Abstract
We analyzed the binding site on Cry1Aa toxin for the Cry1Aa receptor in Bombyx mori, 115-kDa aminopeptidase N type 1 (BmAPN1) (K. Nakanishi, K. Yaoi, Y. Nagino, H. Hara, M. Kitami, S. Atsumi, N. Miura, and R. Sato, FEBS Lett. 519:215-220, 2002), by using monoclonal antibodies (MAbs) that block binding between the binding site and the receptor. First, we produced a series of MAbs against Cry1Aa and obtained two MAbs, MAbs 2C2 and 1B10, that were capable of blocking the binding between Cry1Aa and BmAPN1 (blocking MAbs). The epitope of the Fab fragments of MAb 2C2 overlapped the BmAPN1 binding site, whereas the epitope of the Fab fragments of MAb 1B10 did not overlap but was located close to the binding site. Using three approaches for epitope mapping, we identified two candidate epitopes for the blocking MAbs on Cry1Aa. We constructed two Cry1Aa toxin mutants by substituting a cysteine on the toxin surface at each of the two candidate epitopes, and the small blocking molecule N-(9-acridinyl)maleimide (NAM) was introduced at each cysteine substitution to determine the true epitope. The Cry1Aa mutant with NAM bound to Cys582 did not bind either of the two blocking MAbs, suggesting that the true epitope for each of the blocking MAbs was located at the site containing Val582, which also consisted of 508STLRVN513 and 582VFTLSAHV589. These results indicated that the BmAPN1 binding site overlapped part of the region blocked by MAb 2C2 that was close to but excluded the actual epitope of MAb 2C2 on domain III of Cry1Aa toxin. We also discuss another area on Cry1Aa toxin as a new candidate site for BmAPN1 binding.
Bacillus thuringiensis, a gram-positive bacterium, produces various insecticidal proteins called Cry toxins which kill only target insects. This bacterium is used as a microbial insecticide and for the genetic development of insect-resistant plants.
Cry toxins are expressed in inclusion bodies as protoxins (70 to 140 kDa) during sporulation. When a protoxin is ingested by target insects, it is solubilized in the insect midgut and digested by proteolytic enzymes in the insect (2). After enzymatic activation, the toxic protease-resistant fragment, which is the 60- to 65-kDa activated toxin, binds to specific receptors located in the columnar cells of the midgut apical brush border membrane (28). The binding of the toxin to receptor molecules leads to a conformational change in the toxin. This allows the toxin to insert into the plasma membrane and subsequently generate pores or ion channels which lead to cellular swelling and lysis (27, 35, 65, 68). Finally, the intoxicated insects die. The binding of the activated toxin to a specific gut receptor is considered to be one of the key factors for insect toxicity. The insecticidal specificity of Cry toxins seems to be largely dependent on this receptor recognition.
The three-dimensional structures of Cry1Aa trypsin-activated toxin, Cry2A protoxin, Cry3A, and Cry3B have been analyzed by X-ray diffraction crystallography (14, 25, 43, 51). These proteins are comprised of three domains. The Cry1Aa and Cry3A structures have many similar features. The N-terminal domain I is composed of a seven-alpha-helix bundle in which the alpha-5 helix is encircled by the other helices. Domain II consists of three antiparallel beta-sheets with exposed loop regions, and domain III is a beta-sandwich. Domain I is involved in membrane insertion and pore formation (5, 21, 25, 43). Domain II plays a role in binding to the insect brush border membranes (16, 39, 42, 44, 46, 61, 62, 63, 64, 72, 76) or receptor molecules (23). Domain III is involved in several functions, including determination of insect specificity (8, 9, 10, 38, 39, 49), recognition of N-acetylgalactosamine (GalNAc) on receptor molecules by a lectin-like structure (4, 30, 31, 36, 40), proteolytic protection (43), and ion channel conductivity (6, 69, 75).
In initial studies, various aminopeptidase N (APN) isoforms (11, 20, 33, 34, 41, 45, 48, 57, 58, 74, 77) and two cadherin-like proteins (52, 53, 73) from several insect species were identified as candidate receptors for B. thuringiensis Cry toxins. These molecules were selected on the basis of their ability to bind Cry1A toxins, which exhibit toxicity for several lepidopteran insects.
Expression of the cadherin-like protein from Bombyx mori, Bt-R175, on the surface of Sf9 insect cells made these cells sensitive to Cry1Aa toxin (54). Hara et al. observed inhibition of the binding of Cry1Aa and Cry1Ac to BtR175 after pretreatment with anti-BtR175 antibody, as this suppressed the lytic activity of the toxins on collagenase-dissociated B. mori midgut epithelial cells (26). The disruption of a cadherin-like protein gene by retrotransposon-mediated insertion has been linked to high levels of resistance to Cry1Ac toxin in Heliothis virescens (12). These findings suggest that the cadherin-like protein plays an important role in Cry toxin susceptibility and that the cadherin-like protein is the functional Cry toxin receptor in the insect midgut at the genetic and cellular levels.
On the other hand, expression of an APN in Drosophila made larvae sensitive to Cry1Ac toxin (19). As determined by RNA interference technology, the silencing of midgut APN in Spodoptera litura resulted in reduced sensitivity to Cry1C toxin (60). These results based on in vivo experiments suggest that APN plays an important role in Cry toxin susceptibility. The 170-kDa APN from H. virescens plays a role in pore formation in membrane vesicles (48). Also, the 120-kDa APN from Manduca sexta mediates channel formation in planar lipid bilayers (70). These results, which were obtained with reconstituted membranes, suggest that APNs function as Cry1 toxin receptors and are involved in the lytic activity of Cry1 toxins. Taken together, the data from in vivo and in vitro experiments provide evidence that APN plays an important role in the toxicity and lytic activity of the Cry toxins.
In ligand blots and surface plasmon resonance experiments, GalNAc inhibited Cry1Ac binding to the M. sexta 120-kDa APN and to the H. virescens 170-kDa APN (15, 48). These results suggest that Cry1Ac toxin contains a lectin-like domain that recognizes part of the GalNAc on APN. It is also notable that the Cry1Ac domain III 509QNR-AAA511 mutant lost the ability to bind APN. Based on homology modeling studies with Cry1Aa and Cry1Ac, the mutated amino acid residues are located around the cavity in domain III which forms the GalNAc binding pocket (31). The mutant toxin exhibited 10- and 22-fold reductions in binding affinity to brush border membrane vesicles but only two- and fourfold reductions in toxicity for M. sexta and H. virescens. The loss of binding affinity relative to toxicity (40) suggests that the interaction between the binding pocket in domain III of Cry1Ac and the GalNAc on APN may not be critical for Cry1Ac toxicity. Cry1Ac toxicity may therefore be mediated by another binding site which is independent of GalNAc recognition.
Surface plasmon resonance studies have shown that Cry1Aa and Cry1Ab toxins bind at single sites on purified M. sexta APN and H. virescens APN, whereas Cry1Ac binds at two distinct sites on these insect APNs, only one of which involves GalNAc recognition (48, 50). The X-ray crystal structure of Cry1Aa revealed that the cavity forming the GalNAc binding pocket in Cry1Ac does not exist in the structure of Cry1Aa (25). According to amino acid residue alignment analysis, the amino acid residues forming this cavity in Cry1Ac are not conserved in Cry1Aa and Cry1Ab. The binding of Cry1Aa and Cry1Ab to M. sexta APN and H. virescens APN is not inhibited by preincubation with GalNAc (48, 50). One of the two Cry1Ac binding sites on these insect APNs is shared with Cry1Aa and Cry1Ab (48). These results might indicate that GalNAc recognition is not essential for the three Cry1A toxin-receptor interactions, leading to the hypothesis that the three Cry1A toxins have alternative binding sites that can bind to APN independent of the GalNAc moiety. This hypothetical APN binding site on Cry1Aa toxin seems to be essential for the toxin-receptor interaction and toxicity because Cry1Aa toxin binds only at a single site on these APNs. Furthermore, it is also likely that this APN binding site is important for the toxicities of Cry1Ab and Cry1Ac.
Previous studies have used alanine scanning or other point mutations to identify amino acid residues that affect the toxin-receptor interaction and toxicity. Amino acid substitution methods have been employed frequently for analysis of receptor binding sites and are effective for evaluating the contribution of an amino acid side chain to the protein interaction. However, mutations may have unexpected effects. For example, when point mutations are introduced at an amino acid located at a distance from the binding site, the electrostatic potential and conformation of the binding site may be changed. For this reason, amino acid substitution cannot be the sole method for determining the binding site, and the results may not indicate any amino acid residues that are important for the toxin-receptor interaction (32).
Identifying the receptor binding site on the Cry1Aa toxin involved in the toxin-receptor interactions could provide key insights into the mechanism of insecticidal specificity. This is one of our aims, as it should make it possible to improve the activity or specificity of Cry toxins. Knowledge of the receptor binding site on Cry1Aa toxin should allow improvements in the specificity and toxicity of Cry toxins. In this study, we attempted to determine the binding site on Cry1Aa toxin for the candidate Cry1Aa receptor in B. mori, 115-kDa APN type 1 (BmAPN1) (58) (GenBank accession no. AF084257).
