Abstract
Burkholderia is an important bacterial genus containing species of ecological, biotechnological, and pathogenic interest. With their taxonomy undergoing constant revision and the phenotypic similarity of several species, correct identification of Burkholderia is difficult. A genetic scheme based on the recA gene has greatly enhanced the identification of Burkholderia cepacia complex species. However, the PCR developed for the latter approach was limited by its specificity for the complex. By alignment of existing and novel Burkholderia recA sequences, we designed new PCR primers and evaluated their specificity by testing a representative panel of Burkholderia strains. PCR followed by restriction fragment length polymorphism analysis of an 869-bp portion of the Burkholderia recA gene was not sufficiently discriminatory. Nucleotide sequencing followed by phylogenetic analysis of this recA fragment differentiated both putative and known Burkholderia species and all members of the B. cepacia complex. In addition, it enabled the design of a Burkholderia genus-specific recA PCR that produced a 385-bp amplicon, the sequence of which was also able to discriminate all species examined. Phylogenetic analysis of 188 novel recA genes enabled clarification of the taxonomic position of several important Burkholderia strains and revealed the presence of four novel B. cepacia complex recA lineages. Although the recA phylogeny could not be used as a means to differentiate B. cepacia complex strains recovered from clinical infection versus the natural environment, it did facilitate the identification of clonal strain types of B. cepacia, B. stabilis, and B. ambifaria capable of residing in both niches.
Burkholderia is a genus with complex taxonomy that currently contains 34 validly described species (5), nine of which are a closely related group, known as the Burkholderia cepacia complex (6). Burkholderia species are widely distributed in the natural environment, and although the majority appear to live either freely or as symbionts or commensals with a variety of higher organisms, several species also cause disease (5). Plant-pathogenic species include Burkholderia glumae and Burkholderia plantari, which are important rice pathogens. The genus also includes mammalian primary pathogens such as Burkholderia pseudomallei, the cause of meliodosis in humans, and Burkholderia mallei, which causes glanders in horses; both species have attracted recent interest as potential bioterrorism agents (19).
Many other Burkholderia species are capable of causing opportunistic infections in humans and animals; for example, the B. cepacia complex (6) can cause serious infections in persons with cystic fibrosis (20) and other vulnerable individuals (31), as well as disease in plants (8) and animals (3). In contrast to these detrimental pathogenic properties, several Burkholderia species have considerable commercial and ecological importance. They have been used in agriculture as biopesticides and plant growth promoters (27), and in the bioremediation of major pollutants such as trichloroethylene (30) and polychlorinated biphenyls (26).
The taxonomy and identification of the genus Burkholderia are complex, with new species being described rapidly (5, 6). Closely related species such as the B. cepacia complex are difficult to identify using conventional biochemical and phenotypic tests, and species belonging to other betaproteobacterial genera (including Pandoraea and Ralstonia) may be misidentified as Burkholderia species (6). A polyphasic taxonomic approach (39) utilizing multiple diagnostic tests is often required to identify Burkholderia species accurately.
Although 16S rRNA gene sequence analysis forms an integral part of taxonomical analysis for many bacterial genera (39), its utility in the genus Burkholderia is more limited, especially within the B. cepacia complex, where it cannot be used as a means to accurately distinguish all species (16, 21). The recA gene has been widely applied in bacterial systematics (13) and has proven very useful for the identification of B. cepacia complex species, with phylogenetic analysis of sequence variation within the gene enabling discrimination of all nine current species within the B. cepacia complex (21). However, the PCR primers designed for the original recA-based approach, BCR1 and BCR2, are specific only to members of the B. cepacia complex and fail to amplify this gene from other Burkholderia species (21). While this can be used as a positive means to confirm an isolate's position within the complex, it limits the application of the approach to identify other Burkholderia species in diverse natural habitats.
Given the ecological, biotechnological, and pathogenic importance of these bacteria, there is a clear need for a molecular diagnostic scheme capable of discrimination across all Burkholderia spp. This paper describes the use of genome sequence data from several Burkholderia species genome sequencing projects, in combination with an extensive collection of recA sequences from B. cepacia complex bacteria (21), to develop and evaluate a scheme for identification of all Burkholderia spp. based on the recA gene. New PCR primers were designed to amplify 87% of the recA gene, and novel recA sequence data were obtained from known and unknown species. The large size of this recA PCR product enabled the development of a PCR-restriction fragment length polymorphism method. Once recA sequence information for each Burkholderia species had been obtained, PCR primers specific for the genus were designed and tested. Finally, to further examine the phylogenetic relationships between strains of clinical and environmental origin, the recA genes from a large collection of B. cepacia complex strains were sequenced and compared.
MATERIALS AND METHODS
Bacterial strains, identification, and culture.
Molecular identification approaches were developed and evaluated on the collection of strains listed in Table 1. The panel was selected to be representative of the current species diversity of Burkholderia and contained 28 species from the genus, with the exception of the primary pathogens B. pseudomallei and B. mallei. These Burkholderia species and all B. cepacia complex strains were obtained from the Belgian Co-ordinated Collections of Microorganisms (BCCM), LMG Bacteria collection, the Cardiff University collection (over 800 isolates) (2, 21), and U.S. B. cepacia Complex Research Laboratory and Repository (18) and included representatives of the published strain panels (7, 22). Nine isolates representing putative novel Burkholderia species, a selection of non-Burkholderia control species for PCR, and four isolates of the closely related genus Pandoraea were also included in the test strain panel (Table 1). The Burkholderia ubonensis type strain, which appears to be a new species member of the B. cepacia complex (42), was also included in the study. The ecologically and biotechnologically relevant B. cepacia complex strains examined are described in Table 2. All Burkholderia species were cultured and identified as described previously (6, 21).
TABLE 1.
