Abstract
An operon involved in fructooligosaccharide breakdown was identified in the genome of Bifidobacterium breve UCC2003. This 2.6-kb transcriptional unit was comprised of three genes that encoded a putative permease, a conserved hypothetical protein, and a β-fructofuranosidase. Active transcription of the operon was observed when B. breve UCC2003 was grown on sucrose or Actilight, while transcription appeared to be repressed when the organism was grown on glucose, fructose, a combination of glucose and sucrose, or a combination of fructose and sucrose. The β-fructofuranosidase encoded by this operon was purified and biochemically characterized. The optimum pH and temperature for catalytic activity were determined to be pH 6.0 and 37°C, respectively, and there was a dependence on bivalent cations, particularly manganese. The Km and Vmax values for sucrose hydrolysis were determined to be 25 ± 2 mM and 24 ± 3 μmol min−1 mg−1, respectively. Interestingly, the enzyme was shown to specifically catalyze cleavage of the β(2-1) glycosidic bond between glucose and its neighboring fructose moiety in sucrose and other fructooligosaccharides with a relatively low degree of polymerization, and there was no detectable activity towards the β(2-1) glycosidic bond between two fructose moieties within the same substrate. To our knowledge, such an enzymatic activity has not previously been described in bifidobacteria or other gram-positive bacteria.
Prebiotics are nondigestible oligosaccharides that are not metabolized in the human small intestine and are passed to the colon, where they selectively stimulate growth and/or metabolic activity of beneficial microbial strains residing in the host intestine. The usual targets for prebiotic action are probiotic members of the bacterial genera Bifidobacterium and Lactobacillus (16, 39).
Fructooligosaccharides (FOS) are among the sugars that qualify as prebiotics. The FOS comprise a diverse family of naturally occurring oligosaccharides used commercially in food products and nutritional supplements. They represent the nondigestible oligosaccharides that are most used commercially and are composed of short to medium-size chains (degree of polymerization, 4 to 60) of fructose moieties connected by β(2-1) linkages, which are in turn attached to a terminal glucose unit [also by a β(2-1) bond]. Because of these β(2-1) linkages FOS are resistant to mammalian enzymes and thus are able to reach the colon, where they are reported to serve as a source of highly digestible substrates for bifidobacteria (11, 17, 29, 31). FOS and other oligosaccharides have been shown in vivo to beneficially modulate the composition of the intestinal microbiota by preferentially increasing the numbers of bifidobacteria and lactobacilli at the cost of potentially pathogenic or harmless bacteria (15, 16, 24). Despite the recent interest in FOS utilization, little information is available about the regulation of the metabolic pathways and enzymes responsible for transport and catabolism of such oligosaccharides in bifidobacteria.
Invertase (EC 3.2.1.26, β-fructofuranosidase) was one of the first described biocatalysts and has served as a paradigm for establishing many principles of enzymatic kinetics (30). Invertase catalyzes the hydrolysis of the β(2-1) glycosidic linkage of sucrose, releasing invert sugar, an equimolar mixture of glucose and fructose. Recent research on β-fructofuranosidase activities found in bifidobacteria has demonstrated that it breaks down not only sucrose but also FOS (14, 21, 22, 29, 32, 43). Characterized bifidobacterial β-fructofuranosidases have been shown to display different substrate kinetics and biochemical properties (14, 21, 22, 32). An exoinulinase which exhibited β-fructofuranosidase activity was purified from Bifidobacterium infantis ATCC 15697 (43). This enzyme was shown to represent a monomeric protein (Mr, 70,000) that catalyzes the degradation of both sucrose and inulin. A β-fructofuranosidase was identified from B. infantis and was shown to be composed of three identical subunits (Mr, 75,000) and to be capable of catalyzing the hydrolysis of sucrose, 1-kestose, nystose, inulin, and raffinose (21). The gene structure and enzymatic properties of a β-fructofuranosidase from Bifidobacterium lactis DSM10140T have been described by two groups, which obtained somewhat conflicting outcomes (14, 22). Ehrmann et al. (14) reported that the enzyme had the highest activity with sucrose, whereas Janer et al. (22) reported that the same enzyme cleaved terminal β(2-1) glycosyl linkages between fructose moieties, with the highest level of activity for Raftilose and the lowest level of activity for sucrose.
In this paper, we describe the genetic structure of an operon harboring a gene encoding a novel FOS-degrading enzyme with an affinity for only the β(2-1) glycosyl bonds between glucose and fructose moieties. A transcriptional analysis of this operon and flanking regions is described, as is the biochemical analysis of the FOS-degrading enzyme.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
Bifidobacterium breve UCC2003 (previously designated NCIMB8807), originally isolated from an infant nursling stool (37), was used in this study. Cells were cultured in modified de Man-Rogosa-Sharpe medium (12) made from its individual components and omitting any carbon source, which was supplemented with 1% sucrose as the sole carbon source unless indicated otherwise. de Man-Rogosa-Sharpe medium was supplemented with 0.05% (wt/vol) l-cysteine-HCl, and strains were grown at 37°C under anaerobic conditions that were maintained using an Anaerocult oxygen-depleting system (Merck, Darmstadt, Germany) in an anaerobic chamber. Escherichia coli strains Pet101/DTOPO and BL21 Star (Invitrogen, Carlsbad, Calif.) were used as cloning and expression hosts, respectively, and were routinely cultured aerobically at 37°C in Luria-Bertani medium. When appropriate, this medium was supplemented with 100 μg ml−1 ampicillin for plasmid maintenance and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for induction of target gene expression.
Carbohydrate sources.