To avoid any misinterpretations that might result from the unexpected effects of the methods mentioned above, we analyzed the receptor binding site using monoclonal antibodies (MAbs) that block the binding site for BmAPN1 on the Cry1Aa toxin. We produced a series of MAbs against Cry1Aa and screened them for the ability to block the binding between Cry1Aa and BmAPN1. We obtained two monoclonal antibodies capable of blocking the binding between Cry1Aa and BmAPN1 (blocking MAbs). We used a combination of several approaches to identify the location in the structure of Cry1Aa of the epitopes recognized by the blocking MAbs. We report here the location of the BmAPN1 binding site on the Cry1Aa molecule as determined by examining the region that is blocked by the blocking MAbs.
MATERIALS AND METHODS
Construction of the recombinant Cry1Aa protoxin gene.
The Cry1Aa protoxin gene (encoding Asp2-Glu1176) was amplified by PCR using primers 1Aa-S2 (5′-TGGATAACAATCCGAACATCAATGAATGC-3′) and 1Aa-AS2(XhoI) (5′-TTGCT CGAGACTATTCCTCCATAAGG-3′) with the template pES1 containing the Cry1Aa protoxin gene from B. thuringiensis subsp. kurstaki strain HD-1-Dipel (67). The amplified fragment was digested with NotI and was subcloned into the SmaI-NotI sites of the glutathione S-transferase (GST)-tagged expression vector pGEX-4T-3 (Amersham Pharmacia Biotech). The construct was used to transfect Escherichia coli BL21.
Preparation of the Cry1Aa, Cry1Ab, and Cry1Ac toxins.
Recombinant E. coli expressing the Cry1Aa toxin was constructed as described above. E. coli expressing the Cry1Ab toxin from B. thuringiensis subsp. aizawai strain IPL7 was provided by Sumitomo Chemical (59). These toxins were expressed as insoluble proteins. After the bacteria were disrupted by sonication, the inclusion bodies were separated from the cell debris by centrifugation and washed with 5% Triton X-100 in phosphate-buffered saline (PBS) (10 mM sodium phosphate buffer, pH 7.4, containing 145 mM NaCl).
Cry1Ac crystals were purified using the biphasic system of Goodman et al. (24) from a single-toxin producer, B. thuringiensis HD-73. The inclusion bodies from E. coli and crystals from B. thuringiensis were solubilized and treated with trypsin, and the activated toxins were purified as described elsewhere (55). Protein concentrations were determined by the Bradford method (3) using bovine serum albumin (BSA) as a standard.
Biotinylation of Cry1Aa toxin.
Activated Cry1Aa toxin (600 μg/ml in PBS) was mixed with a 20-fold mass of EZ-LinkSulfo-NHS-LC-LC-biotin (Pierce, Chester, United Kingdom), and the mixture was rotary mixed for 1 h at room temperature. The mixture was then dialyzed against several changes of PBS to remove excess biotin. Dialyzed protein solutions were centrifuged at 10,000 × g for 15 min and stored at 4°C.
Immunization and antibody production.
Ten-week-old BALB/c mice were immunized with activated Cry1Aa as follows: 20 μg immunogen emulsified in complete Freund's adjuvant was injected into the hypodermis on days 1, 14, and 28. The spleens were harvested for fusion on day 31. Spleen cells were fused with a nonsecreting mouse myeloma (P3-X63-Ag8-U1) as described previously (13).
Screening for hybridoma cells producing MAbs directed against Cry1Aa toxin was based on an enzyme-linked immunosorbent assay (ELISA) which evaluated the binding of the antibodies to Cry1Aa toxin from a preparation comparable to that used for the immunization.
Microtiter plates were coated with 100 μl of activated Cry1Aa (600 ng/ml) for 2 h at 4°C and blocked with 150 μl of 2% BSA in PBS. Aliquots of the culture supernatant (100 μl) containing MAbs from each hybridoma were added to the wells and incubated for 2 h at 37°C. Following washing, 100 μl of horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G antibody was added to each well and incubated for 2 h at 37°C. The plates were extensively washed with PBS, and 100 μl of 0.05% diammonium-2,2′-azino-bis(3-ethylbenzothiazoline- 6-sulfonate) (ABTS) in sodium citrate-phosphate buffer (100 mM Na2HPO4 · 2H2O, 80 mM citric acid monohydrate; pH 4.0) containing 0.01% H2O2 was added to each well. After 10 min, the absorbance at 415 nm was determined using a microplate reader (model 550; Bio-Rad).
Ascites fluids were prepared from the hybridomas as previously described (22), and MAbs 1B10 and 2C2 were purified from the ascites fluids by affinity chromatography on protein G-Sepharose HiTrap affinity columns (Pharmacia Biotech). Purified antibodies were conjugated with horseradish peroxidase using the method of Nakane and Pierce (56).
Fab fragments were prepared from MAbs 1B10 and 2C2 by papain digestion using immobilized papain (Pierce) and following the manufacturer's instructions. Fab fragments were separated from Fc fragments and intact molecules by using HiTrap protein A HP (Amersham Biosciences).
Competitive ELISA.
A partial fragment of BmAPN1 (7-kDa BmAPN1) containing the Cry1Aa binding region (Ile135 to Pro198) was expressed in E. coli cells as a GST fusion protein, as described previously (78). The 7-kDa BmAPN1 was expressed as an insoluble protein. The GST-fused 7-kDa BmAPN1 was purified by sonication, centrifugation, and washing with 5% Triton X-100 in MilliQ-treated water. The final pellet, referred to as inclusion bodies, was solubilized in an alkaline solution (50 mM NaOH containing 10 mM dithiothreitol) at room temperature for 2 h. The solubilized protein was neutralized with PBS and dialyzed against several changes of PBS. Protein concentrations were determined by the Bradford method (3) using BSA as a standard.
ELISA plates (96 wells; flat bottoms; Corning) were coated with 1 μM 7-kDa BmAPN1 in PBS (100 μl/well) at 37°C for 2 h. The wells were blocked by incubating the plates with PBS containing 2% BSA (150 μl/well) at 37°C for 2 h and by using an avidin/biotin blocking kit according to the manufacturer's instructions (Vector Laboratories). The wells were washed with PBS three times. Biotinylated Cry1Aa (20 nM) was preincubated with various concentrations (0, 20, 50, 100, 200, and 500 nM) of each MAb at 37°C for 90 min and then added to the wells (100 μl/well). Next, 100 μl horseradish peroxidase-conjugated streptavidin was added to each well, and the plates were incubated at 37°C for 90 min. Bound streptavidin was detected by incubation with an ABTS solution (100 μl/well) at room temperature for 15 min and was measured by determining the optical density at 415 nm using a microtiter plate reader. The results were expressed in terms of B/BMAX, where B and BMAX are the enzymatic activities bound to the solid phase measured at the various concentrations of MAbs and in the absence of MAbs, respectively.
Immobilization of Cry1Aa toxin on the sensing surface of a biosensor.
Single-well cuvettes with carboxylate surfaces were used with the N-hydroxysuccinimide and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide coupling system (Affinity Sensors, Cambridge, United Kingdom). The well of each cuvette was coated with activated Cry1Aa which was immobilized on the carboxylate surface via its amino groups using succinimide ester chemistry, as described below. The immobilization buffer was 10 mM sodium acetate buffer, pH 5.0.
For antigen coupling, the cuvette was washed with PBS containing 0.05% Tween 20, pH 7.4, for 10 min prior to activation of the surface using N-hydroxysuccinimide and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide. The cuvette was then equilibrated using sodium acetate buffer (pH 5.0), and 200 μg/ml of Cry1Aa was added to the well. Eventually, 1.82 ng/mm2 of Cry1Aa molecules was immobilized on the sensing surface via amino groups. The noncoupled ligand was removed by washing with 20 mM HCl, and the unreacted surface was blocked with BSA.
Determination of binding compatibility for each pair of MAbs.
After covalent immobilization of Cry1Aa on the carboxylate sensor surface, the compatible binding of three different MAbs, MAbs 1B10, 2F9, and 2C2, for Cry1Aa molecules was determined using an IAsys resonant mirror optical biosensor (Affinity Sensors, Cambridge United Kingdom). To measure the binding of each MAb individually with Cry1Aa molecules, each of the MAbs was added to the Cry1Aa-immobilized surface at a concentration of 100 μM, and the association was observed for 5 min. Subsequently, each MAb solution was replaced with binding buffer (PBS) containing no MAb, and the dissociation of the binding was observed for 5 min. At the end of this cycle, the PBS in the cuvette was replaced with 20 mM HCl to regenerate the sensor surface.
Next, we determined the binding compatibility of two of the MAbs, MAbs 2F9 and 2C2, for Cry1Aa in the presence of MAb 1B10. MAb 1B10 was added to the Cry1Aa-immobilized surface as the first antibody at a concentration of 100 μM, and the association of MAb 1B10 was observed for 5 min. Each of the MAbs (MAbs 1B10, 2F9, and 2C2) was added as the second antibody at a concentration of 100 μM, and the additive association was observed for 5 min. The MAb solution was replaced with PBS with no MAb, and the dissociation of the binding was observed for 5 min. At the end of this cycle, the PBS in the cuvette was replaced with 20 mM HCl to regenerate the sensor surface. This procedure was repeated using MAb 2F9 as the first antibody to determine the binding compatibility of each of the other two MAbs (MAbs 1B10 and 2C2) for Cry1Aa molecules in the presence of MAb 2F9.
