results showed that OusB is involved in the uptake of several substrates, but the main route for pipecolate, proline, and carnitine to enter the cells is OusA and not OusB.
TABLE 2.
Effect of competitors on GB uptake in E. chrysanthemi 5512
| Competitor | % Inhibitiona with excess of: |
||
|---|---|---|---|
| 10-fold | 50-fold | 100-fold | |
| GB | 87 | 97 | 100 |
| Ectoine | 17 | 35 | 47 |
| Choline | 29 | 43 | 64 |
| Carnitine | 2 | 8 | 12 |
| DMSA | 51 | 71 | 95 |
| DMSP | 21 | 45 | 68 |
| Pipecolate | 9 | 17 | 26 |
| Proline | 4 | 10 | 15 |
Cells were grown in M63 with 0.3 M NaCl. [14C]GB was used at 5.5 μM. The competitors were added at 10-, 50-, and 100-fold excesses. Results are given as the percentage of the control uptake rate in the absence of competitor. The rate of GB transport was 65 nmol/min/mg of protein. Values are means of duplicate determinations; the standard deviation did not exceed 5%.
Since the uptake of these compounds was mediated by the OusB transport system, we analyzed the involvement of its OusX protein component in osmoprotectant binding. Periplasmic fractions from strains 3937 and W245 grown in M63 medium with 0.3 M NaCl were prepared and incubated with labeled osmoprotectants, and the osmoprotectant-binding activity was measured as described in Materials and Methods. Among all assayed radiolabeled osmoprotectants (GB, proline, DMSA, DMSP, carnitine, ectoine, and choline), only GB was bound to periplasmic proteins. With the periplasmic fraction from the W245 strain, no GB-binding activity could be detected (data not shown), suggesting that ousX probably encodes the GB-binding protein of the OusB ABC-type transporter. Moreover, we have tested the ability of unlabeled osmoprotectants to compete with the binding of [14C]GB to the periplasmic protein, OusX. Ectoine, choline, carnitine, and proline in a 1,000-fold excess had no effect on GB binding (Fig. 5B). DMSP and DMSA had no effect on GB binding when they were added in 1- or 10-fold excess, but these two compounds allowed 38 and 69% inhibition, respectively, in a 100-fold excess (Fig. 5B). The inhibition reached 71 and 90%, respectively, when DMSP and DMSA were added in a 1,000-fold excess. The inhibition of GB-binding activity by S-methylated analogues of GB at high competitor/GB ratios is intriguing, since no labeled DMSA or DMSP was bound to periplasmic extracts. Only cold GB allowed 48% binding inhibition at a 1-fold excess and 84% binding inhibition at a 10-fold excess (Fig. 5B).
Uptake parameters and fate of choline in E. chrysanthemi.
Choline is a powerful osmoprotectant in E. coli that is able to convert it into GB through the action of betBA gene products (43, 63). Curiously, choline was a less effective osmoprotectant in E. chrysanthemi (Fig. 4). Since no osmoprotective effect of choline was observed in the double-mutant W246 and because OusB restored the uptake of choline in E. coli MKH13 impaired in proP, proU, and betT transporters (31), we measured [14C]choline uptake activity (50 μM) in wild type and mutant strains. The transport rate of choline decreased slightly in the ousA mutant in comparison to the wild-type strain, but choline uptake was strongly impaired in the ousB mutant (Fig. 6A). The choline transport activity was completely abolished in the ousA ousB mutant, indicating that OusB and OusA are the only choline transporters operating in E. chrysanthemi. The kinetic parameters of choline uptake by OusB were determined showing a Km of 2 μM and a Vmax of 50 nmol/min/mg of protein. These data demonstrated clearly that OusB was also a high-affinity transport system for choline in addition to GB. The fact that no choline uptake activity was detected in the W246 (ousA ousB) strain implied that E. chrysanthemi was defective in BetT-like choline uptake activity. This interpretation was confirmed by an analysis of the E. chrysanthemi 3937 genome (https://asap.ahabs.wisc.edu/annotation). This analysis showed that this bacterium possesses genes homologues to betI, betA, and betB of E. coli, a part of genes encoding the osmoregulatory choline/GB pathway (41), but the betT gene encoding a choline high-affinity transporter in E. coli (41, 43, 63) was missing in E. chrysanthemi 3937.