Neosugar (Actilight), manufactured from sucrose, was supplied by Beghin-Meiji Industries (Neuilly/Seine, France) and had the following composition: oligofructose (GF2 [1-kestose], GF3 [nystose], and GF4 [1F-fructofuranosylnystose]) in prevailing quantities (∼96%, wt/wt), and glucose (G), fructose (F), and sucrose (GF) (<4%, wt/wt). The fructooligosaccharide standard set comprised 1-kestose, nystose, and 1F-fructofuranosylnystose (Wako Chemicals, Germany). Other carbohydrates used were sucrose (Merck); raffinose, Raftilose, and Raftiline (Orafti, Tienen, Belgium); and inulin, stachyose, palatinose, melibiose, melezitose, trehalose, lactose, maltose, and cellobiose (Sigma). A final concentration of 1% (wt/vol) of each of the above carbon sources was used for the experiments described in this paper.
Bioinformatics.
Database searches were performed using nonredundant sequences accessible at the National Center for Biotechnology Information internet site (http://www.ncbi.nlm.nih.gov) using the tBlastN, tBlastX, and BlastP programs (1, 2). Multiple-sequence alignment was performed using the Clustal method of the MEGALIGN program of the DNASTAR software package (DNASTAR, Madison, WI). The biological software programs SignalP (http://www.cbs.dtu.dk/services/SignalP), iPSORT (http://www.hypothesiscreator.net/iPSORT), TMPRED (http://www.ch.embnet.org/software/TMPRED_form.html), and DAS (http://www.sbc.su.se/∼miklos/das/maindas.html) were used to predict the location of proteins.
DNA manipulations.
Primers FruA (5′-CACCATGACTGACTTCACTCCC) and FruB (5′-CTCCAGTCCGATGGACTTCATGTG) were used to amplify the fosC gene from B. breve UCC2003. PCRs were performed using the Tgo I DNA polymerase template PCR system (Roche Diagnostics GmbH, Mannheim, Germany) in accordance with the manufacturer's instructions. PCRs were carried out using a Primus thermal cycler (MWG-BIOTECH AG, Ebersberg, Germany). Plasmid DNA was obtained from E. coli using a QIAprep Spin Plasmid Miniprep kit (QIAGEN GmbH, Hilden, Germany). Large-scale preparation of total DNA from B. breve was performed as described previously (34). Purified DNA was obtained by cesium chloride ultracentrifugation of the preparation (38). The amplified fosC gene was cloned into pET101/D-TOPO, which allowed translational fusion between a C-terminal six-His tag and a target gene (Invitrogen), and was transformed into E. coli BL21 Star for overexpression. The enzyme was purified using a nickel-nitrilotriacetic acid column (QIAGEN GmbH, Hilden, Germany) for purification of six-His-tagged proteins and was dialyzed against 20 mM Tris (pH 7.0) and 1 mM dithiothreitol for 24 h. Samples taken at different stages of the purification process and the purified protein were analyzed by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis (SDS-PAGE) as described by Laemmli et al. (27). Samples were denatured at 100°C for 5 min with an equal volume of sample buffer (50 mM Tris-HCl, 3% SDS, 1% β-mercaptoethanol, 20% glycerol, 0.7% bromophenol blue; pH 6.8) and applied to a 12.5% polyacrylamide gel using a mini-Protean II system (Bio-Rad Laboratories, Richmond, CA). Gels were stained with Coomassie brilliant blue R-250 (Sigma) to visualize proteins.
Enzyme and protein assays.
β-Fructofuranosidase activity was assayed using an adaptation of the dinitrosalicylic acid (DNS) assay (6), which measures the reducing sugars released (in this case glucose and fructose). The modifications to the DNS assay were as follows: a known concentration of enzyme was added to 1.9 ml of 0.3 M sucrose in 0.05 M acetate buffer, pH 6.0. For substrates that already contained a reducing sugar (Raftilose, maltose, cellobiose, lactose, melibiose, and palatinose) the glucose oxidase assay was employed (5), which measured the amount of glucose released. The glucose oxidase assay was also used for the other substrates (nonreducing sugars) to compare the results to the results of the DNS assay. The amount of liberated fructose was determined using a d-fructose determination kit (Sigma). In all assays β-fructofuranosidase activity was expressed in μmol glucose or fructose produced min−1 mg−1 protein. Protein concentrations were determined using the Bradford method (7). The DNS assay was used for measurement of enzymatic activity with sucrose for determination of enzyme properties.
(i) Effects on enzyme activity.
The enzyme activity was measured in two different types of buffers of various pHs, a 0.05 M sodium acetate buffer with the pH ranging from 2 to 5.5 and a 0.05 M sodium phosphate buffer with the pH ranging from 5.5 to 8. Standard enzymatic assays were conducted at temperatures ranging from 4 to 60°C. The enzyme and substrate-buffer solutions were preincubated separately for 20 min at a particular temperature before the reaction was started by adding the buffer and the enzyme together. The activity of the treated enzyme was assayed under the standard assay conditions (37°C and pH 6.0). To determine the dependence of enzyme activity on metal ions, a purified enzyme preparation was dialyzed against 20 mM EDTA (pH 7.0) at 4°C for 12 h, followed by dialysis against sodium phosphate buffer (pH 6.0) for 12 h at 4°C. The enzyme was then incubated for 10 min at 37°C with one of the following compounds present at a concentration of 1 mM in sodium phosphate buffer, pH 6.0: MnCl2, MgCl2, CoCl2, Fe2(SO4)3, KCl, CuSO4, CaCl2, ZnCl2, AgNO3, or HgCl2. The enzymatic activity recovered was measured following addition of the substrate sucrose using the standard conditions (37°C and pH 6.0). The metal content of the purified enzyme was determined by inductively coupled plasma atomic emission spectrometry at the Department of Chemistry, University of Sheffield, England.