Construction of Cry1Aa gene recombinant deletion mutants.
The activated Cry1Aa toxin gene, N615 (encoding Ile11 to Asp615), was amplified by PCR using primers 1Aa-S11(BamHI) (5′-CAATGGATCCATTCCTTATAATTGTTTAAGTAAC-3′) and 1Aa-AS615 (XhoI) (5′-TGCTCTCGAGAAATCATATTCTGCCTCAAAGGT-3′) with the template pES1. Four C-terminal deletions of the Cry1Aa toxin gene, N593 (encoding Ile11 to Gly593), N570 (encoding Ile11 to Phe570), N531 (encoding Ile11 to Thr531), and N513 (encoding Ile11 to Asn513), were amplified by PCR using the sense primer 1Aa-S11(BamHI) (5′-CAATGGATCCATTCCTTATAATTGTTTAAGTAAC-3′) and the antisense primers 1Aa-AS593(XhoI) (5′-AACTCTCGAGCCTGAATTGAAGACATGAGCACT-3′), 1Aa-AS570(XhoI) (5′-CGGACTCGAGAAACCTACAGTCCTAAAGCTTCC-3′), 1Aa-AS531(XhoI) (5′-TAAACTCGAGGTAGAAGCGTAGCGAATTCTTAC-3′), and 1Aa-AS513(XhoI) (5′-TGCACTCGAGTTTACTCTTAAGGTTGAAATCTG-3′), respectively, with the template pES1. Five N-terminal deletions of the Cry1Aa toxin gene, C197 (encoding Thr197 to Asp615), C255 (encoding Tyr255 to Asp615), C354 (encoding Gly354 to Asp615), C456 (encoding His456 to Asp615), and C514 (encoding Ile514 to Asp615), were amplified by PCR using the sense primers 1Aa-S197(BamHI) (5′-TAATGGATCCACTAGGCTTATTGGCAACTATACA-3′), 1Aa-S255 (BamHI) (5′-TAGTGGATCCTATCCAATTCGAACAGTTTCCCAA-3′), 1Aa-S354(BamHI) (5′-CTCAGGATCCGGTTTGGGGATTTTTAGAACATTA-3′), 1Aa-S456(BamHI) (5′-TTCTGGATCCCATCGCAGTGCTGAATTTAATAAT-3′), and 1Aa-S514(BamHI) (5′-AAGAGGATCCATTACTGCACCATTATCACAAAGA-3′), respectively, andthe antisense primer 1Aa-AS615(XhoI) (5′-TGCTCTCGAGAAATCATATTCTGCCTCAAAGGT-3′) with the template pES1. The 10 amplified fragments were double digested with BamHI and XhoI and subcloned into the BamHI and XhoI sites of the GST-tagged expression vector pGEX-4T-3(Amersham Pharmacia Biotech); the constructs were transfected into E. coli BL21. The other two C-terminal deletions of the Cry1Aa toxin gene, N507 (encoding Met1 to Ile507) and N282 (encoding Met1 to Gly282), were amplified by PCR using the sense primer 1Aa-S1(SmaI) (5′-AGGCCCGGGATGGATAACAATCCGAACA-3′) and the antisense primers 1Aa-AS507(NotI) (5′-TCTTGCGGCCGCAATCTGGCCAGGTGAA-3′) and 1Aa-AS282(NotI) (5′-TTCTCGCGGCCGCTCCACGAAAACTACC-3′), respectively, with the template pES1. The amplified fragments were digested with NotI and subcloned into the SmaI-NotI sites of the GST-tagged expression vector pGEX-4T-3; the constructs were transfected into E. coli BL21.
Preparation of activated Cry1Aa toxin and Cry1Aa deletion mutants.
E. coli BL21, transformed with plasmids encoding wild-type activated Cry1Aa toxin (N615) and 11 Cry1Aa deletion mutants, were grown in LB medium (1.5% Polypepton [Wako Pure Chemical Industries, Ltd.], 1% Bacto yeast [Nacalai tesque], 85 mM NaCl [Wako Pure Chemical Industries, Ltd.]) at 37°C to an optical density at 600 nm of 0.6, induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Wako Pure Chemical Industries, Ltd.), and grown for another 4 h at 37°C. The proteins were expressed as insoluble proteins. The bacteria were disrupted by sonication, and the inclusion bodies were separated from the cell debris by centrifugation and washed with 5% Triton X-100 in PBS. The inclusion bodies were treated with sodium dodecyl sulfate (SDS) sample buffer. Protein concentrations were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) using BSA as a standard.
To avoid any effects on antibody binding due to steric hindrance by glutathione S-transferase, two Cry1Aa deletion mutants (C456 and C514) were cleaved with thrombin. E. coli BL21, transformed with the plasmid encoding C456 and C514, was grown in LB medium at 37°C to an optical density at 600 nm of 0.6, induced with 1 mM IPTG, and grown for another for 12 h at 25°C. Each of the GST fusion proteins was produced as a soluble protein, which was separated from the cell debris by sonication and centrifugation. Each GST fusion protein was purified as a single band by sequential use of glutathione-Sepharose 4B (Amersham Pharmacia Biotech AB). GST fusion proteins bound to glutathione-Sepharose beads were incubated at room temperature for 2 h with bovine thrombin (10 cleavage units/mg; Wako Pure Chemical Industries, Ltd). The proteins cleaved from the beads by thrombin were treated with SDS sample buffer. Protein concentrations were analyzed by SDS-PAGE using BSA as a standard. The details of the experimental procedures are included in the manufacturer's instructions (GST gene fusion system; Amersham Pharmacia Biotech AB).
Immunoblotting.
Prepared proteins (1 μg/lane) were subjected to SDS-PAGE (10% polyacrylamide) by the method of Laemmli (37). After SDS-PAGE, the proteins were transferred to Immobilon filters (nitrocellulose membrane; Millipore), blocked with 2% BSA dissolved in 10 mM Tris-HCl, pH 8.3, 150 mM NaCl, 0.05% (vol/vol) Tween 20 (TNT) for 12 h at 4°C, and then incubated for 2 h at room temperature with horseradish peroxidase-conjugated MAb diluted 1:10,000. The membrane was washed three times with TNT. The bands on the blots were visualized using the ECL detection system (Amersham Pharmacia Biotech).
Octapeptide ELISA.
Antibody binding to peptides coupled to a solid phase was determined using PEPSCAN as described by Geysen et al. (18). Eight-by-twelve arrays of plastic pins in which each pin bore a different octapeptide covering the entire domain III of Cry1Aa (amino acid residues 462 to 609) with six-residue overlaps (for example, residues 462 to 469, 464 to 471, etc.) were synthesized using a MULTIPIN peptide synthesis kit according to the manufacturer's instructions (Chiron Technologies Pty. Ltd.). The pins were first incubated in 50 ml of PBS containing 0.1% Tween 20 (PBST) supplemented with 1% BSA and 1% ovalbumin for 2 h at room temperature with gentle rocking to block nonspecific protein binding sites. The pins were washed three times with PBST and then incubated for 90 min at room temperature with horseradish peroxidase-conjugated MAb 1B10 or 2C2. After three washes with 50 ml of PBST, the bound streptavidin was detected by incubation with an ABTS solution for 15 min, and the amount was determined by determining the optical density at 415 nm using a microtiter plate reader.
After each use, the pins were cleaned by ultrasound treatment in a solution containing SDS and 2-mercaptoethanol for 2 h at 65°C to remove the bound antibody. Proper cleaning was confirmed by checking for residual bound streptavidin by incubation with an ABTS solution. The pins were reused approximately 10 times.
Site-directed mutagenesis.
From E. coli harboring the plasmid containing the B. thuringiensis δ-endotoxin-activated Cry1Aa gene (N615, encoding Ile11 to Asp615), the pN615 plasmid was purified using a QIAprep Spin miniprep kit (QIAGEN). Site-directed mutations were introduced into Cry1Aa using pN615 as a template and a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The mutated DNA was used to transform E. coli BL21(DE3) cells. Mutated clones were confirmed by DNA sequencing; sequencing reactions were performed by the method of Sanger et al., and sequences were resolved using a Long-Read Tower sequencer (Amersham Pharmacia Biotech) according to the manufacturer's instructions (66).
Introduction of small blocking molecules into single-Cys-substitution Cry1Aa mutants.
N-(9-Acridinyl)maleimide (NAM) (2.8 mg/ml; DOJINDO) was solubilized in acetone. Single-Cys-substitution Cry1Aa mutants were expressed as insoluble proteins. The bacteria were disrupted by sonication The inclusion bodies were separated from the cell debris by centrifugation, washed with 5% Triton X-100 in PBS, solubilized, and treated with trypsin. The activated toxins were purified as described elsewhere (55). Protein concentrations were determined by the Bradford method (3) using BSA as a standard.