FIG. 6.
Uptake and fate of choline in E. chrysanthemi. (A) Strains 3937 (OusA+ OusB+) (⧫), 5512 (OusA− OusB+) (□), W245 (OusA+ OusB−) (▴), and W246 (OusA− OusB−) (○) were grown in M63 with 0.3 M NaCl to mid-log phase and assayed for the uptake of [14C]choline (188 MBq mmol−1) at a final substrate concentration of 50 μM. (B and C) Fate of [14C]choline supplied to E. chrysanthemi cells. Cells were grown in M63 with 0.3 M NaCl and 100 μM [14C]choline (94 MBq mmol−1); they were withdrawn at the mid-log (B) or stationary (C) phase to analyze the oxidation of choline. Cells were harvested by centrifugation, washed with isotonic minimal medium, and extracted by 80% ethanol. The ethanol soluble fraction was analyzed by paper electrophoresis (2,000 V/cm). Signals corresponding to choline (□) and GB (▪) were analyzed by Instant Imager, and the radioactivity corresponding to each spot was determined by scintillation counting. The results are means of duplicate determinations; the standard error did not exceed 10%.
To investigate the fate of choline in E. chrysanthemi, cells of the wild type and the mutant strains 5512, W245, and W246 were grown in M63 medium with 0.3 M NaCl in the presence of 100 μM [14C]choline. The cells were collected in the mid-logarithmic and stationary phases; the radioactivity of ethanol extracts, insoluble materials, and that remaining in the supernatant was further analyzed. When cells were harvested at mid-log phase, the radioactivities remaining in the external medium were 78, 85, 97, and 100% of the initial radioactivities for the wild type, 5512, W245, and W246 strains, respectively. The imported choline was oxidized partially in wild-type and mutant strains 5512 and W245 (Fig. 6B). The amounts of choline were 237, 243, and 13 nmol/mg of protein in the wild-type, 5512 (ousA), and W245 (ousB) strains, respectively (Fig. 6B), whereas those of synthesized GB from choline were 150, 105, and 64 nmol/mg of protein in the wild-type, 5512, and W245 strains, respectively (Fig. 6B). When cells were collected at the stationary phase, the radioactivities remaining in the supernatant were ca. 5% of the initial radioactivity for the wild-type and 5512 strains; meanwhile 69 and 100% of the initial radioactivity remained in the supernatants of W245 and W246 strains, respectively. The analysis by paper electrophoresis of the ethanol-soluble fractions from wild-type and 5512 strains showed that the imported choline was only partially oxidized into GB even in cells from the stationary phase, whereas in W245 cells most of the imported choline was transformed into GB (Fig. 6C). The intracellular levels of choline reached 220, 342, and 26 nmol/mg of protein in the wild-type, 5512, and W245 strains, respectively (Fig. 6C), and those of GB reached 500, 304, and 174 nmol/mg of protein, respectively, in the same strains (Fig. 6C). Thus, when OusA was the only functional osmoporter, the cells imported less choline, but they transformed most of it into GB. Meanwhile, when OusB was the solely functioning transporter, much more choline was taken up by the cells, but choline oxidation into GB was only partial. These data confirmed that OusB and OusA played a major and a minor role, respectively, in choline importation by E. chrysanthemi, and thus suggested that choline-GB conversion enzymes but not choline transport systems represents the limiting step responsible for the poor efficiency of choline as an osmoprotectant in E. chrysanthemi.