Bacterial strains used for recA PCR primer development and evaluation
| Species | Strain | recA GenBank accession no.a | BUR1 and BUR2 PCR | BUR1 and BUR2 RFLP type | BUR3 and BUR4 PCR |
|---|---|---|---|---|---|
| Pandoraea species | |||||
| Pandoraea pulmonicola | #40 | AY619660 | Positive | 67 | Negative |
| Pandoraea apista | Patient W | AY619657 | Positive | 04 | Negative |
| Pandoraea sputorum | AU0012 | AY619659 | Positive | 16 | Negative |
| Pandoraea pnomenusa | Keulen | AY619658 | Positive | 15 | Negative |
| Known Burkholderia species | |||||
| Burkholderia kururiensis | LMG 19447T | AY619654 | Positive | 80 | Positive |
| Burkholderia caledonica | LMG 19076T | AY619669 | Positive | 78 | Positive |
| Burkholderia phenazinium | LMG 2247T | AY619668 | Positive | — | Positive |
| Burkholderia plantarii | LMG 9035T | AY619655 | Positive | 08 | Positive |
| Burkholderia glathei | LMG 14190T | AY619666 | Positive | 10 | Positive |
| Burkholderia glumae | LMG 2196T | AY619675 | Positive | 89 | Positive |
| Burkholderia gladioli | LMG 2216T | AY619665 | Positive | 70 | Positive |
| Burkholderia fungorum | LMG 16225T | AY619664 | Positive | 12 | Positive |
| Burkholderia caryophylli | LMG 2155T | AY619663 | Positive | 53 | Positive |
| Burkholderia graminis | LMG 18924T | AY619653 | Positive | 77 | Positive |
| Burkholderia caribensis | LMG 18531T | AY619662 | Positive | 18 | Positive |
| Burkholderia thailandensis | LMG 20219T | AY619656 | Positive | 83 | Positive |
| Burkholderia sacchari | LMG 19450T | AY619661 | Positive | 71 | Positive |
| Burkholderia terricola | LMG 20594T | AY619672 | Positive | 85 | Positive |
| Burkholderia tuberum | LMG 21444T | AY619674 | Positive | 86 | Positive |
| Burkholderia phymatum | LMG 21445T | AY619667 | Positive | 87 | Positive |
| Burkholderia xenovorans | LB400T | AAAJ00000000 | Positive | 01 | Positive |
| Burkholderia andropogonis | LMG 2129T | — | Negative | — | Negative |
| Burkholderia hospita | LMG 20598T | — | Positiveb | — | Positive |
| Indeterminate Burkholderia species | |||||
| R-20943 | AY619679 | Positive | 71 | Positive | |
| R-8349 | AY619681 | Positive | 84 | Positive | |
| R-15273 | AY619677 | Positive | 81 | Positive | |
| R-701 | AY619680 | Positive | 90 | Positive | |
| R-15821 | AY619678 | Positive | 01 | Negative | |
| R-13392 | AY619676 | Positive | 77 | Positive | |
| LMG 21262 | AY619673 | Positive | 02 | Positive | |
| LMG 19510 | AY619670 | Positive | 82 | Positive | |
| LMG 20580 | AY619671 | Positive | 84 | Positive | |
| B. cepacia complex species | |||||
| Burkholderia vietnamiensis | LMG 10929T | AF143793 | Positive | 66 | Positive |
| Burkholderia multivorans | C1576 | AF143774 | Positive | 73 | Positive |
| Burkholderia multivorans | LMG 13010T | — | Positive | — | Positive |
| Burkholderia cepacia | ATCC 25416T | AF143786 | Positive | 88 | Positive |
| Burkholderia cenocepacia | J2315/LMG16656T | www.sanger.ac.uk | Positive | 20 | Positive |
| Burkholderia stabilis | LMG 14294T | AF456031 | Positive | 63 | Positive |
| Burkholderia pyrrocinia | LMG 14191T | AF143794 BPP | Positive | 75 | Positive |
| Burkholderia ambifaria | LMG 19182T | AF323985 | Positive | 48 | Positive |
| Burkholderia dolosa | LMG 18943T | AF323971 | Positive | 58 | Positive |
| Burkholderia anthina | LMG 20980T | AF456059 | Positive | 57 | Positive |
| Burkholderia cenocepacia | CFLG | AF456021 | Positive | 25 | Positive |
| Burkholderia ubonensis | LMG 20358T | AY780511 | — | — | — |
| PCR control species | |||||
| Bordetella parapertussis | LMG 14449T | — | Positive | — | Negative |
| Rhizobium vitis | LMG 8750T | — | Negative | — | Negative |
| Xanthomonas sacchari | LMG 471T | — | Positive | — | Negative |
| Ralstonia metallidurans | LMG 1195T | — | Negative | — | Negative |
| Ralstonia gilardii | LMG 5886T | — | Positive | — | Negative |
| Ralstonia eutropha | JMP134/LMG1197 | www.jgi.doe.gov | Positive | — | Negative |
| Neisseria elongata | LMG 5124T | — | Negative | — | Negative |
| Mycobacterium smegmatis | MC2155 | — | Negative | — | Negative |
| Pseudomonas aeruginosa | C3719 | — | Negative | — | Negative |
| Pseudomonas aeruginosa | PAO1 | NC_002156 | Negative | — | Negative |
—, recA sequence or RFLP not available or not determined.
PCR amplification with BUR1 and BUR2 resulted in one product of the correct size, 869 bp, and another of approximately 400 bp which could not be resolved by optimization.
TABLE 2.
Characteristics of selected B. cepacia complex strains
| Species (recA phylogenetic cluster) | Strain name (other strain designations) | Source and relevant information | Reference(s) |
|---|---|---|---|
| B. cepacia (genomovar I type strain cluster) | ATCC 49709 | Grass seed biological control strain | 11 |
| B. cepacia (genomovar I group K) | SAR-1 | Burkholderia species metagenomic strain from Sargasso Sea | 40 |
| B. cepacia (genomovar I group K) | 383 (ATCC 17660; LMG 6991) | Forest soil, Trinidad; genome draft available (http://genome.jgi-psf.org/draft_microbes/bur94/bur94.home.html) | 32 |
| B. cepacia (genomovar I type strain cluster) | ATCC 25416T (LMG 1222) | Onion rot; B. cepacia type strain | 32, 37 |
| B. cepacia (genomovar I type strain cluster) | J1050 | Clinical infection, USA | 28 |
| B. cepacia (genomovar I type strain cluster) | ATCC 17759 (LMG 2161) | Forest soil, Trinidad | 32 |
| B. cepacia (genomovar I type strain cluster) | LMG 14087 | Wound swab, UK | 37 |
| B. cenocepacia (III-B) | M36 | Corn rhizosphere, USA; registered biopesticide withdrawn from commercial use; type Wisconsin strain; encodes B. cenocepacia pathogenicity island | 27 |
| B. cenocepacia (III-B) | BC-1 | Corn rhizosphere, USA; biological control strain; encodes B. cenocepacia pathogenicity island | 24 |
| B. cenocepacia (III-B) | BC-2 | Corn rhizosphere, USA; biological control strain; encodes B. cenocepacia pathogenicity island | 24 |
| B. stabilisa | HI-2482 | Veterinary shampoo contaminant, USA | This study |
| B. stabilisa | LMG 14294 | Cystic fibrosis patient, Belgium | 38 |
| B. vietnamiensisa | G4 (ATCC 53617; R-1808) | Waste water, USA; capable of trichloroethylene degradation; derivative strain ENV435 effective in commercial field test on a contaminated aquifer; genome draft available (http://genome.jgi-psf.org/draft_microbes/bur08/bur08.home.html) | 29, 33 |
| B. ambifariaa | M54 (R-5142) | Corn rhizosphere, USA; registered biopesticide in commercial use; type Wisconsin strain with antifungal properties | 27 |
| B. ambifariaa | J82 (R-5140) | Corn rhizosphere, USA; registered biopesticide in commercial use; type Wisconsin strain with antinematodal properties | 27 |
| B. ambifariaa | BC-F | Corn rhizosphere, USA; U.S. Department of Agriculture biological control strain; production of antifungal agents | 43 |
| B. ambifariaa | AMMDT (LMG 19182T) | Pea rhizosphere, USA; biological control strain and species type strain | 4, 27 |
| B. ambifariaa | AU212 (LMG 19466) | Cystic fibrosis strain isolated from a patient in Wisconsin, USA | 4 |
| B. pyrrociniaa | BC11 | Soil; antifungal-producing biological control strain | 12 |
| B. pyrrociniaa | ATCC 39277 | Corn field soil; production of antifungal agents | 25 |
Species group and recA cluster are identical.