(ii) Kinetic parameters.
Reaction rates were measured separately at pH 6.0 and at 37°C with sucrose at concentrations ranging from 0.02 to 0.4 M. Km and Vmax values were obtained by fitting the data obtained to the Michaelis-Menten equation using the nonlinear regression libraries from R (20).
(iii) HPTLC analysis.
For qualitative determination of enzyme activity, sugars incubated with the purified enzyme or crude extract were analyzed by high-performance thin-layer chromatography (HPTLC). An aliquot (0.5 μl) of the reaction mixture was spotted onto a Silica Gel 60 plate (10 by 10 cm; Merck) with a Nanomat 4 (Camag, Switzerland). The chromatogram was developed with a butanol-acetic acid-water (5:4:1, vol/vol/vol) solvent system in a horizontal developing chamber. Ascending development was repeated twice at room temperature. The plate was allowed to air dry in a hood and then developed by spraying it evenly with 20% (vol/vol) sulfuric acid in ethanol. The plate was dried and heated at 120°C for 10 min to visualize the sugar-containing spots. The plate was scanned immediately and analyzed using ImageJ NIH (National Institutes of Health).
RNA isolation and Northern analysis.
Aliquots of total RNA were isolated from B. breve UCC2003 in the early exponential phase using the Macaloid method (25). RNA samples were treated with a DNase and RNase inhibitor (Roche Diagnostics), denatured at 70°C for 10 min, and loaded along with a formamide-containing dye on a 1.2% formaldehyde agarose gel (4). RNA size standards (Promega) were included to estimate the sizes of the transcripts. Capillary blotting to Hybond-N+ nylon membranes (Amersham, Little Chalfont, Buckinghamshire, United Kingdom) was performed (38). Internal 500-bp fragments, amplified using PCR for each of the identified open reading frames (ORFs) of the fos locus, were used as probes. The probes were radiolabeled with α-32P using a Prime-a-Gene kit (Promega). Overnight hybridization was performed in 0.5 M sodium phosphate (pH 7.0) and 5% SDS buffer at 50°C. Blots were washed with 2× SSC-0.1% SDS, followed by 0.2× SSC-0.1% SDS and 0.1× SSC-0.1% SDS, at temperatures ranging from 55 to 65°C, depending on the stringency required (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
Nucleotide sequence accession numbers.
The sequences of the fos loci have been deposited in the GenBank database under the following accession numbers: AY692230 (lacIfos), AY692231 (fosA), AY692232 (fosB), AY549965 (fosC), and AY692233 (glkA).
RESULTS
Identification and analysis of a B. breve UCC2003 β-fructofuanosidase-encoding gene and flanking regions.
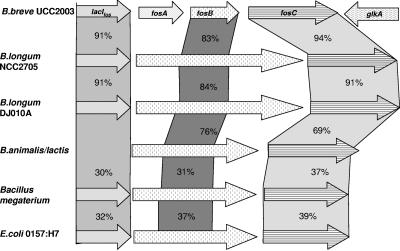
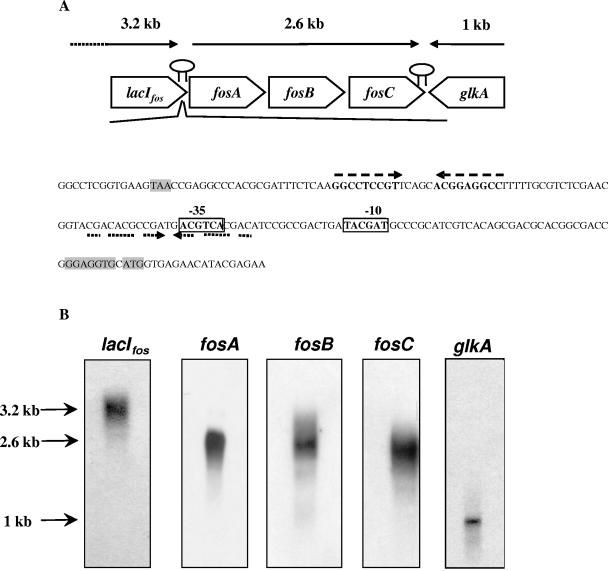
A gene designated fosC, predicted to encode β-fructofuranosidase activity, was identified from the preliminary annotation of the genome sequence of B. breve UCC2003 (S. Leahy, J. Moreno Munoz, M. O'Connell-Motherway, G. F. Fitzgerald, D. Higgins, and D. van Sinderen, unpublished data). At the protein level, the predicted product of fosC displays significant similarity to predicted β-fructofuranosidases encoded in the genome sequences of Bifidobacterium longum strains NCC2705 and DJ010A and, although to a lesser degree, a β-fructofuranosidase from Bifidobacterium animalis (Fig. 1). However, when it was compared to the B. lactis β-fructofuranosidase characterized by Ehrmann et al. (14) and Janer et al. (22), the degree of identity dropped to 46%. When the sequence was aligned with the sequences of other characterized β-fructofuranosidases, residues that represent highly conserved regions and the active site were found (14, 18) (results not shown). Bioinformatic analysis indicated that the B. breve fosC product, similar to previously characterized bifidobacterial β-fructofuranosidases, lacks secretory signals or transmembrane helices and is therefore predicted to be an intracellular enzyme. Immediately upstream from fosC, the fosB and fosA genes were identified (Fig. 1). The predicted product of fosB consists of 157 amino acids (Mw, 17,000) and exhibits similarity to a central region of a hypothetical sucrose permease of B. longum NCC2705, B. animalis, and E. coli 0157:H7 EDL933 (Fig. 1). Furthermore, significant similarity was observed between fosB and the central portion of an E. coli anion sucrose symport system (Fig. 1). The fosB-encoded protein was predicted to have a hydrophilic N terminus and four potential transmembrane helices. This secondary structure is common in proteins that belong to the sugar transport family (25). The fosA gene is predicted to encode a cytosolic protein consisting of 173 amino acids with similarity to members of the COG4283 family, which is a family of uncharacterized