DISCUSSION
In this study, we have exploited the characteristic of the widespread ability of microorganisms to accumulate GB to deliver structurally related compounds with antibacterial activity (12). This allowed the characterization in E. chrysanthemi of an ousB operon encoding a multicomponent, binding-protein-dependent transport system involved in the high-affinity uptake of GB and choline. Bacterial binding-protein-dependent transport systems are members of a superfamily of prokaryotic and eukaryotic transporters known as ATP-binding cassette (ABC) transporters or traffic ATPases (18, 32). The OusB system that belongs to this family is composed of OusV, an ATPase, OusW, an integral inner membrane protein, and OusX a periplasmic substrate-binding protein. The deduced amino acid sequences of the three open reading frames showed high degrees of similarities with ProU from both S. enterica serovar Typhimurium and E. coli (29) and somewhat lower homology with Hut from S. meliloti (6); BusA (also named OpuA) from Lactococcus lactis (9, 50, 67); OpuA, OpuC, and OpuB from B. subtilis (37, 38); and OpuC and Gbu from Listeria monocytogenes (22, 40).
ousB expression (Fig. 2A and 2B), as well as that of ousA (28), is induced by increased medium osmolality. Although the level of ousA transcription is proportionally correlated with the osmolality of the growth medium up to 0.5 M NaCl (28), the expression of ousB does not increase above 0.3 M NaCl (Fig. 2B). In E. chrysanthemi, as well as in E. coli, an osmotic upshift results in an immediate reduction in turgor and a concomitant accumulation of K+-glutamate (21, 25), which functions as a “second messenger” that elicits the secondary responses (such as the osmoprotectant uptake systems) of the cell to adapt to high-osmolality conditions (7). The osmotic activation of the transcription of ousB from E. chrysanthemi, like that of proU from E. coli (45), is reversed by the presence of GB in the growth medium regardless of medium osmolality. In E. chrysanthemi, as in E. coli, GB triggers an efflux of K+ and consequently reduces the intracellular concentration of K+-glutamate (25, 45, 64). Since K+-glutamate, the “secondary messenger,” is the critical determinant for regulating the transcriptional level of proU in E. coli (45, 64), it may also play a similar role in E. chrysanthemi and reduce the expression of ousB in the presence of GB.
OusB and OusA are two osmoregulated transporters, which mediate the uptake of all assayed osmoprotectants in E. chrysanthemi. The characterization of the double-mutant strain W246 (ousA ousB) showed that no additional osmoprotectant transporter of physiological relevance operates in E. chrysanthemi (Fig. 5). OusA and OusB differ in their contribution to the overall GB accumulation by the cell, as reflected by their different Km and Vmax parameters. The ABC-type OusB system allows the bacterial cell to acquire GB from the environment even when this osmoprotectant is present at very low concentrations, but the OusA system is certainly the predominant GB transporter of E. chrysanthemi at high GB concentrations, even if it exhibits a relatively low affinity toward GB.
Four types of osmoprotectants transport systems have been reported until now in bacteria. One group is related to the binding protein-dependent ProU from E. coli and serovar Typhimurium (29, 62), and this class comprises E. chrysanthemi OusB; B. subtilis OpuA, OpuB, and OpuC (36-38); L. lactococcus BusA (50); L. monocytogenes Gbu and OpuC (22, 40); and S. meliloti Hut (6). The second group is the major facilitator superfamily, which is represented by the single-component ProP transporter from E. coli, serovar Typhimurium (11, 16, 52), and C. glutamicum (52), as well as E. chrysanthemi OusA (28). The third group comprises the betaine-choline-carnitine transporter family, which includes BetT and CaiT from E. coli (20, 41), OpuD from B. subtilis (36), BetM and EctM from Marinococcus halophilus (68), ButA from Tetragenococcus halophila (3), BetS from S. meliloti (8), BetP and EctP from C. glutamicum (51, 52), and BetL from L. monocytogenes (60). The latter group corresponds to the tripartite ATP-independent-periplasmic transporter family, represented by only one example, the Tea uptake system from Halomonas elongata (30). Most osmoporters of the first and second groups display a broad substrate specificity such as that of ProU and ProP from E. coli, whereas those belonging to the third and fourth groups generally have a stricter substrate specificity.