Chromosomal DNA extraction.
DNA was prepared for PCR amplification from overnight cultures as described previously (21). Rapid DNA extraction by boiling in the presence of a chelating resin was also carried out as follows. Bacteria from 1 ml of a liquid culture were harvested by centrifugation and resuspended in 100 μl of a sterile solution containing 5% Chelex 100 (Sigma Aldrich, Gillingham, United Kingdom). The suspension was boiled for 5 min and immediately placed on ice for a further 5 min, and then this procedure was repeated. The final supernatant was recovered after centrifugation and stored at −20°C. Before use DNA was diluted in sterile water, and approximately 20 ng of template DNA was incorporated into each PCR.
PCR analysis.
PCR was performed as described previously (21) using QIAGEN reagents (QIAGEN Ltd., Crawley, United Kingdom). Each 25-μl PCR contained the following: 1 U Taq polymerase, 250 μM of each deoxynucleoside triphosphate, 1x PCR buffer (including 1.5 mM MgCl2), 10 pmol of each appropriate oligonucleotide primer, and 10 to 50 ng of template DNA.
PCR of B. cepacia complex recA genes was carried out using primers BCR1 and BCR2 as previously described (21). New primers for specific amplification and sequencing of Burkholderia species recA were designed: BUR1, GATC GA(AG)AAGCAGTTCGGCAA, and BUR2, TTGTCCTTGCCCTG(AG)C CGAT, amplifying an 869-bp fragment; and BUR3, GA(AG) AAG CAG TTC GGC AA, and BUR4, GAG TCG ATG ACG ATC AT, amplifying a 385-bp fragment. OligoCheck (1; http://www.cf.ac.uk/biosi/research/biosoft) was used to assist in primer design and analyze primers BUR1 and BUR2 by rating the likelihood of publicly available bacterial recA sequences being amplified. Thermal cycling was carried out in a Flexigene thermal cycler (Techgene Ltd., Cambridge, United Kingdom) for 30 cycles of 30 s at 94°C, annealing for 30 s at 60°C, and extension at 72°C for 45 s, with a final 5-min extension at 72°C. Approximately 2 μl of each PCR product was visualized by agarose gel electrophoresis as described previously (21).
Restriction fragment length polymorphism analysis.
Restriction fragment length polymorphism (RFLP) fingerprinting of the recA amplicons of B. cepacia complex bacteria were performed as previously described (21). RFLP analysis of Burkholderia species and other strains (Table 1) was performed on 5 μl of PCR product in a mixture containing the appropriate restriction enzyme buffer and restriction endonuclease as outlined by the manufacturer (Promega, Southampton, United Kingdom) and incubated at 37°C for 4 h. Analysis of the BUR1 and BUR2 recA amplicons was performed with HaeIII. RFLP patterns obtained from recA were recorded and compared using computer software (GeneSnap, GeneTools, and GeneDirectory, Syngene, Cambridge, United Kingdom). RFLP pattern similarity was calculated as a Dice coefficient at 1% tolerance and clustered using the unweighted pair-group method average. RFLP patterns with a similarity index of 0.75 or higher were clustered as a single group. Macrorestriction and pulsed-field gel electrophoresis (PFGE) analysis was performed as described previously (38); PFGE fingerprints were also compared and analyzed using computer software as described above.
Nucleotide sequence analysis.
All recA PCR products were sequenced directly using the appropriate primers as described previously (21) as well as those developed in this study (BUR1 and BUR2). Sequencing reactions were prepared using Applied Biosystems Big Dye Terminator ready reaction mix version 3.1 in accordance with the manufacturer's instructions and analyzed using Applied Biosystems ABI-Prism 3100 genetic analyzer capillary electrophoresis system running Applied Biosystems Performance Optimized Polymer 6 (POP-6). Raw sequences from both strands of the PCR products were aligned, and a consensus sequence was derived using the CAP contig assembly program within the BioEdit software (10). Analysis also involved the use of Basic Local Alignment Search Tool (BLAST; www.ncbi.nlm.nih.gov) to establish the correct gene identity.
Phylogenetic analysis.
Multiple nucleotide sequence alignments spanning 760 nucleotides of the recA gene were constructed using CLUSTAL W (34). Phylogenetic and molecular evolutionary analyses were conducted using genetic-distance-based neighbor-joining algorithms within MEGA version 2.1 (http://www.megasoftware.net/). Gaps and missing data were completely deleted in MEGA before trees were constructed using the Jukes and Cantor matrix model with random sequence input order and 1,000 data sets examined by bootstrapping. All trees were rooted with the Pseudomonas aeruginosa PAO1 recA gene.
Nucleotide sequence accession numbers and aligned sequence sets.
Novel recA nucleotide sequences were determined for 28 Burkholderia strains, four Pandoraea strains, and the B. ubonensis type strain (GenBank accession numbers are listed in Table 1). Sequences were also determined for 106 B. cepacia complex strains representative of the diversity seen in the recA gene RFLP, and these have been submitted under accession numbers AF143782, AF143797, AF143800, AF456003 to AF456015, AF456017, AF456018, AF456020, AF456023, AF456024, AF456026 to AF456028, AF456035, AF456036, AF456038, AF456039 to AF456050, AF456052, AF456054, AF456056, AF456057, AF456062 to AF456064, AF456066, AF456067, AF456069 to AF456083, AF456085 to AF456124, and AY753187. Phylogenetic analysis was performed on the latter novel sequences, 52 previously published recA genes (4, 21, 35, 36, 41, 42), and four sequences obtained from the following genome sequence projects: B. cenocepacia strain J2315 (NC_004503; www.sanger.ac.uk/Projects /B_cenocepacia/), B. vietnamiensis strain G4 (NZ_AAEH00000000; http: //genome.jgi-psf.org/draft_microbes/bur08/bur08.home.html), B. cepacia strain ATCC 17660 (strain 383; http://genome.jgi-psf.org/draft_microbes/bur94/bur94.home.html), and the Burkholderia Sargasso Sea Metagenome (40) strain SAR-1 (NS_000028; www.ncbi.nlm.nih.gov/genomes/static/es.html). Aligned sequence sets of all the recA sequences used in this study are available from ftp://cepacia.bios.cf.ac.uk
RESULTS
Design of PCR probes and amplification of Burkholderia species recA gene.
Data for the design of a Burkholderia genus-specific recA PCR were obtained from the alignment of 10 published recA genes spanning the B. cepacia complex (21) and genomic regions spanning the recA genes from the B. xenovorans LB400T, B. cenocepacia J2315, and B. pseudomallei K96243 genome sequence projects. Few primer sites capable of amplification across the genus were identified in silico either within or just outside the recA coding sequence. Sites that facilitated amplification of a large (869-bp) internal fragment suitable for both RFLP and sequence analysis were selected and used to design primers BUR1 (21-mer; spanning bases 72 to 92 relative to the B. cenocepacia J2315 genome recA gene [all subsequent primer positions are given relative to this sequence]) and BUR2 (20-mer; bases 819 to 938).