conserved proteins for which no function has been assigned, although genes encoding COG4283 members are frequently associated with genes that are predicted to be involved in transport (results not shown). Upstream from fosA, an ORF designated lacIfos, encoding a probable transcriptional regulator, was identified. At the protein level, the lacIfos gene product displays significant similarity to members of the GalR-LacI family of bacterial transcriptional regulators, particularly those found in the related strains B. longum NCC2705 and B. longum DJ010A (Fig. 1). The lacIfos product also showed identity to proven sucrose regulators, such as the sucrose operon repressor (Csc regulatory protein) of E. coli (level of identity, 46%; accession no. P40715) and Shigella flexneri (level of identity, 46%; accession no. NP_708235). A helix-turn-helix region (pfam00354) was identified within the N-terminal portion of LacIfos, which is a region found in the GalR-LacI family implicated in DNA binding, while the C-terminal end of LacIfos contains an expected sugar-binding domain (pfam00532). Immediately downstream of fosC, a putative glucokinase gene (glkA) was identified in an orientation opposite that of fosC, while a putative rho-independent terminator structure with a ΔG of −30.2 kcal/M was identified between fosC and glkA (data not shown). Also, between lacIfos and fosA two inverted repeats were identified. The first repeat was a perfect repeat (ΔG = −16.9 kcal/M) and was followed by a stretch of consecutive thymines, which may represent a Rho-independent transcriptional terminator, while the second repeat was an imperfect repeat (ΔG = −5.6 kcal/M) and may be involved in transcriptional regulation (Fig. 2A) (see below).
FIG. 1.
Comparison of various presumed or characterized fos operons. The putative function of the protein is indicated in the arrows for B. breve UCC2003. Related proteins were compared to B. breve UCC2003 proteins and are linked by bars indicating the different levels of amino acid identity, expressed as percentages.
FIG. 2.
(A) Sequence of the promoter region between lacIfos and fosA. The start and stop codons and putative ribosome binding site are shaded. The putative −10 and −35 regions are enclosed in boxes, the putative terminator between lacIfos and fosA is indicated by two dashed arrows, and the putative regulatory region involving the inverted repeat is indicated by two dotted arrows. The terminator is indicated by hairpin structures. (B) Northern analysis of RNA isolated from B. breve UCC2003 grown to the mid-logarithmic phase on sucrose and hybridized with 500-bp probes specific for lacIfos, fosA, fosB, fosC, and glkA.
Transcriptional analysis of the fos operon.
In order to elucidate the manner in which the assumed β-fructofuranosidase-encoding fosC gene is transcribed, Northern blot analysis was performed using PCR-generated probes corresponding to the central region of each of the five ORFs described above. Blots representing the total RNA isolated from sucrose-grown cells of B. breve strain UCC2003 were hybridized with these probes. The probes corresponding to fosA, fosB, and fosC (Table 1) all hybridized with a 2.6-kb transcript, indicating that these genes were transcribed as a tricistronic operon (Fig. 2B). Northern blot analysis using a probe corresponding to lacIfos revealed a 3.2 kb transcript, which showed that it was not transcribed with the other genes in this operon, but its proximity to the fos operon indicated that it might be involved in the regulation of the operon. The probe corresponding to the predicted glkA glucokinase-encoding gene was transcribed as a monocistronic operon with a 1-kb transcript (Fig. 2B). A schematic representation of the fos locus and its transcriptional patterns are shown in Fig. 2A.
TABLE 1.
Primers used to amplify internal fragments of the genes described in this paper that were used as probes for Northern hybridization
| Gene | Forward primer | Reverse primer |
|---|---|---|
| lacIfos | CCACCGGCGCCGATCAGG | AGGTCGGCGGCGACGGCTG |
| fosA | CAGGAGCTCAAAGAGGAG | CTGAACGATCCGAACGGG |
| fosB | TGTCCAGTTCGTCTACGC | ATGAAGACGATGAGCACC |
| fosC | GTGGCTGTTCTCCTCCAA | TCGGTGGCGTGAATCTTC |
| glkA | CCGCTACGACGCCAATGG | CCACCTGATCCGCGCCGC |
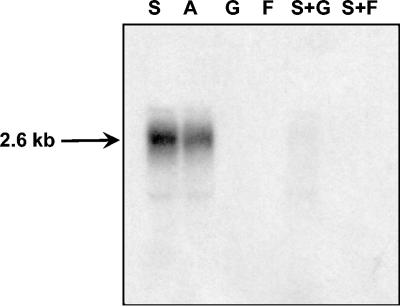
A 2.6-kb transcript was also observed with RNA isolated from cells grown in Actilight, but no transcriptional signals were observed using RNA isolated from cells grown in glucose, fructose, a combination of sucrose and glucose, or a combination of sucrose and fructose (Fig. 3). These findings show that the fos operon is subject to a dual transcriptional control mechanism that is dependent on carbohydrate availability.
FIG. 3.