OusB behaves like ProU from E. coli; it mediates the uptake of all assayed osmoprotectants but only GB can bind to OusX, the periplasmic binding protein. In E. coli, GB and proline betaine are the only substrates, which can bind to the periplasmic protein encoded by proX (4, 27, 31, 57). The high-resolution structure of E. coli ProX in complex with GB and proline betaine was determined (57), and it showed that three tryptophan residues (Trp65, Trp140, and Trp188), which form a rectangular aromatic box, are involved in the cation-π interaction between the ProX protein and the positive charge of the quaternary amine of its ligands (57). These tryptophan residues and the entire motif 136CXPGWGC142 are strictly conserved in OusX and may also be involved in the GB binding activity of OusX. However, unlike the OusA and ProP systems, which transport GB with a relatively low affinity and exhibit Km values of 50 (28) and 44 (11) μM, respectively, OusB like ProU transports GB with a higher affinity and exhibits Km values of 1.6 and 1.3 μM (10), respectively. The E. chrysanthemi OusA (28) and OusB transport systems (the present study) proved to be functional in E. coli and to be related to the ProP and ProU osmoporters, respectively. The components of OusB system have similar sequences to those of ProU from E. coli, with a high degree of conservation of residues, which is accompanied by a common physiological function and kinetic properties. In contrast, there still remain some differences related to the physiological functions of osmoporters in E. chrysanthemi and E. coli. This concerns the uptake of choline, which is transported into the E. coli cells via the high- and low-affinity systems, BetT and ProU, respectively (41, 42), whereas in E. chrysanthemi choline is transported with a high and a low affinity through OusB and OusA, respectively (Fig. 2 and 3). These observations suggest that E. chrysanthemi lacks a BetT-like protein, which was confirmed by searching for bet genes in the whole genome sequence of E. chrysanthemi strain 3937 (https://asap.ahabs.wisc.edu/annotation). This bacterium has conserved betI, betA, and betB genes, but it lacks a betT homologue, probably as the result of a deletion event. In E. coli choline enters the cells via BetT and ProU transporters and is then oxidized into glycine betaine aldehyde, and in GB by a membrane-bound, FAD-containing choline dehydrogenase (BetA), which can oxidize both choline and glycine betaine aldehyde (2, 43, 63). A second, highly substrate-specific cytoplasmic enzyme (BetB) converts glycine betaine aldehyde into GB (41, 43). The E. coli strains that synthesize BetA but not BetB were protected against osmotic stress by choline. In this respect, they behaved similarly to the strains that synthesized both BetA and BetB dehydrogenases (2). The efficient osmoprotective effect of choline in E. coli is due essentially to the efficient choline and glycine betaine aldehyde oxidization activity of BetA and to the presence of functional BetT or ProU transport systems. The determination of BetA and BetB dehydrogenases activities in correlation with the lack of BetT-like transporter in E. chrysanthemi will probably provide an explanation for the partial conversion of choline into GB and for the lower osmoprotective efficiency of choline in E. chrysanthemi.
The data presented here showed that E. chrysanthemi possesses two osmoprotectant uptake systems and demonstrated the kinetic properties and physiological functions of the individual transporters.
Acknowledgments
We thank E. Bremer and S. Reverchon-Pescheux for providing the E. coli MKH13 and E. chrysanthemi A3779 strains, respectively. We also thank P. Uriac for the gift of BGB and T. Bernard and J.-A. Pocard for helpful discussions. C. Monnier, M. C. Savary, and M. Uguet are acknowledged for technical assistance.