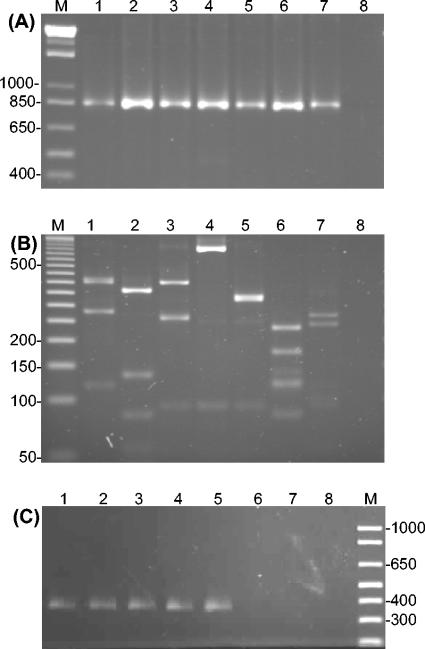
Prior to laboratory testing, the OligoCheck software was used to examine the performance of primers in silico against all available recA sequences. A panel of nine non-Burkholderia control strains, each containing fewer than five putative mismatches to the PCR primers, were selected from the OligoCheck output for testing (Table 1). BUR1 and BUR2 produced an 869-bp amplicon from all Burkholderia species (Fig. 1A) except B. andropogonis (Table 1). Four of the control species, Bordetella parapertussis, Xanthomonas sacchari, Ralstonia gilardii, and Ralstonia eutropha, also produced the same-sized amplicon; the remaining control strains were negative (Table 1). Because of its broad specificity for Burkholderia species, the PCR employing BUR1 and BUR2 was subsequently used as a means to test a recA-based RFLP approach and to obtain further nucleotide sequence information for specific primer design.
FIG. 1.
Burkholderia recA gene PCR analysis. Panel A shows PCR products obtained with primers BUR1 and BUR2 from Burkholderia and control species as follows in each lane: 1, B. stabilis LMG14294; 2, B. caryophylli LMG2155T; 3, B. fungorum LMG16225T; 4, B. graminis LMG18924T; 5, B. plantarii LMG9035T; 6, Bordetella parapertussis LMG14449; 7, Ralstonia eutropha JMP134; 8, negative control. Panel B shows the RFLP analysis of the BUR1 and BUR2 recA amplicon performed by digestion of PCR products with the enzyme HaeIII. Samples in each lane are as follows: 1, B. stabilis LMG14294; 2, B. caryophylli LMG2155T; 3, B. fungorum LMG16225T; 4, B. graminis LMG18924T; 5, B. plantarii LMG9035T; 6, Bordetella parapertussis LMG14449; 7, Ralstonia eutropha JMP134; 8, negative control. Panel C shows the specific amplification of the Burkholderia recA gene with primers BUR3 and BUR4 with the following species run in each lane as follows: 1, B. stabilis LMG14294; 2, B. caryophylli LMG2155T; 3, B. fungorum LMG16225T; 4, B. graminis LMG18924T; 5, B. plantarii LMG9035T; 6, Bordetella parapertussis LMG14449; 7, Ralstonia eutropha JMP134; 8, H2O negative control. Molecular size markers are shown in lane M for all panels, and the sizes of relevant bands are indicated in bp.
RFLP analysis of Burkholderia species recA gene.
BUR1 and BUR2 PCR amplification and digestion with HaeIII (21) were applied to analyze the Burkholderia genus. RFLP analysis with other enzymes was not performed since previous work had already shown HaeIII to be the most discriminatory enzyme for restriction of B. cepacia complex recA genes (21). A total of 103 isolates representative of both Burkholderia species (Table 1) and RFLP pattern diversity with the B. cepacia complex (21; E. Mahenthiralingam, unpublished data) were examined; 81 unique and 13 shared RFLP patterns were identified. Examples of unique RFLP types from B. cepacia complex members, Burkholderia species, and two non-Burkholderia species bacteria are shown in Fig. 1B. Shared RFLP types are described in Table 3.
TABLE 3.
Burkholderia and Pandoraea isolates found to possess matching recA RFLP types
| Species | Strain name | HaeIII RFLP type |
|---|---|---|
| B. xenovorans | LB400T | 01 |
| “Burkholderia” sp. nov. | R-15821 | 01 |
| P. pulmonicola | BCC0150 | 15 |
| P. pnomenusa | BCC0580 | 15 |
| B. caribiensis | LMG 18531 | 18 |
| B. cepacia | BCC0679 | 18 |
| B. cepacia complex unknown | AU0553 | 38 |
| B. cepacia complex unknown | BCC0095 | 38 |
| B. pyrrocinia | LMG 21823 | 38 |
| B. vietnamiensis | CLO | 45 |
| B. ubonensis | R-11767 | 45 |
| B. ambifaria | MVP/C1 64 | 46 |
| B. cenocepacia | MDII 367 | 46 |
| B. cenocepacia | MVP/C1 73 | 62 |
| B. cenocepacia | BELF 2 | 62 |
| B. stabilis | LMG 14294 | 63 |
| B. cenocepacia | PC184 | 63 |
| B. gladioli | LMG 2216 | 70 |
| B. gladioli | BCC0238 | 70 |
| B. sacchari | LMG 19450 | 71 |
| “Burkholderia” sp. nov. | R-20943 | 71 |
| B. graminis | LMG 18924 | 77 |
| “Burkholderia” sp. nov. | R-13392 | 77 |
| “Burkholderia” sp. nov. | R-8349 | 84 |
| “Burkholderia” sp. nov. | LMG 20580 | 84 |
Five known species and several strains representing putative novel species possessed overlapping RFLP patterns (Table 3). Therefore, although the majority of strains and species analyzed (86%) possessed unique patterns, the discriminatory power of the recA-RFLP analysis was limited. However, it could serve as a useful primary screen in a Burkholderia species identification scheme in an analogous fashion to its use for determining species within the B. cepacia complex (21, 42). Further resolution was achieved with nucleotide sequence analysis of Burkholderia recA.
Development of Burkholderia-specific recA PCR.
To confirm the sequence variations detected by RFLP analysis of the amplified recA fragment and facilitate the design of Burkholderia-specific PCR primers, the 869-bp BUR1 and BUR2 recA amplicons of 16 Burkholderia strains, nine indeterminate Burkholderia species strains, the B. ubonensis type strain, and four Pandoraea strains (Table 1) were sequenced. These novel recA sequences were analyzed by alignment with published B. cepacia complex sequences and genomic recA sequences.
A single Burkholderia genus-specific adenosine was identified at base 445 in the recA gene and used to design the downstream primer BUR4 (17-mer; bases 445 to 461). The upstream primer, BUR3 (17-mer; bases 76 to 92), was designed to prime off the same base as BUR1 but was shorter to facilitate potential future use in a nested PCR priming off a primary amplicon resulting from BUR1 and BUR2 PCR. BUR3 and BUR4 produced a 385-bp amplicon for all Burkholderia species examined (examples shown in Fig. 1C) except Burkholderia androprogonis and novel Burkholderia sp. strain R-15821. The recA sequence of the last strain was subsequently shown to be more closely related to that of Bordetella (see below; Table 1 and Fig. 2). BUR3 and BUR4 PCR did not produce amplicons with non- closely Burkholderia-related species, including those that had originally amplified with BUR1 and BUR2 (Table 1).