Northern analysis of RNA isolated from B. breve UCC2003 grown to the mid-logarithmic phase on either sucrose (lane S), Actilight (lane A), glucose (lane G), fructose (lane F), sucrose and glucose (lane S+G), or sucrose and fructose (lane S+F). The blot was hybridized with a 500-bp probe specific for fosC. The estimated size of the transcripts is indicated.
Cloning, expression, and purification.
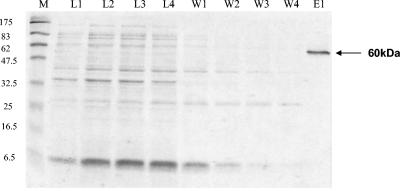
The PCR product encompassing fosC was obtained and cloned into an E. coli His-tagged expression system (see Materials and Methods). The cloned fosC gene product containing a His tag attached to its C terminus was purified from the soluble cell extract fraction of E. coli BL21 Star harboring recombinant plasmid pFruH using metal chelate affinity chromatography. SDS-PAGE analysis of purified FosC-His revealed a single band at an apparent molecular mass of approximately 60 kDa (Fig. 4), which is in agreement with the expected size calculated from the recombinant gene sequence (58.36 kDa).
FIG. 4.
SDS-PAGE analysis of purified B. breve UCC2003 β-fructofuranosidase. Proteins were denatured, separated on a 12.5% polyacrylamide gel, and stained with Coomassie blue R250. Lane M, molecular mass marker; lanes L1 to L4, lysate fractions; lanes W1 to W4, wash fractions; lane E1, purified β-fructofuranosidase.
FosC represents a bifidobacterial β-fructofuranosidase with novel glycolytic specificity.
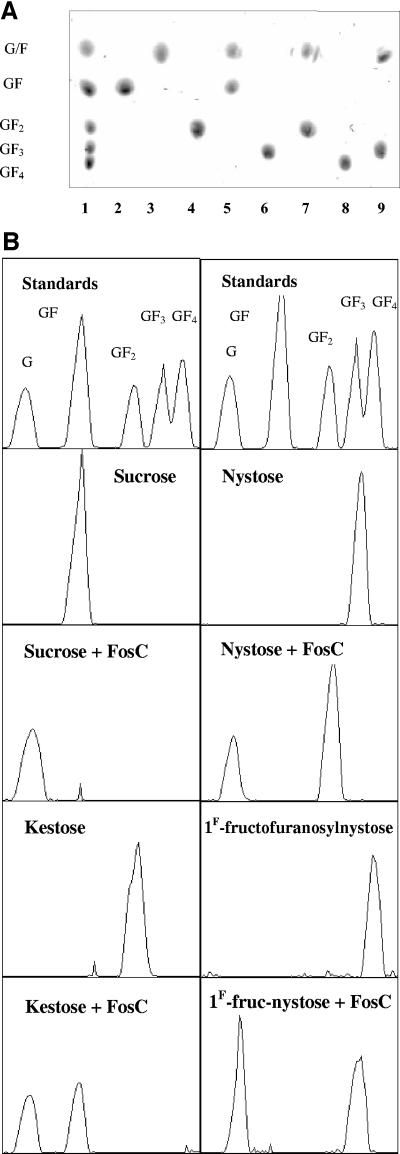
Purified B. breve FosC-His was shown to liberate glucose only from linear β(2-1)-linked carbohydrates like sucrose, 1-kestose, nystose, 1F-fructofuranosylnystose, Actilight, Raftilose, Raftiline, and inulin with relative activities of 100%, 91%, 90%, 85%, 84%, 69%, 28%, and 27%, respectively (Table 2). HPTLC analysis indicated that FosC specifically hydrolyzes the β(2-1) bond between glucose and fructose moieties. However, the Rf values of glucose and fructose obtained using HPTLC, as well as the Rf values of their oligomers (e.g., levanbiose and sucrose, levantriose and 1-kestose), were similar and therefore it was not possible to distinguish one from another using HPTLC. Therefore, in order to verify the assumed specificity, liberated d-glucose and d-fructose were measured in each of the reactions described above. When incubated with FosC-His, sucrose was hydrolyzed to glucose and fructose; 1-kestose was hydrolyzed to levanbiose (two fructose molecules, F2) and glucose; nystose was hydrolyzed to levantriose (F3) and glucose; and 1F-fructofuranosylnystose was hydrolyzed to levantetraose (F4) and glucose (Fig. 5). The HPTLC results combined with measurement of liberated d-glucose and d-fructose confirmed that the enzyme cleaved only the β(2-1) glycosidic bond between glucose and fructose moieties, leaving only a chain of fructose molecules. Substrates containing linkages other than β(2-1) linkages, such as lactose, cellobiose, stachyose, palatinose, melbiose, melezitose, trehalose, and maltose, were shown not to be degraded by the enzyme (results not shown). Also, raffinose, which contains an internal β(2-1) glycosidic bond, was not degraded by the enzyme.
TABLE 2.
Relative activities of β-fructofuranosidase and growth in each of the substrates relative to growth in sucrose of B. breve UCC2003a
| Substrate | Structure | Relative activity (%) | Growth |
|---|---|---|---|
| Sucrose | Glcβ(2-1)Fru | 100 | + |
| 1-Kestose | Glcβ(2-1)Frun (n = 2) | 91 | + |
| Nystose | Glcβ(2-1)Frun (n = 3) | 90 | + |
| 1F-Fructofurano- sylnystose | Glcβ(2-1)Frun (n = 4) | 85 | + |
| Actilight | Glcβ(2-1)Frun (n = 2 to 4) | 84 | + |
| Raftilose | Glcβ(2-1)Frun (n = 1 to >20) | 69 | +/− |
| Raftiline | Glcβ(2-1)Frunβ(2-1)Fru (n = 1 to >60) | 28 | − |
| Inulin | Glcβ(2-1)Frunβ(2-1)Fru (n = 1 to >60) | 27 | − |
+, growth; +/−, poor growth; −, no growth.