Financial support for this study was provided by the Centre National de la Recherche Scientifique and the Ministère de la Recherche et de l'Education Nationale.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andresen, P. A., I. Kaasen, O. B. Styrvold, G. Boulnois, and A. R. Strøm. 1988. Molecular cloning, physical mapping, and expression of the bet genes governing the osmoregulatory choline-glycine betaine pathway of Escherichia coli. J. Gen. Microbiol. 134:1737-1746. [DOI] [PubMed] [Google Scholar]
- 3.Baliarda, A., H. Robert, M. Jebbar, C. Blanco, and C. Le Marrec. 2003. Isolation and characterization of ButA, a secondary glycine betaine transport system operating in Tetragenococcus halophila. Curr. Microbiol. 47:347-351. [DOI] [PubMed] [Google Scholar]
- 4.Barron, A., J. U. Jung, and M. Villarejo. 1987. Purification and characterization of a glycine betaine binding protein from Escherichia coli. J. Biol. Chem. 262:11841-11846. [PubMed] [Google Scholar]
- 5.Bernard, T., M. Jebbar, Y. Rassouli, S. Himdi-Kabbab, J. Hamelin, and C. Blanco. 1993. Ectoine accumulation and osmotic regulation in Brevibacterium linens. J. Gen. Microbiol. 139:129-138. [Google Scholar]
- 6.Boncompagni, E., L. Dupont, T. Mignot, M. Østerås, A. Lambert, M. C. Poggi, and D. Le Rudulier. 2000. Characterization of a Sinorhizobium meliloti ATP-binding cassette histidine transporter also involved in betaine and proline uptake. J. Bacteriol. 182:3717-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Booth, I. R., and C. F. Higgins. 1990. Enteric bacteria and osmotic stress: intracellular potassium glutamate as a secondary signal of osmotic stress? FEMS Microbiol. Rev. 75:239-246. [DOI] [PubMed] [Google Scholar]
- 8.Boscari, A., K. Mandon, L. Dupont, M. C. Poggi, and D. Le Rudulier. 2002. BetS is a major glycine betaine/proline betaine transporter required for early osmotic adjustment in Sinorhizobium meliloti. J. Bacteriol. 184:2654-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouvier, J., P. Bordes, Y. Romeo, A. Fourcans, I. Bouvier, and C. Gutierrez. 2000. Characterization of OpuA, a glycine-betaine uptake system of Lactococcus lactis. J. Mol. Microbiol. Biotechnol. 2:199-205. [PubMed] [Google Scholar]
- 10.Cairney, J., I. R. Booth, and C. F. Higgins. 1985. Osmoregulation of gene expression in Salmonella typhimurium: proU encodes an osmotically induced betaine transport system. J. Bacteriol. 164:1224-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cairney, J., I. R. Booth, and C. F. Higgins. 1985. Salmonella typhimurium proP gene encodes a transport system for the osmoprotectant betaine. J. Bacteriol. 164:1218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosquer, A., M. Ficamos, M. Jebbar, J. C. Corbel, G. Choquet, C. Fontenelle, P. Uriac, and T. Bernard. 2004. Antibacterial activity of glycine betaine analogues: involvement of osmoporters. Bioorg. Med. Chem. Lett. 14:2061-2065. [DOI] [PubMed] [Google Scholar]
- 13.Csonka, L. N. 1981. Proline over-production results in enhanced osmotolerance in Salmonella typhimurium. Mol. Gen. Genet. 182:82-86. [DOI] [PubMed] [Google Scholar]
- 14.Csonka, L. N., and W. Epstein. 1996. Osmoregulation, p. 1210-1223. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 15.Csonka, L. N., and A. D. Hanson. 1991. Prokaryotic osmoregulation: genetics and physiology. Annu. Rev. Microbiol. 45:569-606. [DOI] [PubMed] [Google Scholar]
- 16.Culham, D. E., B. Lasby, A. G. Marangoni, J. L. Milner, B. A. Steer, R. W. Van Nues, and J. M. Wood. 1993. Isolation and sequencing of Escherichia coli gene proP reveals unusual structural features of the osmoregulatory proline/betaine transporter, ProP. J. Mol. Biol. 229:268-276. [DOI] [PubMed] [Google Scholar]
- 17.Dattananda, C. S., and J. Gowrishankar. 1989. Osmoregulation in Escherichia coli: complementation analysis and gene-protein relationships in the proU locus. J. Bacteriol. 171:1915-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davidson, A. L., and J. Chen. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73:241-268. [DOI] [PubMed] [Google Scholar]