FIG. 2.
Phylogenetic analysis of the Burkholderia genus using novel Burkholderia recA sequences. A phylogenetic tree comparing representatives from each species is shown; 27 novel Burkholderia recA genes and four novel Pandoraea recA genes are highlighted. Bootstrap values over 70% and genetic distance scale are indicated. The species name and strain number are shown, with the accession number in brackets. The tree was rooted with the P. aeruginosa PAO1 recA gene taken from the complete genome sequence as a representative member of a species outside the betaproteobacteria group. The following recA sequences were also included in the phylogenetic analysis as indicators of the ability of the analysis to differentiate among genera closely related to Burkholderia: Ralstonia eutropha JMP135, Bordetella avium AY124330, Bordetella hinzii AY124331, Bordetella parapertussis AF399659, Bordetella pertussis X53457, Xanthomonas axonopodis AE011806, and Neisseria meningitidis NC_0031112. Sequences from the species comprising the B. cepacia complex were included.
Phylogenetic analysis of Burkholderia species recA gene.
A phylogenetic tree constructed with novel recA sequences from Burkholderia species and Pandoraea species (Table 1) is shown in Fig. 2 to illustrate the diversity of the Burkholderia genus. The B. cepacia complex formed a distinct phylogenetic cluster. Burkholderia thailandensis, B. pseudomallei, and B. mallei (the B. pseudomallei group) formed a cluster, which, although distinct, was more closely related to the B. cepacia complex than other Burkholderia species. The Pandoraea genus also formed a distinct cluster consistent with their recent separation from the Burkholderia genus. All the indeterminate Burkholderia species (Table 1) strains clustered within the Burkholderia genus cluster except strain R-15821, which was more closely related to Bordetella species.
To test if the same cluster assignments were made using sequence obtained from the smaller BUR3 and BUR4 amplicon, the sequences were trimmed to the 300 bases within the 385-bp amplicon produced by these primers and subjected to an identical analysis. The trimmed sequences produced a tree with the same topology and clusters that had been observed with the 800-base sequences derived from the 865-bp BUR1 and BUR2 amplicon (data not shown).
Phylogenetic diversity of the B. cepacia complex.
Although the recA gene has proven to be a valuable tool in determining the taxonomy of the B. cepacia complex, species such as B. cenocepacia and B. cepacia are split into distinct phylogenetic clusters when analyzed in this way. Two recA lineages were originally observed in B. cepacia genomovars III, III-A, and III-B (21), and subsequently clusters III-C and III-D were reported when the formal name B. cenocepacia was proposed for this genomovar (36). Similarly, B. cepacia strains also divide into two lineages by phylogenetic polymorphism in the recA gene; one cluster includes the type strain (ATCC 25416T) (21), while the second group was named group K based on the most common recA RFLP found within that cluster (42). Further novel recA phylogenetic groups have also been observed for other strains. However, the significance of these clusters was difficult to interpret within a phylogeny based solely on the B. cepacia complex (E.M., unpublished data), and so further analysis with the new data obtained in this study was carried out to resolve phylogenetic relationships on a broader scale.
A phylogenetic tree comprising a subset of 101 B. cepacia complex strain recA genes and including 28 novel Burkholderia and Pandoraea sequences (Table 1) is shown in Fig. 3. Seventeen phylogenetically distinct clusters were observed for B. cepacia complex species. As observed with the Burkholderia genus phylogeny (Fig. 2), the B. pseudomallei group clustered adjacent to the B. cepacia complex. B. gladioli and B. plantarii, and B. glathei and B. carophylli also formed distinct clusters. All the remaining Burkholderia species formed a diverse separate group.
FIG. 3.
Phylogenetic analysis of B. cepacia complex strains using novel Burkholderia recA sequences. A phylogenetic tree comparing 160 B. cepacia complex strains, 28 Burkholderia, and four Pandoraea recA sequences is shown as a composite of phylogenetically distinct sequence clusters. Bootstrap values and genetic distance scale are indicated. The species or group name for each cluster is shown in bold. Additional information for one or more reference or interesting strains within the group is also shown. The number of sequences deriving from environmental strains versus the total number of sequences in the group is shown in brackets.
Within the B. cepacia complex, all B. stabilis, B. pyrrocinia, B. anthina, B. vietnamiensis, B. dolosa, B. ambifaria, and B. multivorans strains and the B. ubonensis type strain formed single discrete phylogenetic clusters. As shown previously (21), B. cepacia strains were split into sublineages (Fig. 3), while B. cenocepacia III-A, III-B, III-C, and III-D were all distinct. Three B. cepacia clusters were observed, one containing the type strain, ATCC 25416T, one composed of RFLP type K strains (42), and another composed of RFLP type AW strains that previously belonged to B. cepacia (9) (Fig. 3). However, it must be noted that B. cepacia RFLP group K cluster was the least robust group within the phylogenetic analysis (bootstrap value 30) (Fig. 3), suggesting that further species diversity may be present. Several strains which previously were difficult to assign (E.M., unpublished work), fell into distinct branches of the new tree. B. cepacia complex group 1 strains were closely related to B. cenocepacia III-A, while B. cepacia complex group 2 strains clustered between B. cenocepacia III-D and B. anthina (Fig. 3). Again all the clusters remained intact when the recA sequences were trimmed to 300 bp within the BUR3 and BUR4 amplicon (data not shown).
Phylogenetic positions of clinical and environmental strains.
Of the 101 B. cepacia strains analyzed phylogenetically (Fig. 3), 41 were from the environment and 60 were from human sources. No clinical strains were present in B. cepacia type AW, B. pyrrocinia, and B. cenocepacia III-C clusters, and no environmental strains were present in the B. cenocepacia III-D cluster. All the remaining B. cepacia complex phylogenetic clusters contained strains from both sources, with the exception of B. ubonensis, which comprised the single type strain (Fig. 3).
The phylogenetic positions of several environmentally, biotechnologically and clinically relevant strains were also resolved (Table 2). Strains with documented biological control activities were found to be present in the B. cepacia type strain cluster, B. cenocepacia III-B, B. ambifaria, and B. pyrrocinia (Table 2). Interestingly, all three B. cenocepacia III-B biological control strains contained the B. cenocepacia pathogenicity island (2) (Table 2), including strain M36, which was originally registered for commercial use in the United States and has now been withdrawn (23). One other registered biopesticide strain, M54 (27), was assigned to the B. ambifaria cluster, where the majority of strains with biological control activity were found (Table 2). The recA genes drawn from Burkholderia genomic resources clustered as follows (Fig. 1; Table 2): strain G4, a well-known bioremediation strain, clustered with B. vietnamiensis, as expected; and strain 383, one of the isolates from the pioneering study of Stanier et al. (32), clustered within the group K lineage of B. cepacia, as did the metagenomic Sargasso Sea strain SAR-1 (40).