The trypsin-digested, single-Cys-substitution Cry1Aa mutants (60 μg/ml in PBS) were mixed with the NAM solution (protein/NAM ratio, 1:20 [mol/mol]), and the mixture was rotary mixed at room temperature for 12 h. The mixture was then dialyzed against several changes of PBS to remove unreacted NAM. Dialyzed protein solutions were centrifuged at 10,000 × g for 15 min and stored at 4°C.
Immunodot blotting.
Prepared proteins (100 ng/μl) were dotted onto a nitrocellulose membrane by using a digital micropipette (Nichiryo). The membrane was allowed to air dry and was tested for MAb reactivity by immunoblotting.
RESULTS
MAbs that block the binding of Cry1Aa toxin to BmAPN1.
Seven monoclonal antibodies (MAbs 1B10, 1E10, 1G10, 2A11, 2C2, 2F9, and 3C7) were raised against Cry1Aa toxin in a BALB/c mouse. These seven anti-Cry1Aa MAbs were tested for the ability to block the binding of Cry1Aa to BmAPN1 in a competitive ELISA. The ability of each MAb to block the binding of Cry1Aa toxin to a 7-kDa fragment of BmAPN1 containing the Cry1Aa binding region (78) was investigated by preincubating each MAb with biotinylated Cry1Aa before the reaction with the 7-kDa BmAPN1 fragment.
Of the seven MAbs, only MAbs 1B10 and 2C2 blocked the binding of Cry1Aa toxin to 7-kDa BmAPN1; the other MAbs (MAbs 1E10, 1G10, 2A11, 2F9, and 3C7) had little or no effect on the binding of Cry1Aa toxin to 7-kDa BmAPN1 (Fig. 1). The concentration dependence of the blocking effects of the two blocking MAbs was determined in a similar experiment; MAbs 1B10 and 2C2 blocked the binding of Cry1Aa toxin to 7-kDa BmAPN1 in a dose-dependent manner (Fig. 2). The blocking effects of the Fab fragments prepared from MAbs 1B10 and 2C2 were determined in a similar experiment. The Fab fragment of MAb 2C2 blocked the binding of Cry1Aa toxin to 7-kDa BmAPN1 in a dose-dependent manner, similar to the intact molecule, while the Fab fragment of MAb 1B10 had no effect on the binding of Cry1Aa toxin to 7-kDa BmAPN1. As intact molecules, both of the blocking antibodies competed with BmAPN1 for binding to Cry1Aa. However, only the Fab fragments of MAb 2C2 competed with BmAPN1 for binding to Cry1Aa; the Fab fragment of MAb 1B10 did not compete (Fig. 2). These results suggest that the surface of Cry1Aa blocked by MAb 2C2 overlaps the binding site for BmAPN1 on Cry1Aa toxin, whereas the surface on Cry1Aa blocked by MAb 1B10 does not appear to overlap but rather is located close to the BmAPN1 binding site on Cry1Aa toxin.
FIG. 1.
Inhibition of Cry1Aa binding to BmAPN1 by monoclonal antibodies. Biotinylated Cry1Aa was preincubated with or without each MAb at a concentration of 500 nM for 90 min and then added to wells coated with 7-kDa BmAPN1. Then it was incubated for 90 min with horseradish peroxidase-conjugated streptavidin. Bound streptavidin was detected after incubation with an ABTS solution for 15 min by determining the optical density at 415 nm using a microtiter plate reader.
FIG. 2.
Dose-dependent inhibition of Cry1Aa binding to BmAPN1 by blocking antibodies. Biotinylated Cry1Aa was preincubated with various concentrations of intact MAbs and Fab fragments of MAbs for 90 min and then added to wells coated with 7-kDa BmAPN1. Then it was incubated for 90 min with horseradish peroxidase-conjugated streptavidin. Bound streptavidin was detected after incubation with an ABTS solution for 15 min by determining the optical density at 415 nm using a microtiter plate reader.
Determination of binding compatibility for MAbs 1B10, 2C2, and 2F9.
To investigate the relative locations of the epitopes for MAbs 1B10, 2C2, and 2F9 on the surface of Cry1Aa, tests were performed using an IAsys resonant mirror optical biosensor. The same quantity of each of the MAbs (MAbs 1B10, 2C2, and 2F9) was bound individually to Cry1Aa immobilized on a cuvette surface, as demonstrated by the equal heights of the sensorgrams. To test the binding competition among the three MAbs, a solution of MAb 1B10, 2C2, or 2F9 was added to the cuvette after MAb 1B10 had been bound to the immobilized Cry1Aa, and the binding of the added antibody to the surface was determined. The results are shown in Fig. 3A. When additional MAb 1B10 was added, almost no further binding to the immobilized Cry1Aa occurred, suggesting that the MAb 1B10 epitopes on the surface had been largely saturated by the initial solution of MAb 1B10. This type of binding curve, which demonstrates that there was no additional antibody binding, would be observed if a second antibody (in this case, the same antibody, MAb 1B10) competed for the same binding site as the first MAb. When MAb 2F9 was added, the amount of the bound antibody was twofold greater than the amount observed with MAb 1B10 alone, indicating that the binding of MAb 2F9 was not blocked by MAb 1B10. This type of binding curve would be observed if a second antibody (in this case, a different antibody, MAb 2F9) did not compete for the same binding site as the first MAb. The binding curve for the added MAb 2C2 was similar to that for the additional MAb 1B10, suggesting that MAbs 1B10 and 2C2 compete for binding to Cry1Aa and that the binding sites for MAbs 1B10 and 2C2 on Cry1Aa overlap or are close together.
FIG. 3.
Determination of binding compatibility using the IAsys optical sensor. The tests were performed using Cry1Aa covalently immobilized on the carboxylated surface of a cuvette and the IAsys optical sensor. The molecules were injected successively. (A) First MAb 1B10 was added to block its epitope on the immobilized Cry1Aa, and then MAbs 1B10, 2F9, and 2C2 were added. (B) First MAb 2F9 was added to block its epitope on the immobilized Cry1Aa, and then MAbs 1B10, 2F9, and 2C2 were added.
Using the same method, the binding of MAbs 1B10 and 2C2 to immobilized Cry1Aa was determined after MAb 2F9 had first been allowed to bind to the immobilized Cry1Aa. The results are shown in Fig. 3B. When additional MAb 2F9 was added, almost no additional antibody was bound to the immobilized Cry1Aa, indicating that the epitopes on the surface had been fully saturated by the initial MAb 2F9. When MAb 1B10 was added, the antibody binding was twofold greater than that seen with MAb 2F9 alone, indicating that MAb 2F9 did not block the binding of MAb 1B10 and that these two antibodies do not compete for binding to Cry1Aa. The binding curve for MAb 2C2 was similar to that for the additional MAb 2F9 following the initial MAb 2F9 binding. This result shows that MAbs 2F9 and 2C2 compete for binding to Cry1Aa and suggests that the areas on Cry1Aa blocked by MAbs 1B10 and 2C2 overlap but are distinct.
Binding of MAbs to Cry1Aa toxin domains.
The domain similarities between the Cry1Aa, Cry1Ab, and Cry1Ac toxins are shown in Fig. 4A. In domain I, three toxins have the same sequence, with the exception of only six different amino acid residues between Cry1Aa and Cry1Ab, three different amino acid residues between Cry1Aa and Cry1Ac, and three different amino acid residues between Cry1Ab and Cry1Ac. The domain II sequence of Cry1Aa toxin exhibits only 69% identity with the domain II sequences of Cry1Ab and Cry1Ac, whereas Cry1Ab and Cry1Ac have the same sequence, with the exception of only two amino acid residues. In domain III, Cry1Aa and Cry1Ab are identical, but each exhibits only 38% identity with Cry1Ac.
FIG. 4.
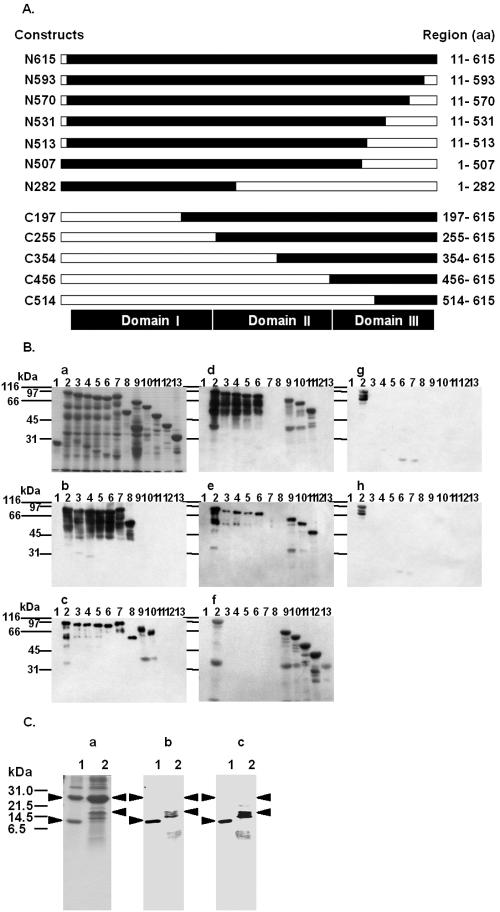
Immunoblot analysis of the reactivity of each MAb with the three toxins. (A) Schematic representation of the domain similarities of Cry1Aa, Cry1Ab, and Cry1Ac. (B) Approximately 1 μg of Cry1Aa (lane 1), Cry1Ab (lane 2), and Cry1Ac (lane 3) was subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and probed with MAbs 3C7 (blot b), 1G10 (blot c), 1B10 (blot d), 2C2 (blot e), 1E10 (blot f), 2F9 (blot g), and 2A11 (blot h). The proteins on the gels were visualized by Coomassie brilliant blue staining (gel a).