FIG. 5.
(A) HPTLC analysis of the reaction products produced by FosC. Lane 1, standard sugars; lane 2, sucrose; lane 3, sucrose plus FosC; lane 4, kestose; lane 5, kestose plus FosC; lane 6, nystose; lane 7, nystose plus FosC; lane 8, 1F-fructofuranosylnystose; lane 9, 1F-fructofuranosylnystose plus FosC. (B) Chromatograms of the reaction products produced by FosC.
The end products of native FosC activity derived from crude cell extracts were also analyzed by HPTLC. When sucrose, 1-kestose, nystose, and 1F-fructofuranosylnystose were incubated with crude extract from B. breve UCC2003, the end products obtained were identical to those obtained with the purified recombinant FosC (data not shown).
Biochemical characteristics of FosC-His activity. (i) pH and temperature.
The optimum pH for B. breve UCC2003 β-fructofuranosidase activity was determined to be in the slightly acidic range, pH 6.0, and there was a marked decrease in β-fructofuranosidase activity at pH values below 5.5 and above 6.5. The thermostability of the purified enzyme was determined at temperatures ranging from 4 to 60°C in 0.05 M acetate buffer (pH 6.0) and 0.3 M sucrose. The FosC enzyme exhibited the highest activity at 37°C, which corresponds to the optimal growth temperature of this bacterium.
(ii) Catalytic properties.
Kinetic analysis using Michaelis-Menten kinetics indicated that the purified enzyme had an apparent Km for sucrose of 25 ± 2 μmol min−1 mg−1 and a Vmax of 24 ± 3 mM.
(iii) Metal cofactors.
Following dialysis of the purified enzyme against a buffer containing EDTA to remove all traces of bivalent metal ions, the enzyme was shown to have lost all activity. Following incubation with different bivalent metal ions, it was shown that 1 mM Mn2+ restored 96% of the total activity (Table 3). Other metal ions only partially restored the activity, whereas Hg2+, Ag+, or Na+ did not restore enzymatic activity. The metal content of the purified recombinant β-fructofuranosidase was also determined using atomic emission analysis (data not shown), and the results were in agreement with the metal dependency of the enzyme.
TABLE 3.
Effects of metal cofactors on the relative activity of β-fructofuranosidase from B. breve UCC2003
| Metala | Relative activity (%) |
|---|---|
| Mn2+ | 96 ± 1 |
| Co2+ | 40 ± 3 |
| K+ | 24 ± 1 |
| Fe2+ | 23 ± 2 |
| Ca2+ | 21 ± 2 |
| Cu2+ | 18 ± 3 |
| Mg2+ | 11 ± 1 |
| Zn2+ | 10 ± 2 |
The enzyme was dialyzed against EDTA and placed in a solution containing a metal at a concentration of 1 mM. No activity was found with Ag+, Hg2+, and Na+.
DISCUSSION
The ability of bifidobacteria to ferment fructooligosaccharides is well established (23, 28), although our knowledge about the biochemistry and genetics of the enzymes involved, particularly the regulation of transcription or expression of the gene, is still limited. The ability of β-fructofuranosidase to catalyze the hydrolysis of a variety of sugars (not just sucrose) and sugars derived from a diet of fruits and vegetables, such as bananas, leeks, onions, and wheat, is an important finding for explaining the survival and growth of Bifidobacterium spp. in the gastrointestinal tract. The β-fructofuranosidase activity corresponding to the fosC gene of B. breve UCC2003 was shown to specifically cleave the β(2-1) glycosyl linkages between glucose and fructose moieties of sucrose and FOS, while it did not catalyze cleavage of the same linkage between two fructose molecules in these substrates (Fig. 5). This specific β-fructofuranosidase activity has not been reported previously for the other bifidobacterial β-fructofuranosidases that have been characterized (14, 22, 32, 43) or for other gram-positive bacteria (10). It has been reported that long-chain FOS, such as inulin, stimulate growth of Bifidobacterium spp. (29), although the substrate uptake system by which this occurs has not been elucidated yet. FosC exhibits relatively low enzyme activity towards inulin, which also does not support growth of B. breve UCC2003. Poor growth on inulin has also been reported for other bifidobacteria (14, 22), suggesting that inulin, which is sold as an effective prebiotic, is not a suitable growth substrate for at least a number of bifidobacterial strains.
A gene encoding a putative repressor protein displaying significant similarity to the GalR-LacI family of bacterial transcriptional regulators was identified upstream of the gene encoding the conserved hypothetical protein. Analogous to other GalR-LacI-like regulatory proteins, B. breve UCC2003 LacIfos may in the absence of its substrate(s) (i.e., sucrose or other FOS) repress transcription by binding to an operator site within the promoter region of the fos operon. An analysis of the sequence between the lacIfos and fosA genes resulted in identification of an imperfect inverted repeat overlapping the putative −35 region of the predicted promoter of the fos operon which could act as a recognition site (Fig. 2A).
The genetic organization of the fos operon in B. breve UCC2003 was highly homologous to a region in B. longum NCC2705 predicted to be involved in FOS metabolism, although the latter organism lacks a fosA homologue (Fig. 1). The deduced fosA product of B. breve UCC2003 resembles a conserved hypothetical protein (Chp) which may have an accessory transport function, a notion that was deduced from the observation that genes surrounding other Chp-encoding genes are also associated with transporters.