Genetic identity of B. cepacia complex strains.
Several strains were found to possess the same recA RFLP type and identical recA sequences, yet were of distinct environmental or clinical origin. To resolve their clonality to the strain type level, macrorestriction and PFGE fingerprinting was performed, resulting in the definition of four strain types, each comprising a strain from an environmental and a clinical source (Fig. 4). Each strain pair possessed almost identical genomic fingerprints (all Dice coefficients of similarity >0.93 for each pair) clearly defining each pair as a distinct genetic strain type. Pairs were found within the following species (see Table 2 and Fig. 4): B. cepacia strains, ATCC 17759 (environmental, isolated before 1966), and LMG 14087 (clinical, isolated in 1988 from a wound swab in the United Kingdom); ATCC 25416T (environmental, isolated in the 1940s) and J1050 (clinical); B. ambifaria strains AMMDT (environmental) and AU0212 (clinical); and B. stabilis strains HI-2482 (environmental) and LMG 14294 (clinical, isolated from sputum from a patient with cystic fibrosis in Belgium in 1993).
FIG. 4.
PFGE fingerprinting of environmental and clinical B. cepacia complex strains. Macrorestriction analysis with SpeI of the following strains is shown in each lane as follows (given, respectively, for each strain pair): 1 and 2, B. cepacia strains ATCC 17759 and LMG 14087; 3 and 4, B. cepacia strains ATCC 25416T and J1050; 5 and 6, B. ambifaria strains AMMDT and AU0212; 7 and 8, B. ambifaria strains M54 and J82; 9, B. cenocepacia strain M36; 10 and 11, B. stabilis strains HI-2482 and LMG 14294. Molecular size markers were run in lane M, and the sizes of relevant fragments are indicated in kb.
Strain AMMDT, a well-characterized biocontrol isolate (4, 27), was almost identical to strain AU0212 recovered from a cystic fibrosis patient, differing only in one macrorestriction fragment (Fig. 4). Interestingly, both strains had the same geographic origin, Wisconsin, even though their sources were distinct, soil and cystic fibrosis sputum. Genomic fingerprinting of the three B. cepacia complex biopesticide strains registered for commercial use (Table 2) (27) also revealed an interesting feature of these isolates in that they were all registered as type Wisconsin, a designation derived from their phenotypic features and source. However, it is clear that this designation was not related to their genotype or even species identity. Strain M36 was a member of B. cenocepacia III-B (Fig. 3; Table 3) and possessed a macrorestriction profile clearly distinct from strains M54 and J82 (Fig. 4). Moreover, the latter biocontrol isolates were in fact exactly the same B. ambifaria strain type (Fig. 4), even though each possess slightly different biopesticidal properties (Table 2) (27).
DISCUSSION
We have developed a genetic identification approach for the entire Burkholderia genus that can discriminate between members of the closely related B. cepacia complex. Sequence polymorphism within the recA gene has proven very useful in defining the taxonomy of the B. cepacia complex (6, 21), but the original approach could not be applied to Burkholderia species outside the complex. By designing new Burkholderia species recA PCRs, we have demonstrated that RFLP analysis of the gene can be used to differentiate the majority of Burkholderia strains. However, this pattern-matching technique was still limited in its ability to completely discriminate between all species and required comparison to known reference strains for preliminary grouping of strains. The Burkholderia-specific recA primers and an extensive set of reference recA sequences provided by this study have facilitated further definition of species diversity within both Burkholderia and the B. cepacia complex and were also used to examine the phylogenetic and genotypic relationships of strains from clinical versus environmental sources.
The BUR1 and BUR2 primers enabled amplification of an 869-bp recA fragment, but were not absolutely specific to Burkholderia and cross-reacted with control strains of the genera Bordetella, Pandoraea, Ralstonia, and Xanthomonas. However, nucleotide sequence deriving from analysis of novel Burholderia and Pandoraea recAs enabled the design of primers BUR3 and BUR4, which were found to be specific for the genus. Only B. andropogonis repeatedly failed to produce correct amplification products with both the BUR1 and BUR2, and BUR3 and BUR4 primers. Southern hybridization of B. andropogonis chromosomal DNA demonstrated that DNA homologous to the recA gene of B. cenocepacia J2315 was present, suggesting that the lack of amplification had not resulted from gene deletion in this isolate (data not shown).
Phylogenetic analysis of eight of the nine strains representing putative novel Burkholderia species (Table 1) corroborated the results of the phenotypic data assigning them to the genus (Fig. 2). Only strain R-15281 clustered outside the genus and adjacent to members of Bordetella. Two of the novel Burkholderia species strains, R-15273 and LMG 21262, possessed identical recA genes, suggesting they were members of the same novel species group that was closely related to B. fungorum by phylogenetic analysis (Fig. 2). The remaining six Burkholderia species strains were all highly distinct according to the bootstrap values observed in the phylogenetic analysis, suggesting they were members of novel Burkholderia species. These findings were consistent with the analysis of their whole-cell protein profiles (P.V., unpublished data). Two clusters within the B. cepacia complex were only weakly grouped by the phylogenetic analysis, B. cepacia group K and B. cenocepacia group III-B (Fig. 3; bootstrap values of 30 and 34, respectively). All other B. cepacia complex species recA genes fell into clusters associated with bootstrap values of greater than 70, consistent with their recognition as distinct species. The considerable polymorphism observed within the recA genes of B. cepacia complex group K and B. cenocepacia III-B indicates that further species diversity may be present within these groups.
The topology of the Burkholderia genus recA phylogeny (Fig. 2) was congruent with the 16S rRNA gene-based phylogenetic tree presented by Coenye and Vandamme (5) with respect to the following: (i) all Burkholderia species were distinct from other genera; (ii) all taxonomically classified Burkholderia species formed separate arms within both trees; (iii) species within the B. cepacia complex formed a distinct cluster; and (iv) the B. cepacia complex cluster was most closely linked to a group containing B. mallei, B. pseudomallei, and B. thailandensis. Clusters which were divergent in phylogenetic analysis based on the recA gene and 16S rRNA gene gene (5) were as follows: (i) B. plantari and B. glumae formed a distinct cluster in both trees, however, this group also contained B. gladioli in the recA analysis (Fig. 2); (ii) B. fungorum, B. caledonica, B. graminis, and B. caryophylli, which formed a distinct cluster in the 16S rRNA gene tree, were not closely associated according to recA; and (iii) the positions of all the remaining Burkholderia species examined were not conserved between the recA and 16S rRNA gene analyses. The major advantage recA phylogenetic analysis provides over the 16S rRNA gene for classification of Burkholderia is the greater degree of resolution among closely related species within the genus, such as the B. cepacia complex.