We used immunoblot analysis to determine which Cry1Aa toxin domain bound each MAb. We tested the binding of seven MAbs to membrane blots of the three toxins. MAbs 1B10, 2C2, and 1E10 recognized Cry1Aa and Cry1Ab but not Cry1Ac, suggesting that these MAbs bind to domain III of Cry1Aa toxin (Fig. 4B, blots d, e, and f). MAbs 2A11 and 2F9 recognized only Cry1Aa and not Cry1Ab or Cry1Ac, suggesting that these MAbs bind to domain II of Cry1Aa toxin (Fig. 4B, blots g and h). MAbs 3C7 and 1G10 recognized all three toxins, indicating that these MAbs bind to regions conserved among all three toxins (Fig. 4B, blots b and c).
Binding of MAbs to a set of Cry1Aa toxin in-frame deletion mutants.
The toxin derivatives used in this study were wild-type activated Cry1Aa (N615), Cry1Aa with six C-terminal deletions (N593, N570, N531, N513, N507, and N282), and Cry1Aa with five N-terminal deletions (C197, C255, C354, C456, and C514), which are schematically shown in Fig. 5A. All of the derivatives were constructed as glutathione S-transferase fusion proteins; nonfused glutathione S-transferase was used as a negative control.
FIG. 5.
Immunoblot analysis of the reactivity of each MAb with Cry1Aa toxin deletion mutants. (A) Schematic diagram of Cry1Aa toxin deletion mutants. The open bars indicate the deleted portions of the full-length Cry1Aa toxin. The designation of each deletion mutant is indicated on the left, and the expressed regions are indicated on the right. aa, amino acids. (B) GST (lane 1) and approximately 2 μg of each of the 12 GST-fused Cry1Aa toxin deletion mutants, N615 (lane 2), N593 (lane 3), N570 (lane 4), N531 (lane 5), N513 (lane 6), N507 (lane 7), N282 (lane 8), C197 (lane 9), C255 (lane 10), C354 (lane 11), C456 (lane 12), and C514 (lane 13), were subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and probed with MAbs 3C7 (blot b), 1G10 (blot c), 1B10 (blot d), 2C2 (blot e), 1E10 (blot f), 2F9 (blot g), and 2A11 (blot h). The proteins on the blots were visualized by Coomassie brilliant blue staining (gel a). (C) Two Cry1Aa toxin C-terminal deletion mutants, C514 and C456, were expressed in E. coli at 25°C for 12 h. GST fusion proteins were purified from bacterial lysates by using glutathione-Sepharose 4B. Fusion proteins containing a thrombin recognition site were cleaved while they were bound to glutathione-Sepharose 4B. Approximately 1 μg of C514 (lane 1) and C456 (lane 2) in solution after enzymatic treatment was subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and probed with MAbs 1B10 (blot b) and 2C2 (blot c). The proteins on the gels were visualized by Coomassie brilliant blue staining (gel a). The arrowheads indicate the positions of the 26-kDa glutathione S-transferase (gel a, lanes 1 and 2), the 11-kDa thrombin fragment of C514 protein (lane 1), and the 17-kDa thrombin fragment of C456 (lane 2).
To determine the structure involved in antigen recognition by each anti-Cry1Aa MAb, we investigated the pattern of immunoreactivity against a set of Cry1Aa toxin in-frame deletion mutants. This analysis was performed using seven MAbs and membrane blots of the Cry1Aa derivatives (Fig. 5B). The deletion mutant proteins were separated on an SDS-polyacrylamide gel, transferred to a nitrocellulose membrane, and tested for reactivity with each MAb by immunoblotting. Interestingly, the seven MAbs revealed several different immunoblot patterns, as shown in Fig. 5B. An evaluation of these results for all the MAbs tested is summarized in Table 1.
TABLE 1.
Immunoreactivity of MAbs against wild-type (N615) and deletion mutant proteins of Cry1Aa
| Cry1Aa constructa | Reactivity of MAbsb
|
||||||
|---|---|---|---|---|---|---|---|
| 3C7 | 1G10 | 1B10 | 2C2 | 1E10 | 2F9 | 2A11 | |
| C-terminal deletions | |||||||
| N615 | + | + | + | + | + | + | + |
| N593 | + | + | + | + | − | − | − |
| N570 | + | + | + | + | − | − | − |
| N531 | + | + | + | + | − | − | − |
| N513 | + | + | + | + | − | − | − |
| N507 | + | + | − | − | − | − | − |
| N282 | + | + | − | − | − | − | − |
| N-terminal deletions | |||||||
| C197 | − | + | + | + | + | − | − |
| C255 | − | + | + | + | + | − | − |
| C354 | − | − | + | + | + | − | − |
| C456 | − | − | +c | +c | + | − | − |
| C514 | − | − | +c | +c | + | − | − |
Cry1Aa deletion mutants are illustrated in Fig. 5A.
+, immunoreactive; −, not immunoreactive.
These analyses were performed after thrombin cleavage of GST fusion deletion mutant proteins.
MAbs 1B10 and 2C2 reacted with four of the C-terminal deletion mutants (N593, N570, N531, and N513) but not with N507 and N282, and they reacted with three of the N-terminal deletion mutants (C197, C255, and C354) but not with C456 and C514 (Fig. 5B, blots d and e). These two MAbs also reacted with N513 (amino acids 11 to 513) but not with N507 (amino acids 1 to 507), suggesting that the region between amino acids 508 and 513 may play an important role in the recognition by MAbs 1B10 and 2C2. However, this interpretation is inconsistent with the finding that MAbs 1B10 and 2C2 did not react with C456 (amino acids 456 to 615). We propose that steric hindrance owing to the GST fused at the N terminus of these recombinant proteins might have prevented the binding of these MAbs to C456 and C514.
To test this hypothesis, C456 and C514 were cleaved by thrombin in order to remove GST from the recombinant proteins, and the cleaved proteins were tested by immunoblotting to determine their reactivity with MAbs 1B10 and 2C2. MAbs 1B10 and 2C2 reacted with C514 and C456 without GST (Fig. 5C, blots b and c). Thus, C456 and C514 contained the epitopes for MAbs 1B10 and 2C2, and the results suggest that the epitopes consist of two regions, one between amino acids 508 to 513 and the other between amino acids 514 to 615.
MAb 1E10 reacted with all of the N-terminal deletion mutants but did not react with any of the C-terminal deletions (Fig. 5B, blot f). The lack the C-terminal 11 amino acids (amino acids 594 to 615) in all of the C-terminal deletion mutants may explain their failure to be recognized by MAb 1E10, suggesting that these 11 amino acids play an important role in the recognition by MAb 1E10. An alternative explanation is that the lack of the C-terminal 11 amino acids influenced the general structure of domain III, in which this sequence is located. In any event, this result confirmed that the epitope of MAb 1E10 is localized in domain III.
MAb 1G10 reacted with all of the C-terminal deletions and with only two of the Cry1Aa N-terminal deletions (C197 and C255); MAb 1G10 did not react with C354, C456, or C514. As MAb 1G10 reacted with N282 (amino acids 1 to 282) and C255 (amino acids 255 to 615) but not with C354 (amino acids 354 to 615), the data suggested that the region between amino acids 255 and 282 may be important in the recognition by MAb 1G10 (Fig. 5B, blot c). This result indicates that MAb 1G10 recognizes an epitope located in domain II.
MAb 3C7 reacted with all of the C-terminal deletions but not with any N-terminal deletions (Fig. 5B, blot b); it did not react with C197, which lacked the region between amino acids 11 and 196, indicating that the C-terminal 86 amino acids are important for recognition by MAb 3C7. This region is localized in domain I, suggesting that the epitope for MAb 3C7 is located in domain I.
MAbs 2F9 and 2A11 reacted with wild-type activated Cry1Aa (N615; full length) but did not react with any of the C- or N-terminal deletions (Fig. 5B, blots g and h). Despite the results shown in gels g and h of Fig. 4 suggesting that the epitopes of MAbs 2F9 and 2A11 are localized in domain II, MAbs 2F9 and 2A11 did not react with the deletion proteins containing domain II, which included the five C-terminal deletions N593, N570, N531, N513, and N507 and the two N-terminal deletions C197 and C255. One possible explanation for this finding is that the complete conformation of the antigen is essential for recognition by MAbs 2F9 and 2A11 and that the C- and N-terminal deletions altered the structure of domain II. Another possible explanation is that, although the epitopes for MAbs 2F9 and 2A11 are mainly localized in domain II, parts of the structure of domain I or III may be required for recognition by MAbs 2F9 and 2A11. In either case, the C- and N-terminal deletions might have disrupted the epitopes for MAbs 2F9 and 2A11 such that the MAbs could not maintain sufficient affinity for the mutants. In any event, the epitopes for MAbs 2F9 and 2A11 appear to be localized in domain II.