- 19.Dinnbier, U., E. Limpinsel, R. Schmid, and E. P. Bakker. 1988. Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch. Microbiol. 150:348-357. [DOI] [PubMed] [Google Scholar]
- 20.Eichler, K., F. Bourgis, A. Buchet, H. P. Kleber, and M. A. Mandrand-Berthelot. 1994. Molecular characterization of the cai operon necessary for carnitine metabolism in Escherichia coli. Mol. Microbiol. 13:775-786. [DOI] [PubMed] [Google Scholar]
- 21.Epstein, W. 1986. Osmoregulation by potassium transport in Escherichia coli. FEMS Microbiol. Rev. 39:73-78. [Google Scholar]
- 22.Fraser, K. R., D. Harvie, P. J. Coote, and C. P. O'Byrne. 2000. Identification and characterization of an ATP binding cassette l-carnitine transporter in Listeria monocytogenes. Appl. Environ. Microbiol. 66:4696-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galinski, E. A., H. P. Pfeiffer, and H. G. Trüper. 1985. 1,4,5,6-tetrahydro-2-methyl-4-pyrimidine-carboxylic acid: a novel cyclic amino acid from halophilic phototrophic bacterium Ectothiorhodospira. Eur. J. Biochem. 149:135-139. [DOI] [PubMed] [Google Scholar]
- 24.Giaever, H. M., O. B. Styrvold, I. Kaasen, and A. R. Strøm. 1988. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J. Bacteriol. 170:2841-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goude, R., S. Renaud, S. Bonnassie, T. Bernard, and C. Blanco. 2004. Glutamine, glutamate and α-glucosylglycerate are the major osmotic solutes accumulated by Erwinia chrysanthemi strain 3937. Appl. Environ. Microbiol. 70:6535-6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gouesbet, G., M. Jebbar, S. Bonnassie, N. Hugouvieux-Cotte-Pattate, S. Himdi-Kabbab, and C. Blanco. 1995. Erwinia chrysanthemi at high osmolarity: influence of osmoprotectants on growth and pectate lyase production. Microbiology 141:1407-1412. [DOI] [PubMed] [Google Scholar]
- 27.Gouesbet, G., M. Jebbar, R. Talibart, T. Bernard, and C. Blanco. 1994. Pipecolic acid is an osmoprotectant for Escherichia coli taken up by the general osmoporters ProU and ProP. Microbiology 140:2415-2422. [DOI] [PubMed] [Google Scholar]
- 28.Gouesbet, G., A. Trautwetter, S. Bonnassie, L. F. Wu, and C. Blanco. 1996. Characterization of the Erwinia chrysanthemi osmoprotectant transporter gene ousA. J. Bacteriol. 178:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gowrishankar, J. 1989. Nucleotide sequence of the osmoregulatory proU operon of Escherichia coli. J. Bacteriol. 171:1923-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grammann, K., A. Volke, and H. J. Kunte. 2002. New type of osmoregulated solute transporter identified in halophilic members of the bacteria domain: TRAP transporter TeaABC mediates uptake of ectoine and hydroxyectoine in Halomonas elongata DSM 2581T. J. Bacteriol. 184:3078-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haardt, M., B. Kempf, E. Faatz, and E. Bremer. 1995. The osmoprotectant proline betaine is a major substrate for the binding-protein-dependent transport system ProU of Escherichia coli K-12. Mol. Gen. Genet. 246:783-786. [DOI] [PubMed] [Google Scholar]
- 32.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 33.Jebbar, M., G. Gouesbet, S. Himdi-Kabbab, C. Blanco, and T. Bernard. 1995. Osmotic adaptation in Brevibacterium linens: differential effects of proline and glycine betaine on cytoplasmic osmolyte pool. Arch. Microbiol. 163:380-386. [Google Scholar]
- 34.Jebbar, M., R. Talibart, K. Gloux, T. Bernard, and C. Blanco. 1992. Osmoprotection of Escherichia coli by ectoine: uptake and accumulation characteristics. J. Bacteriol. 174:5027-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jebbar, M., C. von Blohn, and E. Bremer. 1997. Ectoine functions as an osmoprotectant in Bacillus subtilis and is accumulated via the ABC-transport system OpuC. FEMS Microbiol. Lett. 154:325-330. [Google Scholar]