Although RFLP analysis of the recA gene did not discriminate between all species of Burkholderia, sequence analysis of the BUR1 and BUR2 amplicon was sufficient to separate all strains, including their differentiation from members of the B. cepacia complex (Fig. 2 and 3). In particular, the degree of resolution of the recA phylogenetic trees for members of the B. cepacia complex was much greater than observed with 16S rRNA gene analysis (6). However, the presence of discrete recA lineages within species such as B. cenocepacia and B. cepacia adds an additional level of complexity. A promising finding of the study was that analysis of just a 300-bp region of recA sequence within the BUR3 and BUR4 amplicon produced phylogenetic trees with the same topology and discrimination as the nearly full-length trees derived from analysis of the BUR1 and BUR2 PCR. These data suggest that the BUR3 and BUR4 PCR could provide a useful and rapid non-culture-based approach to explore the Burkholderia diversity in various environments.
Genomic resources for Burkholderia have increased substantially in the last few years, and we have shown that by phylogenetic analysis of whole-genome sequence derived recAs the classification of both cultured and uncultured genomic strains can be clarified. The genome strains B. cepacia ATCC 17760 (strain 383) (32) and Burkholderia Sargasso Sea strain SAR-1 reconstructed from metagenomic data (38) were both found to be members of B. cepacia group K. The recA gene of SAR-1 was identical to that of strain R-12710 (data not shown), cultivated from sheep with mastitis (3), and another marine isolate (obtained from the Sea of Japan) also belongs to B. cepacia group K (P.V., unpublished data). While the presence of B. cepacia complex members in river water has been reported (15), their isolation from marine environments is thought to be rare. The novel primers developed in this study can be applied to investigate this area of scientific interest.
Phylogenetic analysis of Burkholderia recA genes also demonstrated that, like other taxonomic criteria, it could not be used as a means to distinguish environmental from clinical strains. Strains from both sources were found throughout the extensive phylogenetic tree derived from this study (Fig. 3). Although no clinical strains were found in four clusters and no environmental strains found in one other B. cepacia complex cluster (Fig. 3), these clusters contained small numbers of strains. It is likely that as more B. cepacia complex strains are examined, so that the number of strains in these clusters is increased, environmental and clinical stains will be identified for each cluster.
Analysis of the recA gene of B. cepacia complex strains in this study resulted in the identification of several clonal pairs of strains each from distinct environmental and clinical sources (Fig. 4). Such pairs were found for B. cepacia, B. stabilis, and B. ambifaria and clearly showed that genotypically identical B. cepacia complex strains can be isolated from both human infections and natural or environmental sources. This work extends the work of LiPuma and colleagues (17), who demonstrated that an epidemic cystic fibrosis strain of B. cenocepacia was identical to a strain found in the soil. B. ambifaria strain AMMDT is the first strain with known biopesticidal properties (4, 27) to have an essentially clonal relative, strain AU0212, cultured from a patient with cystic fibrosis (Fig. 4). In conclusion, it appears that all B. cepacia complex bacteria are capable of colonizing a wide range of habitats, and in the case of infectious niches, this trait appears more dependent on the vulnerability of the host than the taxonomic or phylogenetic classification of the strain.
B. cepacia complex strains with useful biotechnological properties were also found throughout the recA phylogeny. B. cepacia, B. cenocepacia, B. ambifaria, and B. pyrrocinia strains with biopesticidal or antifungal properties were identified (Table 2). The ability to obtain accurate species information using recA sequence data may prove vital in the characterization of future biotechnologically useful strains, especially since detailed understanding of the strain taxonomy forms one of the major risk assessment criteria for commercial registration of bacteria (27). In addition, our study has shown that classification of biopesticidal strains based on phenotypic criteria alone is not adequate. The two type Wisconsin strains of B. ambifaria, M54 and J82, which remain registered for commercial use were found to be genotypically identical (Fig. 4) even though they possess different biopesticidal properties (Table 2) (27). This kind of phenotypic variability within a single clone has also be observed in isolates from cystic fibrosis patient infections (14). Although the B. cepacia complex has been highlighted as a group of bacterial species which require risk assessment as biological control agents (27), the case for this evaluation is not limited to these organisms.
In summary, the recA-based approach developed in this study provides molecular tools for the identification of Burkholderia species that will help to enable researchers to keep pace with the ever-increasing ecological, pathogenic, and genomic interest in the genus.
Acknowledgments
G.W.P. is grateful for Ph.D. funding from the Natural Environment Research Council (NER/S/A/2002/10347A) and their Environmental Genomics Program (NER/T/S/2001/0299). Determination of the B. cepacia complex recA gene sequences was performed via funding from the Cystic Fibrosis Trust (project grant PJ472). T.C. and P.V. are indebted to the Fund for Scientific Research-Flanders (Belgium) for a position as postdoctoral fellow and research grants, respectively. J.J.L. is supported by a grant from the Cystic Fibrosis Foundation (U.S.).
We acknowledge K. E. Ashelford for assistance with in silico primer analysis.
REFERENCES
- 1.Ashelford, K. E., N. Banning, J. C. Fry, and A. Weightman. Assessing the effectiveness of commonly used 16S rRNA universal primer sets to target intended taxa: an in silico study. Manuscript in preparation.
- 2.Baldwin, A., P. A. Sokol, J. Parkhill, and E. Mahenthiralingam. 2004. The Burkholderia cepacia epidemic strain marker is Part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia. Infect. Immun. 72:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berriatua, E., I. Ziluaga, C. Miguel-Virto, P. Uribarren, R. Juste, S. Laevens, P. Vandamme, and J. R. Govan. 2001. Outbreak of Subclinical Mastitis in a Flock of Dairy Sheep Associated with Burkholderia cepacia Complex Infection. J. Clin. Microbiol. 39:990-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coenye, T., E. Mahenthiralingam, D. Henry, J. J. LiPuma, S. Laevens, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Microbiol. 51:1481-1490. [DOI] [PubMed] [Google Scholar]
- 5.Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5:719-729. [DOI] [PubMed] [Google Scholar]
- 6.Coenye, T., P. Vandamme, J. R. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coenye, T., P. Vandamme, J. J. LiPuma, J. R. Govan, and E. Mahenthiralingam. 2003. Updated version of the Burkholderia cepacia complex experimental strain panel. J. Clin. Microbiol. 41:2797-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engledow, A. S., E. G. Medrano, E. Mahenthiralingam, J. J. LiPuma, and C. F. Gonzalez. 2004. Involvement of a plasmid-encoded type IV secretion system in the plant tissue watersoaking phenotype of Burkholderia cenocepacia. J. Bacteriol. 186:6015-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillis, M., T. Vanvan, R. Bardin, M. Goor, P. Hebbar, A. Willems, P. Segers, K. Kersters, T. Heulin, and M. P. Fernandez. 1995. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of The genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int. J. Syst. Bacteriol. 45:274-289. [Google Scholar]