Binding of blocking antibodies to octapeptides.
We investigated the linear amino acid sequences recognized by the two blocking MAbs. Octapeptides were synthesized covering the entire domain III of the Cry1Aa sequence (amino acids 462 to 609) with six-residue overlaps (for example, residues 462 to 469, 464 to 471, etc.); each octapeptide was attached to a plastic pin in an 8-by-12 array of pins. Both of the blocking MAbs, MAbs 1B10 and 2C2, were able to bind to several octapeptides coupled to pins, as detected by ELISA. MAb 1B10 bound mainly to a single peptide, 582VFTLSAHV589 (Fig. 6A), while MAb 2C2 bound to three peptides, 520QRYRVRIR527, 570FTTPFNFS577, and 582VFTLSAHV589 (Fig. 6B). Thus, both antibodies bound to 582VFTLSAHV589.
FIG. 6.
Analysis of the binding of MAbs to octapeptides. Eight-by-twelve arrays of plastic pins were constructed, and each pin bore a different octapeptide; together, the pins covered the entire domain III of the Cry1Aa sequence (amino acids 462 to 609) with six-residue overlaps. The pins were incubated for 90 min at room temperature with horseradish peroxidase-conjugated MAbs 1B10 (A) and 2C2 (B). Bound streptavidin was detected after incubation with an ABTS solution for 15 min by determining the optical density at 415 nm (OD415) using a microtiter plate reader. To measure the binding of the MAbs to the pins, the background value was first calculated by determining the measured value for the control pin bearing an octaglycine peptide (GGGGGGGG). The background value was subtracted from each value. For MAbs 1B10 and 2C2, the assay was carried out twice, and nearly the same results were obtained each time for each MAb. The peptide coupled to each pin is indicated at the bottom by the N-terminal amino acid number of the octapeptide; the sequence of the octapeptide consisted of this residue and the subsequent seven residues in the linear sequence.
Location of candidate epitope sequences for blocking MAbs in the three-dimensional structure of Cry1Aa and effect of small blocking molecules on MAb binding to Cry1Aa.
We used three approaches to epitope mapping, two involving immunoblotting and one involving PEPSCAN, to identify four regions of Cry1Aa at which the blocking MAbs bound. However, because the results were obtained using fragments of proteins or short peptides, it is unclear whether these regions play a role in the binding of the blocking MAbs to the intact antigen. In order to examine the locations of these candidate epitopes within the structure of Cry1Aa, these regions were mapped on the three-dimensional model of Cry1Aa (Fig. 7). Two candidate epitopes were determined; one site consisted of 508STLRVN513 and 582VFTLSAHV589, and the other site consisted of 520QRYRVRIR527 and 570FTTPFNFS577. As shown in Fig. 7, 508STLRVN513 and 582VFTLSAHV589 are located adjacent to each other, in the brown and red regions; 520QRYRVRIR527 and 570FTTPFNFS577 are located adjacent to each other, in the blue and magenta regions.
FIG. 7.
Epitopes of blocking antibodies in a three-dimensional model of Cry1Aa. (A) The amino acid residues in domains I, II, and III are pink, light green, and light blue, respectively. The single octapeptide to which MAb 1B10 bound, 582VFTLSAHV589, is red. The three octapeptides to which MAb 2C2 bound, 520QRYRVRIR527, 570FTTPFNFS577, and 582VFTLSAHV589 are blue, magenta, and red, respectively. In addition, Arg521 (green) and Val582 (yellow), each of which was mutated to cysteine, are shown. The peptide 508STLRVN513 (brown) adjoins the 582VFTLSAHV589 site in the three-dimensional structure; this region is expected to be involved in the binding of the blocking MAbs. The candidate binding sites for MAbs 1B10 and 2C2 are indicated by blue and red circles. (B) Panel A rotated 72° about the y axis and 45° about the x axis. (C) Panel A rotated 90° about the x axis.
The epitope for MAb 1B10 might be the single site consisting of 508STLRVN513 and 582VFTLSAHV589. However, the candidate epitopes for MAb 2C2 could be 508STLRVN513 and 582VFTLSAHV589 or 520QRYRVRIR527 and 570FTTPFNFS577. Todetermine the true epitopes for MAb 2C2, we introduced two cysteine substitutions, at R521C and V582C in the two candidate epitopes on the Cry1Aa toxin, to act as sites for small blocking molecules (NAM). In Fig. 7, Arg521 is indicated by green spheres, and Val582 is indicated by yellow spheres.
The single-Cys-substitution mutants of Cry1Aa were constructed as GST fusion proteins, which were purified from E. coli, solubilized, activated by trypsin, and analyzed by SDS-PAGE (Fig. 8). Almond and Dean reported that poor expression can be correlated for an unstable or misfolded protein (1), but this finding is not applicable to our results because we found that both of the mutant proteins (26-kDa GST plus 60-kDa mutant toxins, resulting in 86-kDa proteins) were expressed (Fig. 8, lanes 1 and 3) as strongly as the wild-type Cry1Aa (data not shown). After tryptic digestion, each mutant toxin yielded a 60-kDa trypsin-resistant core fragment (Fig. 8, lanes 2 and 4). According to protein sequencing results, the N-terminal amino acid sequences of the tryptic core fragments of the two mutant proteins were similar to those of the wild-type Cry1Aa, starting at Ile29 (data not shown). Finally, the wild-type Cry1Aa and the mutant proteins were similarly processed into stable 60-kDa toxin molecules upon treatment with trypsin, suggesting that the substitutions did not cause any structural alterations to the protein.
FIG. 8.
Coomassie brilliant blue R250-stained SDS-PAGE comparing the yields of the GST-toxin constructs and the stabilities of the mutant toxin constructs in the presence of trypsin. Two mutant toxin constructs, R521C and V582C, were solubilized and activated by trypsin treatment. The details are described in Materials and Methods. Solubilized GST-R521C (lane 1), trypsin-activated R521C (lane 2), solubilized GST-V582C (lane 3), and trypsin-activated V582C (lane 4) were subjected to SDS-PAGE. The proteins on the gel were visualized by Coomassie brilliant blue staining.
The mutant proteins expressed in E. coli have four Cys residues (amino acids 84, 137, 168, and 177) in the GST region and only two Cys residues (amino acids 15 and 521 or 582) in the Cry1Aa toxin region; however, the GST region and the N-terminal part of the Cry1Aa toxin region containing five of the six Cys residues were removed by tryptic digestion. Thus, each of the two single-Cys-substitution mutants of Cry1Aa had only one Cys residue (amino acid 521 or 582) after digestion with trypsin. We introduced a different small blocking molecule, NAM (molecular weight, 274.27), at two candidate epitopes on Cry1Aa. NAM contains a maleimide group, which can specifically react with the thiol group of Cys in protein molecules to covalently bind it to the protein. When NAM was reacted with the single-Cys-substitution mutants, it specifically bound to the Cys residues of the R521C and V582C mutants of Cry1Aa. NAM molecules introduced on the surface of Cry1Aa at either of these two sites may block MAb binding at the site owing to steric hindrance.
We used immunoblot analysis to determine which site on the surface of the Cry1Aa toxin bound each MAb. In this analysis we examined the reactivities of the seven MAbs with wild-type Cry1Aa and Cry1Ac, the two single-Cys-substitution mutants of Cry1Aa (R521C and V582C), and the two NAM-modified Cys substitution mutants of Cry1Aa (R521C-NAM and V582C-NAM). The six toxin constructs were dotted onto a nitrocellulose membrane and probed with each of the seven horseradish peroxidase-conjugated MAbs. The bound MAbs were detected by ECL. One advantage of this method was that it allowed investigation of true epitopes because it used the antigen protein in its correctly folded, native structural conformation with no denaturation.
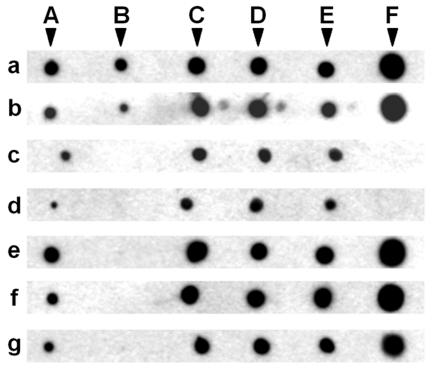
MAbs 3C7 and 1G10 recognized all six toxin constructs (Fig. 9, blots a and b). MAb 1E10, with an epitope in domain III, and MAbs 2F9 and 2A11, with epitopes in domain II, recognized all the toxin constructs but one, Cry1Ac toxin (Fig. 9, blots e, f, and g). Similarly, both blocking MAbs, MAbs 1B10 and 2C2, recognized Cry1Aa toxin but not Cry1Ac toxin. In this assay, both blocking MAbs recognized V582C but not V582C-NAM, suggesting that the presence of NAM on V582C prevented the two blocking MAbs from binding to Cry1Aa but that the Cys substitution did not affect the binding of these MAbs (Fig. 9, blots c and d). All of the MAbs except MAbs 1B10 and 2C2 recognized V582C-NAM, showing that NAM did not affect the binding of the other antibodies (Fig. 9, blots a, b, e, f, and g). In addition, both blocking MAbs recognized R521C-NAM, indicating that the effect of NAM on the binding of these MAbs was site specific for V582C (Fig. 9, blots c and d). These results suggest that the true epitopes of MAbs 1B10 and 2C2 are located at the site containing Val582.