- 36.Kappes, R. M., B. Kempf, and E. Bremer. 1996. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 178:5071-5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kappes, R. M., B. Kempf, S. Kneip, J. Boch, J. Gade, J. Meier-Wagner, and E. Bremer. 1999. Two evolutionarily closely related ABC transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol. 32:203-216. [DOI] [PubMed] [Google Scholar]
- 38.Kempf, B., and E. Bremer. 1995. OpuA, an osmotically regulated binding protein-dependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis. J. Biol. Chem. 270:16701-16713. [DOI] [PubMed] [Google Scholar]
- 39.Kempf, B., and E. Bremer. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170:319-330. [DOI] [PubMed] [Google Scholar]
- 40.Ko, R., and L. T. Smith. 1999. Identification of an ATP-driven, osmoregulated glycine betaine transport system in Listeria monocytogenes. Appl. Environ. Microbiol. 65:4040-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamark, T., I. Kaasen, M. W. Eshoo, P. Falkenberg, J. McDougall, and A. R. Strøm. 1991. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol. Microbiol. 5:1049-1064. [DOI] [PubMed] [Google Scholar]
- 42.Lamark, T., O. B. Styrvold, and A. R. Strøm. 1992. Efflux of choline and glycine betaine from osmoregulating cells of Escherichia coli. FEMS Microbiol. Lett. 96:149-154. [DOI] [PubMed] [Google Scholar]
- 43.Landfald, B., and A. R. Strøm. 1986. Choline-glycine betaine pathway confers a high level of osmotic tolerance in Escherichia coli. J. Bacteriol. 165:849-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 45.Lucht, J. M., and E. Bremer. 1994. Adaptation of Escherichia coli to high osmolarity environments: osmoregulation of the high-affinity glycine betaine transport system ProU. FEMS Microbiol. Rev. 14:3-20. [DOI] [PubMed] [Google Scholar]
- 46.Martinez, E., B. Bartolomé, and F. de la Cruz. 1988. pACYC184-derived cloning vectors containing the multiple cloning site and lacZα reporter gene of pUC8/9 and pUC18/19 plasmids. Gene 68:159-162. [DOI] [PubMed] [Google Scholar]
- 47.May, G., E. Faatz, J. M. Lucht, M. Haardt, M. Bolliger, and E. Bremer. 1989. Characterization of the osmoregulated Escherichia coli proU promoter and identification of ProV as a membrane-associated protein. Mol. Microbiol. 3:1521-1531. [DOI] [PubMed] [Google Scholar]
- 48.McLaggan, D., J. Naprstek, E. T. Buurman, and W. Epstein. 1994. Interdependence of K+ and glutamate accumulation during osmotic adaptation of Escherichia coli. J. Biol. Chem. 269:1911-1917. [PubMed] [Google Scholar]
- 49.Nasser, W., M. Faelen, N. Hugouvieux-Cotte-Pattat, and S. Reverchon. 2001. Role of the nucleoid-associated protein H-NS in the synthesis of virulence factors in the phytopathogenic bacterium Erwinia chrysanthemi. Mol. Plant-Microbe Interact. 14:10-20. [DOI] [PubMed] [Google Scholar]
- 50.Obis, D., A. Guillot, J. C. Gripon, P. Renault, A. Bolotin, and M. Y. Mistou. 1999. Genetic and biochemical characterization of a high-affinity betaine uptake system (BusA) in Lactococcus lactis reveals a new functional organization within bacterial ABC transporters. J. Bacteriol. 181:6238-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peter, H., A. Burkovski, and R. Krämer. 1996. Isolation, characterization, and expression of the Corynebacterium glutamicum betP gene, encoding the transport system for the compatible solute glycine betaine. J. Bacteriol. 178:5229-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peter, H., B. Weil, A. Burkovski, R. Krämer, and S. Morbach. 1998. Corynebacterium glutamicum is equipped with four secondary carriers for compatible solutes: identification, sequencing, and characterization of the proline/ectoine uptake system, ProP, and the ectoine/proline/glycine betaine carrier, EctP. J. Bacteriol. 180:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pichereau, V., J.-A. Pocard, J. Hamelin, C. Blanco, and T. Bernard. 1998. Differential effects of dimethylsulfoniopropionate, dimethylsulfonioacetate, and other S-methylated compounds on the growth of Sinorhizobium meliloti at low and high osmolarities. Appl. Environ. Microbiol. 64:1420-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Resibois, A., M. Colet, M. Faelen, E. Schoonejans, and A. Toussaint. 1984. ΦEC2, a new generalized transducing phage of Erwinia chrysanthemi. Virology 137:102-112. [DOI] [PubMed] [Google Scholar]