- 10.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 11.Jayaswal, R. K., M. Fernandez, R. S. Upadhyay, L. Visintin, M. Kurz, J. Webb, and K. Rinehart. 1993. Antagonism of Pseudomonas cepacia against phytopathogenic fungi. Curr. Microbiol. 26:17-22. [DOI] [PubMed] [Google Scholar]
- 12.Kang, Y., R. Carlson, W. Tharpe, and M. A. Schell. 1998. Characterization of genes involved in biosynthesis of a novel antibiotic from Burkholderia cepacia BC11 and their role in biological control of Rhizoctonia solani. Appl. Environ. Microbiol. 64:3939-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlin, S., G. M. Weinstock, and V. Brendel. 1995. Bacterial classifications derived from recA protein sequence comparisons. J. Bacteriol. 177:6881-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsen, G. Y., T. L. Stull, and J. L. Burns. 1993. Marked phenotypic variability in Pseudomonas cepacia isolated from a patient with cystic fibrosis. J. Clin. Microbiol. 31:788-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leff, L. G., R. M. Kernan, J. V. McArthur, and L. J. Shimkets. 1995. Identification of aquatic Burkholderia (Pseudomonas) cepacia by hybridization with species-specific rRNA gene probes. Appl. Environ. Microbiol. 61:1634-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LiPuma, J. J., B. J. Dulaney, J. D. McMenamin, P. W. Whitby, T. L. Stull, T. Coenye, and P. Vandamme. 1999. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J. Clin. Microbiol. 37:3167-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LiPuma, J. J., T. Spilker, T. Coenye, and C. F. Gonzalez. 2002. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet 359:2002-2003. [DOI] [PubMed] [Google Scholar]
- 18.LiPuma, J. J., T. Spilker, L. H. Gill, P. W. Campbell 3rd, L. Liu, and E. Mahenthiralingam. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am. J. Respir. Crit. Care Med. 164:92-96. [DOI] [PubMed] [Google Scholar]
- 19.Lowe, P., C. Engler, and R. Norton. 2002. Comparison of automated and nonautomated systems for identification of Burkholderia pseudomallei. J. Clin. Microbiol. 40:4625-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahenthiralingam, E., A. Baldwin, and P. Vandamme. 2002. Burkholderia cepacia complex infection in patients with cystic fibrosis. J. Med. Microbiol. 51:533-538. [DOI] [PubMed] [Google Scholar]
- 21.Mahenthiralingam, E., J. Bischof, S. K. Byrne, C. Radomski, J. E. Davies, Y. Av-Gay, and P. Vandamme. 2000. DNA-Based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahenthiralingam, E., and P. Sayre. 2003. Burkholderia cepacia complex, p. 41-44. In M. D. Licker (ed.), McGraw-Hill yearbook of science and technology. McGraw-Hill, New York, NY.
- 24.Mao, W., R. D. Lumsden, J. A. Lewis, and P. K. Hebbar. 1998. Seed treatment using pre-infiltration and biocontrol agents to reduce damping-off of corn caused by species of Pythium and Fusarium. Plant Dis. 82:294-299. [DOI] [PubMed] [Google Scholar]
- 25.Meyers, E., G. S. Bisacchi, L. Dean, W. C. Liu, B. Minassian, D. S. Slusarchyk, R. B. Sykes, S. K. Tanaka, and W. Trejo. 1987. Xylocandin: a new complex of antifungal peptides. I. Taxonomy, isolation and biological activity. J. Antibiot. (Tokyo) 40:1515-1519. [DOI] [PubMed] [Google Scholar]
- 26.Mondello, F. J. 1989. Cloning and expression in Escherichia coli of Pseudomonas strain LB400 genes encoding polychlorinated biphenyl degradation. J. Bacteriol. 171:1725-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parke, J. L., and D. Gurian-Sherman. 2001. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 39:225-258. [DOI] [PubMed] [Google Scholar]
- 28.Rabkin, C. S., W. R. Jarvis, R. L. Anderson, J. Govan, J. Klinger, J. LiPuma, W. J. Martone, H. Monteil, C. Richard, S. Shigeta, et al. 1989. Pseudomonas cepacia typing systems: collaborative study to assess their potential in epidemiologic investigations. Rev. Infect. Dis. 11:600-607. [DOI] [PubMed] [Google Scholar]
- 29.Shields, M. S., S. O. Montgomery, S. M. Cuskey, P. J. Chapman, and P. H. Pritchard. 1991. Mutants of Pseudomonas cepacia G4 defective in catabolism of aromatic compounds and trichloroethylene. Appl. Environ. Microbiol. 57:1935-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shields, M. S., M. J. Reagin, R. R. Gerger, R. Campbell, and C. Somerville. 1995. Tom, a new aromatic degradative plasmid from Burkholderia (Pseudomonas) cepacia G4. Appl. Environ. Microbiol. 61:1352-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Speert, D. P., M. Bond, R. C. Woodman, and J. T. Curnutte. 1994. Infection with Pseudomonas cepacia in chronic granulomatous disease: role of nonoxidative killing by neutrophils in host defense. J. Infect. Dis. 170:1524-1531. [DOI] [PubMed] [Google Scholar]
- 32.Stanier, R. Y., N. J. Palleroni, and M. Doudoroff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 33.Steffan, R. J., K. L. Sperry, M. T. Walsh, S. Vainberg, and C. W. Condee. 1999. Field-scale evaluation of in situ bioaugmentation for remediation of chlorinated solvents in groundwater. Environ. Sci. Technol. 33:2771-2781. [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vandamme, P., D. Henry, T. Coenye, S. Nzula, M. Vancanneyt, J. J. LiPuma, D. P. Speert, J. R. Govan, and E. Mahenthiralingam. 2002. Burkholderia anthina sp. nov. and Burkholderia pyrrocinia, two additional Burkholderia cepacia complex bacteria, may confound results of new molecular diagnostic tools. FEMS Immunol. Med. Microbiol. 33:143-149. [DOI] [PubMed] [Google Scholar]
- 36.Vandamme, P., B. Holmes, T. Coenye, J. Goris, E. Mahenthiralingam, J. J. LiPuma, and J. R. Govan. 2003. Burkholderia cenocepacia sp. nov.-a new twist to an old story. Res. Microbiol 154:91-96. [DOI] [PubMed] [Google Scholar]
- 37.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J. R. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 38.Vandamme, P., E. Mahenthiralingam, B. Holmes, T. Coenye, B. Hoste, P. De Vos, D. Henry, and D. P. Speert. 2000. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV). J. Clin. Microbiol. 38:1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandamme, P., B. Pot, M. Gillis, P. de Vos, K. Kersters, and J. Swings. 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 60:407-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 41.Vermis, K., T. Coenye, J. J. LiPuma, E. Mahenthiralingam, H. J. Nelis, and P. Vandamme. 2004. Proposal to accommodate Burkholderia cepacia genomovar VI as Burkholderia dolosa sp. nov. Int. J. Syst. Evol. Microbiol. 54:689-691. [DOI] [PubMed] [Google Scholar]
- 42.Vermis, K., T. Coenye, E. Mahenthiralingam, H. J. Nelis, and P. Vandamme. 2002. Evaluation of species-specific recA-based PCR tests for genomovar level identification within the Burkholderia cepacia complex. J. Med. Microbiol. 51:937-940. [DOI] [PubMed] [Google Scholar]
- 43.Zhou, H., F. Yao, D. P. Roberts, and T. G. Lessie. 2003. AHL-deficient mutants of Burkholderia ambifaria BC-F have decreased antifungal activity. Curr. Microbiol. 47:174-179. [DOI] [PubMed] [Google Scholar]