FIG. 9.
Immunodot blot analysis with MAbs 3C7, 1G10, 1B10, 2C2, 1E10, 2F9, and, 2A11 using Cry1Aa, Cry1Ac, R521C, R521C-NAM, V582C, and V582C-NAM as the antigens. The following six toxin constructs were dotted onto a nitrocellulose membrane at a concentration of 100 ng/μl: Cry1Aa (spot A), Cry1Ac (spot B), R521C (spot C), R521C-NAM (spot D), V582C (spot E), and V582C-NAM (spot F). These constructs were probed with the following seven horseradish peroxidase-conjugated MAbs: 3C7 (blot a) 1G10 (blot b), 1B10 (blot c), 2C2 (blot d), 1E10 (blot e), 2F9 (blot f), and 2A11 (blot g). Bound MAb was detected using the ECL system.
DISCUSSION
The Cry toxins produced by B. thuringiensis have insecticidal specificity. In order to understand the specific binding between a Cry toxin and its receptor, it is important to identify the receptor binding site on the Cry toxin, as the insecticidal specificity of the Cry toxin seems to be largely dependent on receptor recognition. It is possible that the binding sites for APN which are common among Cry1Aa, Cry1Ab, and Cry1Ac and which are not blocked by GalNAc are true functional receptor binding sites, but this is not known yet. In order to understand the mechanism of the specific recognition between a Cry toxin and APN, it is necessary to determine the receptor binding site on Cry1Aa toxin. Knowledge of the receptor binding site on Cry1Aa toxin would allow improvements in the specificity and toxicity of Cry toxins.
We wanted to determine the location of the APN binding site on the Cry1Aa toxin molecule in order to understand the mechanism of receptor recognition by Cry toxins. Cry1Aa, unlike Cry1Ac, has only one APN binding site (48), making Cry1Aa more convenient for determination of the APN binding site involved in toxicity. As the three-dimensional structure of Cry1Aa is known, it is possible to consider the APN binding site three dimensionally.
In this study, we produced a series of monoclonal antibodies against Cry1Aa in order to obtain molecules competing with BmAPN1 for binding to Cry1Aa (blocking MAbs). We investigated the location on the structure of Cry1Aa of the epitopes recognized by the blocking MAbs in order to identify the location of the BmAPN1 binding site. This approach was expected to produce results that would compensate for and be more useful than the results obtained by previous approaches involving amino acid substitutions and domain exchange.
Epitope mapping of blocking MAbs.
The immunoblot analyses using three Cry1A toxins suggested that the epitopes for the blocking MAbs, MAbs 1B10 and 2C2, are localized in domain III of Cry1Aa (Fig. 4B, blots d and e). The immunoblot analyses using deletion mutants of Cry1Aa toxin further indicated that the region between amino acids 508 and 513 plays an important role in recognition by these MAbs and that the region between amino acids 514 and 615 contains the epitopes for MAbs 1B10 and 2C2 (Table 1). The binding assays using a series of octapeptides suggested that the candidate epitope for MAb 1B10 is a single peptide, 582VFTLSAHV589 (Fig. 6A), and that the candidate epitope for MAb 2C2 consists of three peptides, 520QRYRVRIR527, 570FTTPFNFS577, and 582VFTLSAHV589 (Fig. 6B). The locations of these candidate epitopes were mapped on the three-dimensional model of the Cry1Aa molecule (Fig. 7); 508STLRVN513 and 582VFTLSAHV589 are located adjacent to each other, as are 520QRYRVRIR527 and 570FTTPFNFS577, making each region a candidate epitope. Two candidate epitopes were determined such that one site consisted of 508STLRVN513 and 582VFTLSAHV589 and the other site consisted of 520QRYRVRIR527 and 570FTTPFNFS577. It isdifficult to believe that a single epitope contains these two regions, as the two regions are distant from each other. The epitope for MAb 1B10 might be the site consisting of 508STLRVN513 and 582VFTLSAHV589, but there is uncertainty concerning whether the epitope for MAb 2C2 is the candidate site consisting of 508STLRVN513 and 582VFTLSAHV589 or the site consisting of 520QRYRVRIR527 and 570FTTPFNFS577.
We constructed two Cry1Aa cysteine substitution mutants, R521C and V582C. NAM was introduced on the surface at the site of each cysteine substitution. The intention was to block the binding of the MAbs by producing steric hindrance on the surface at these two sites. Our results showed that the NAM molecules bound to Cys582 blocked the binding of the two blocking MAbs (Fig. 9, blots c and d), suggesting that the epitopes for MAbs 1B10 and 2C2 are located at the site containing Val582, which also consists of 508STLRVN513 and 582VFTLSAHV589.
Of the seven MAbs raised against Cry1Aa, only MAbs 1B10 and 2C2 blocked the binding of Cry1Aa toxin to 7-kDa BmAPN1 (Fig. 1). However, the finding that the Fab fragments of MAb 2C2 blocked the binding of Cry1Aa toxin to 7-kDa BmAPN1 in a dose-dependent manner, while the Fab fragments of MAb 1B10 did not affect the binding (Fig. 2), suggests that the surfaces occupied by these blocking MAbs differ from each other and that the surface area blocked by MAb 2C2 probably overlaps the BmAPN1 binding site on Cry1Aa toxin. It is likely that the surface area blocked by MAb 1B10 does not overlap the BmAPN1 binding site; instead, it might be located near this site on Cry1Aa.
MAb 2C2, but not MAb 1B10, competes with MAb 2F9 for binding to Cry1Aa (Fig. 3A and B), suggesting that the epitopes for MAbs 2C2 and 2F9 are located near each other. Indeed, the epitope for MAb 2F9 is probably in domain II of Cry1Aa toxin (Fig. 4B, blot g), and the region blocked by MAb 2C2 is at the site containing Val582 and consisting of 508STLRVN513 and 582VFTLSAHV589, which is located close to domain II.
Location of the APN binding site.
The BmAPN1 binding site appears to overlap the putative region blocked by MAb 2C2, which is indicated in Fig. 7. However, the results also suggest that the BmAPN1 binding site does not overlap the region containing 508STLRVN513 and 582VFTLSAHV589, as the Fab fragments of MAb 1B10 did not block the binding of Cry1Aa toxin to 7-kDa BmAPN1 (Fig. 2).
There are two regions to consider, the region containing residues 500 to 507 and 479 to 487 in domain II and the region consisting of 508STLRVN513 and 582VFTLSAHV589. The region containing residues 500 to 509 is located within a prominent loop in the so-called hypervariable region of Cry1Aa (17) and is adjacent to the region containing 508STLRVN513 and 582VFTLSAHV589 in the three-dimensional structure of Cry1Aa. Most of the sequence differences among the closely related Cry1A toxins, each of which has a unique specificity profile for lepidoptera, occur within the hypervariable region (29). Site-directed mutagenesis studies of 22 different mutations in the region encoding residues 500 to 509 of Cry1Ac revealed that changing only two adjacent serine residues, Ser503 and Ser504, resulted in greatly decreased toxicity for M. sexta larvae and poor binding to a 110-kDa protein from M. sexta brush border membrane vesicles. Therefore, the loop containing these two serines must be involved in the formation of a specific toxin recognition domain (2).
Shinkawa et al. previously reported that the Cry1Ac and Cry9Da toxins bound to the same 120-kDa APN from B. mori that Cry1Aa bound to (71), leading to the hypothesis that some of the five structures conserved among these three toxins might be part of the receptor binding site. One of the five conserved structures, the region consisting of Asp409, Leu411, Pro416, Arg424, and Thr269, is in domain II and is distant from the epitope for MAb 2C2; therefore, this region is inappropriate as a BmAPN1 binding site. Two of the five conserved structures, one region consisting of Arg521, Tyr522, Arg523, Arg525, and Phe570 and another region consisting of Gln262, Arg430, His456, Arg457, Gln472, and Lys477, are somewhat close to the epitope for MAb 2C2, but both regions are localized on faces of the three-dimensional structure of Cry1Aa that are different from the surface blocked by MAb 2C2. There is no possibility that MAb 2C2 blocks the binding of BmAPN1 to Cry1Aa at either of these two regions, and therefore, neither region is suitable as a binding site for BmAPN1. Another of the five conserved regions, the region consisting of Leu263, Arg265, Glu266, Tyr268, Glu288, Ile291, Gly490, and Pro491, also seems to be inappropriate, because it is distant from the epitope for MAb 2C2 and it localizes on a different face of Cry1Aa than the face with the area that is blocked by MAb 2C2.