Transcriptional analysis of the B. breve UCC2003 fos operon showed that induction of the operon occurs in the presence of sucrose and Actilight, which is a source of short-chain FOS; however, when B. breve UCC2003 was grown on a mixture of sucrose and glucose or a mixture of sucrose and fructose, transcription of the fos operon appeared to be repressed, indicating that there was a form of glucose- or fructose-dependent transcriptional control (Fig. 3). Therefore, induction occurred when the substrates sucrose and Actilight were present, but only in the absence of glucose or fructose. This suggests that there is a regulated mechanism for preferred carbohydrate utilization that depends on substrate availability (40). Such hierarchical control of carbohydrate metabolic pathways can be achieved through inhibition of expression of genes encoding enzymes that are involved in transport and metabolism of less preferred carbohydrates (9), inhibition of the activity of enzymes that effect the uptake or production of the transcriptional inducer (inducer exclusion) (13), and stimulation of the efflux of an intracellular inducer (i.e., the carbohydrate or the phosphorylated derivative) (inducer expulsion) (36). The inducer exclusion and expulsion mechanisms result in lowering of the intracellular inducer concentration and thereby indirectly affect gene expression (35).
Catabolism of sucrose makes fructose and glucose readily available in the glucose fermentation pathway, where glucose and fructose are converted to fructose 6-phosphate for utilization in the fructose 6-phosphate shunt that is typical of bifidobacteria (39). Catabolic repression of genes involved in carbohydrate metabolism has been reported in Bifidobacterium spp. (41). Nevertheless, the mechanism by which this catabolic repression occurs remains to be determined. Recent work conducted with high-G+C-content bacteria (e.g., Streptomyces coelicolor) has shown that the presence of HPr (phosphocarrier protein) is not necessary for general carbon regulation (33). Moreover, catabolic repression acting independently of a phosphoenolpyruvate phosphotransferase system (PTS) system was demonstrated in both high-G+C-content (S. coelicolor) and low-G+C-content (Staphylococcus xylosus and Bacillus megaterium) gram-positive bacteria (8, 19, 42). In each case, a central function in catabolic repression was attributed to an ROK (repressor, ORF, kinase) family glucose kinase, namely, the glkA product (3, 26, 42). For example, in S. xylosus, the cloned glkA gene was shown to fully restore catabolite repression in the mutant strains in trans (42). Therefore, glucose may also enter the cell by a non-PTS transport system or systems. The efficiency with which PTS transport and non-PTS transport take up glucose may strongly depend on the growth conditions and the sugar concentrations. Interestingly, located downstream of B. breve UCC2003 fosC is a gene which is predicted to encode a glucokinase (Fig. 2A) exhibiting 44% identity to the GlkA protein belonging to the ROK family of S. coelicolor.
Although carbohydrate metabolism and its control in Bifidobacterium spp. are far from being understood, characterization of the fos operon is a significant step forward which should contribute to future research in this area.
Acknowledgments
We are grateful to Beghin-Meiji Industries (Neuilly/Seine, France) and Orafti (Tienen, Belgium) for the generous donation of specialty sugar products.
This work was financially supported by the Higher Education Authority Programme for Research in Third Level Institutions (HEA PRTLI and HEA PRTLIII programs), the Science Foundation Ireland Centre for Science Engineering and Technology, and the Food Institutional Research Measure (01/R&D/C/159) of the Department of Agriculture and Food.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angell, S., E. Schwarz, and M. J. Bibb. 1992. The glucose kinase gene of Streptomyces coelicolor A3(2): its nucleotide sequence, transcriptional analysis and role in glucose repression. Mol. Microbiol. 6:2833-2844. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Green Publishing Associates and Wiley-Interscience, New York, N.Y.
- 5.Bergmeyer, H. U., and E. Bernt. 1974. Determination with glucose oxidase and peroxidase, p. 1205-1212. In H. U. Bergmeyer (ed.), Methods of enzymatic analysis, 2nd ed. Academic Press, New York, N.Y.
- 6.Blatch, G. L., and D. R. Woods. 1993. Molecular characterization of a fructanase produced by Bacteroides fragilis BF-1. J. Bacteriol. 175:3058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Bruckner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209:141-148. [DOI] [PubMed] [Google Scholar]
- 9.Cohn, M., and K. Horibata. 1959. Physiology of the inhibition by glucose of the induced synthesis of β-galactosidase enzyme system of Escherichia coli. J. Bacteriol. 78:624-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuezzo de Gines, S., M. C. Maldonado, and G. Font de Valdez. 2000. Purification and characterization of invertase from Lactobacillus reuteri CRL 1100. Curr. Microbiol. 40:181-184. [DOI] [PubMed] [Google Scholar]
- 11.Cummings, J. H., G. T. Macfarlane, and G. R. Gibson. 1989. Quantitative estimate of fermentation in the hindgut of man. Acta Vet. Scand. 86:76-82. [PubMed] [Google Scholar]