- 55.Richarme, G., and A. Kepes. 1983. Study of binding protein-ligand interaction by ammonium sulfate-assisted adsorption on cellulose esters filters. Biochim. Biophys. Acta 742:16-24. [DOI] [PubMed] [Google Scholar]
- 56.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 57.Schiefner, A., J. Breed, L. Bosser, S. Kneip, J. Gade, G. Holtmann, K. Diederichs, W. Welte, and E. Bremer. 2004. Cation-π interactions as determinants for binding of the compatible solutes glycine betaine and proline betaine by the periplasmic ligand-binding protein ProX from Escherichia coli. J. Biol. Chem. 279:5588-5596. [DOI] [PubMed] [Google Scholar]
- 58.Simon, R., M. O'Connell, M. Labes, and A. Pühler. 1986. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 118:640-659. [DOI] [PubMed] [Google Scholar]
- 59.Simon, R., J. Quandt, and W. Klipp. 1989. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in gram-negative bacteria. Gene 80:161-169. [DOI] [PubMed] [Google Scholar]
- 60.Sleator, R. D., C. G. Gahan, T. Abee, and C. Hill. 1999. Identification and disruption of BetL, a secondary glycine betaine transport system linked to the salt tolerance of Listeria monocytogenes LO28. Appl. Environ. Microbiol. 65:2078-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sleator, R. D., and C. Hill. 2002. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26:49-71. [DOI] [PubMed] [Google Scholar]
- 62.Stirling, D. A., C. S. Hulton, L. Waddell, S. F. Park, G. S. Stewart, I. R. Booth, and C. F. Higgins. 1989. Molecular characterization of the proU loci of Salmonella typhimurium and Escherichia coli encoding osmoregulated glycine betaine transport systems. Mol. Microbiol. 3:1025-1038. [DOI] [PubMed] [Google Scholar]
- 63.Styrvold, O. B., P. Falkenberg, B. Landfald, M. W. Eshoo, T. Bjornsen, and A. R. Strøm. 1986. Selection, mapping, and characterization of osmoregulatory mutants of Escherichia coli blocked in the choline-betaine pathway. J. Bacteriol. 165:856-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sutherland, L., J. Cairney, M. J. Elmore, I. R. Booth, and C. F. Higgins. 1986. Osmotic regulation of transcription: induction of the proU betaine transport gene is dependent on accumulation of intracellular potassium. J. Bacteriol. 168:805-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Touzé, T., G. Gouesbet, C. Boiangiu, M. Jebbar, S. Bonnassie, and C. Blanco. 2001. Glycine betaine loses its osmoprotective activity in a bspA strain of Erwinia chrysanthemi. Mol. Microbiol. 42:87-99. [DOI] [PubMed] [Google Scholar]
- 67.van der Heide, T., and B. Poolman. 2000. Osmoregulated ABC-transport system of Lactococcus lactis senses water stress via changes in the physical state of the membrane. Proc. Natl. Acad. Sci. USA 97:7102-7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vermeulen, V., and H. J. Kunte. 2004. Marinococcus halophilus DSM 20408(T)encodes two transporters for compatible solutes belonging to the betaine-carnitine-choline transporter family: identification and characterization of ectoine transporter EctM and glycine betaine transporter BetM. Extremophiles 8:175-184. [DOI] [PubMed] [Google Scholar]
- 69.Whatmore, A. M., J. A. Chudek, and R. H. Reed. 1990. The effect of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J. Gen. Microbiol. 136:2527-2535. [DOI] [PubMed] [Google Scholar]
- 70.Wood, J. M., E. Bremer, L. N. Csonka, R. Krämer, B. Poolman, T. van der Heide, and L. T. Smith. 2001. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130:437-460. [DOI] [PubMed] [Google Scholar]