- 12.De Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 13.Dills, S. S., A. Apperson, M. R Schmidt, and M. H Saier, Jr. 1980. Carbohydrate transport in bacteria. Microbiol. Rev. 44:385-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehrmann, M. A., M. Korakli, and R. F. Vogel. 2003. Identification of the gene for β-fructofuranosidase of Bifidobacterium lactis DSM10140(T) and characterization of the enzyme expressed in Escherichia coli. Curr. Microbiol. 46:391-397. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, G. R., E. R. Beatty, X. Wang, and J. H. Cummings. 1995. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology 108:975-982. [DOI] [PubMed] [Google Scholar]
- 16.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 17.Gilliland, S. E. 1990. Health and nutritional benefits from lactic acid bacteria. FEMS Microbiol. Rev. 87:175-188. [DOI] [PubMed] [Google Scholar]
- 18.Henrisaat, B. 1991. A classification of glycosyl hydrolases based on amino acids sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huynh, P. L., Jankovic, N. F. Schnell, and R. Bruckner. 2000. Characterization of an HPr kinase mutant of Staphylococcus xylosus. J. Bacteriol. 182:1895-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ihaka, R., and R. Gentleman. 1996. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5:299-314. [Google Scholar]
- 21.Imamura, L., K. Hisamitsu, and K. Kobashi. 1994. Purification and characterization of β-fructofuranosidase from Bifidobacterium infantis. Biol. Pharm. Bull. 17:596-602. [DOI] [PubMed] [Google Scholar]
- 22.Janer, C., L. M. Rohr, C. Pelaez, M. Laloi, V. Cleusix, T. Requena, and L. Meile. 2004. Hydrolysis of oligofructoses by the recombinant β-fructofuranosidase from Bifidobacterium lactis. Syst. Appl. Microbiol. 27:279-285. [DOI] [PubMed] [Google Scholar]
- 23.Kaplan, H., and R. W. Hutkins. 2000. Fermentation of fructooligosaccharides by lactic acid bacteria and bifidobacteria. Appl. Environ. Microbiol. 66:2682-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleessen, B., L. Hartmann, and M. Blaut. 2001. Oligofructose and long-chain inulin: influence on the gut microbial ecology of rats associated with a human faecal flora. Br. J. Nutr. 86:291-300. [DOI] [PubMed] [Google Scholar]
- 25.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterisation of the nisin gene cluster nisABTCIPR of Lactococcus lactis, requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 26.Kwakman, J. H. J. M., and P. W. Postma. 1994. Glucose kinase has a regulatory role in carbon catabolite repression in Streptomyces coelicolor. J. Bacteriol. 176:2694-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 28.Marx, S. P., S. Winkler, and W. Hartmeier. 2000. Metabolization of beta-(2,6)-linked fructose-oligosaccharides by different bifidobacteria. FEMS Microbiol. Lett. 182:163-169. [DOI] [PubMed] [Google Scholar]
- 29.McKellar, R. C., and H. W. Modler. 1989. Metabolism of fructo-oligosaccharides by Bifidobacterium spp. Appl. Microbiol. Biotechnol. 31:537-541. [Google Scholar]
- 30.Michaelis, L., and M. L. Menten. 1913. Die Kinetik der Invertinwirkung. Biochem. Z. 49:334-336. [Google Scholar]
- 31.Modler, H. W., R. C. Mckellar, and M. Yaguchi. 1990. Bifidobacteria and bifidogenic factors. Can. Inst. Food Sci. Technol. J. 23:29-41. [Google Scholar]
- 32.Muramatsu, K., S. Onodera, M. Kikuchi, and N. Shiomi. 1992. The production of β-fructofuranosidase from Bifidobacterium spp. Biosci. Biotechnol. Biochem. 56:1451-1454. [Google Scholar]
- 33.Nothaft, H., S. Parche, A. Kamionka, and F. Titgemeyer. 2003. In vivo analysis of HPr reveals a fructose-specific phosphotransferase system that confers high-affinity uptake in Streptomyces coelicolor. J. Bacteriol. 185:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Riordan, K. 1998. Studies on antimicrobial activity and genetic diversity of Bifidobacterium species: molecular characterization of a 5.75 kb plasmid and a chromosomally encoded recA gene homologue from Bifidobacterium breve. Ph.D. thesis. Department of Microbiology, National University of Ireland, Cork, Ireland.
- 35.Poolman, B. 2002. Transporters and their roles in LAB cell physiology, p. 147-164. Proceedings of the Seventh Symposium on Lactic Acid Bacteria: Genetics, Metabolism and Applications, September 1-5, 2002. Kluwer Academic Publishers, Egmond aan Zee, The Netherlands.
- 36.Reizer, J., and C. Panos. 1980. Regulation of beta-galactosidase phosphate accumulation in Streptpcoccus pyogenes by an expulsion mechanism. Proc. Natl. Acad. Sci. USA 77:5497-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reuter, G. 1963. Vergleichenden Untersuchung uber die Bifidus-Flora im Sauglings und Erwachsenenstuhl. Zentlb. Bakteriol. Parasitenkd. Infektionskrh. Hyg. Abt. 1 Orig. 191:486-507. [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Tannock, G. W. 2002. Prebiotic oligosaccharides: evaluation of biological activities and potential future developments, p. 107-148. In G. W. Tannock (ed.), Probiotics and prebiotics: where are we going? Caister Academic Press, Wymondham, United Kingdom.
- 40.Titgemeyer, F., and W. Hillen. 2002. Global control of sugar metabolism: a gram-positive solution. Antonie Leeuwenhoek 82:59-71. [PubMed] [Google Scholar]
- 41.Trindade, M. I., V. R. Abratt, and S. J. Reid. 2003. Induction of sucrose utilization genes from Bifidobacterium lactis by sucrose and raffinose. Appl. Environ. Microbiol. 69:24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner, E., S. Marcandier, O. Egeter, J. Deutscher, F. Gotz, and R. Bruckner. 1995. Glucose kinase-dependent catabolite repression in Staphylococcus xylosus. J. Bacteriol. 177:6144-6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warchol, M., S. Perrin, J. P. Grill, and F. Schneider. 2002. Characterization of a purified β-fructofuranosidase from Bifidobacterium infantis ATCC 15697. Lett. Appl. Microbiol. 35:462-467. [DOI] [PubMed] [Google Scholar]