The last of the conserved structures to consider, the region consisting of Arg292, Phe294, His295, Leu296, His433, Val487, and Leu481, is localized between domains II and III (Fig. 10). This region might be appropriate as one of the regions constituting the BmAPN1 binding site because it is close to and localizes on the same face as the area that is blocked by MAb 2C2.
FIG. 10.
Proposed APN binding site on a three-dimensional model of Cry1Aa. (A) The amino acid residues of domains I, II, and III are pink, light green, and light blue, respectively. The residues involved in binding of the blocking MAbs are indicated as follows: 582VFTLSAHV589, red;508STLRVN513, brown; and V582, yellow. The conserved region in Cry1Aa, Cry1Ac, and Cry9Da, consisting of R292, P294, H295, L296, H433, V487, and L481, is blue and is localized between domains II and III. The amino acid residues G484, R500, and G505, which are blue, are conserved in the three toxins and are localized in a hypervariable loop of Cry1Aa (17). (B) Panel A rotated 72° about the y axis and 45° about the x axis.
Based on the considerations described above, it is possible that the BmAPN1 binding site is composed of the two regions mentioned above, the region consisting of Arg292, Pro294, His295, Leu296, His433, Val487, and Leu481 and the region that is localized between domain II and the region consisting of 508STLRVN513 and 582VFTLSAHV589 (Fig. 10). The region that is localized between domain II and the region consisting of 508STLRVN513 and 582VFTLSAHV589 includes four conserved amino acids, G484, V487, R500, and G505, which might be especially important (Fig. 10) because they are localized on domain III of Cry1Aa toxin and the BmAPN1 binding site should be on domain III.
More than 340 Cry toxin genes have been cloned from B. thuringiensis, and they have been classified into 46 classes based on their primary sequences (7). As the crystal structures of Cry1Aa, Cry3A, Cry3B, and Cry2Aa have been reported to be similar despite the fact that the molecules belong to different classes (15, 25, 43, 51), the Cry1 toxins with similar primary sequences might have similar three-dimensional structures. Most Cry1 toxins show toxicity for several lepidopteran insects, and various APNs from several insect species have been identified as candidate receptors for Cry1 toxins (11, 20, 33, 34, 41, 45, 48, 57, 58, 74, 77). We hypothesized that the receptor binding sites on Cry1 toxins have two basic features: a highly conserved structure, because the Cry1 toxins have similar primary sequences and three-dimensional structures and can recognize similar proteins (APNs) found in the midguts of several lepidopteran insects, and a nonconserved structure, because the Cry1 toxins also exhibit highly specific insecticidal activity and can distinguish host species in the lepidopteran spectrum. The BmAPN1 binding site proposed in this study is consistent with the viewpoint that the binding site should consist of both the highly conserved and nonconserved structures.
Acknowledgments
This work was supported by grant-in-aid for scientific research 15380228 from the Ministry of Education, Science, and Culture of Japan.
REFERENCES
- 1.Almond, B. D., and D. H. Dean. 1993. Structural stability of Bacillus thuringiensis δ-endotoxin homolog-scanning mutants determined by susceptibility to proteases. Appl. Environ. Microbiol. 59:2442-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aronson, A. I., D. Wu, and C. Zhang. 1995. Mutagenesis of specificity and toxicity regions of a Bacillus thuringiensis protoxin gene. J. Bacteriol. 177:4059-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Burton, S. L., D. J. Ellar, J. Li, and D. J. Derbyshire. 1999. N-Acetylgalactosamine on the putative insect receptor aminopeptidase N is recognised by a site on the domain III lectin-like fold of a Bacillus thuringiensis insecticidal toxin. J. Mol. Biol. 287:1011-1022. [DOI] [PubMed] [Google Scholar]
- 5.Chen, X. J., A. Curtiss, E. Alcantara, and D. H. Dean. 1995. Mutations in domain I of Bacillus thuringiensis δ-endotoxin CryIAb reduce the irreversible binding of toxin to Manduca sexta brush border membrane vesicles. J. Biol. Chem. 270:6412-6419. [DOI] [PubMed] [Google Scholar]
- 6.Chen, X. J., M. K. Lee, and D. H. Dean. 1993. Site-directed mutations in a highly conserved region of Bacillus thuringiensis δ-endotoxin affect inhibition of short circuit current across Bombyx mori midguts. Proc. Natl. Acad. Sci. USA 90:9041-9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crickmore, N., D. R. Zeigler, E. Schnepf, J. van Rie, D. Lereclus, J. Baum, A. Bravo, and D. H. Dean. 2004. Bacillus thuringiensis toxin nomenclature. [Online.] http://www.lifesci.sussex.ac.uk/Home/Neil_Crickmore/Bt/
- 8.de Maagd, R. A., M. S. G. Kwa, H. van der Klei, T. Yamamoto, B. Schipper, and J. M. Vlak. 1996. Domain III substitution in Bacillus thuringiensis δ-endotoxin CryIA(b) results in superior toxicity for Spodoptera exigua and altered membrane protein recognition. Appl. Environ. Microbiol. 62:1537-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Maagd, R. A., M. Weemen-Hendriks, W. Stiekema, and D. Bosch. 2000. Bacillus thuringiensis δ-endotoxin Cry1C domain III can function as a specificity determinant for Spodoptera exigua in different, but not all, Cry1-Cry1C hybrids. Appl. Environ. Microbiol. 66:1559-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Maagd, R. A., P. L. Bakker, L. Masson, M. J. Adang, S. Sangadala, W. Stiekema, and D. Bosch. 1999. Domain III of the Bacillus thuringiensis δ-endotoxin Cry1Ac is involved in binding to Manduca sexta brush border membranes and to its purified aminopeptidase N. Mol. Microbiol. 31:463-471. [DOI] [PubMed] [Google Scholar]
- 11.Denolf, P., K. Hendrickx, J. Vandamme, S. Jansens, M. Peferoen, D. Degheele, and J. van Rie. 1997. Cloning and characterization of Manduca sexta and Plutella xylostella midgut aminopeptidase N enzymes related to Bacillus thuringiensis toxin binding proteins. Eur. J. Biochem. 248:748-761. [DOI] [PubMed] [Google Scholar]
- 12.Gahan, L. J., F. Gould, and D. G. Heckel. 2001. Identification of a gene associated with Bt resistance in Heliothis virescens. Science 293:857-860. [DOI] [PubMed] [Google Scholar]
- 13.Galfre, G., S. C. Howe, C. Milstein, G. W. Butcher, and J. C. Howard. 1977. Antibodies to major histocompatibility antigens produced by hybrid cell lines. Nature 266:550-552. [DOI] [PubMed] [Google Scholar]
- 14.Galitsky, N., V. Cody, A. Wojtczak, D. Ghosh, J. R. Luft, W. Pangborn, and L. English. 2001. Structure of the insecticidal bacterial δ-endotoxin Cry3Bb1 of Bacillus thuringiensis. Acta Crystallogr. Sect. D Biol. Crystallogr. 57:1101-1109. [DOI] [PubMed] [Google Scholar]
- 15.Garczynski, S. F., J. W. Crim, and M. J. Adang. 1991. Identification of putative insect brush border membrane-binding molecules specific to Bacillus thuringiensis δ-endotoxin by protein blot analysis. Appl. Environ. Microbiol. 57:2816-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge, A. Z., D. Rivers, R. Milne, and D. H. Dean. 1991. Functional domains of Bacillus thuringiensis insecticidal crystal proteins. J. Biol. Chem. 266:17954-17958. [PubMed] [Google Scholar]
- 17.Geiser, M., S. Schweitzer, and C. Grimm. 1986. The hypervariable region in the genes coding for entomopathogenic crystal proteins of Bacillus thuringiensis: nucleotide sequence of the kurhd1 gene of subsp. kurstaki HD1. Gene 48:109-118. [DOI] [PubMed] [Google Scholar]
- 18.Geysen, H. M., R. H. Meloen, and S. J. Barteling. 1984. Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc. Natl. Acad. Sci. USA 81:3998-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gill, M., and D. J. Ellar. 2002. Transgenic Drosophila reveals a functional in vivo receptor for the Bacillus thuringiensis toxin Cry1Ac1. Insect Mol. Biol. 11:619-625. [DOI] [PubMed] [Google Scholar]
- 20.Gill, S. S., E. A. Cowles, and V. Francis. 1995. Identification, isolation, and cloning of a Bacillus thuringiensis Cry1Ac toxin-binding protein from the midgut of the lepidopteran insect Heliothis virescens. J. Biol. Chem. 270:27277-27282. [DOI] [PubMed] [Google Scholar]
- 21.Gill, S. S., E. A. Cowles, and P. V. Pietrantonio. 1992. The mode of action of Bacillus thuringiensis endotoxins. Annu. Rev. Entomol. 37:615-636. [DOI] [PubMed] [Google Scholar]
- 22.Goding, J. W. 1980. Antibody production by hybridomas. J. Immunol. Methods 39:285-308. [DOI] [PubMed] [Google Scholar]
- 23.Gomez, I., J. Miranda-Rios, E. Rudino-Pinera, D. I. Oltean, S. S. Gill, A. Bravo, and M. Soberon. 2002. Hydropathic complementarity determines interaction of epitope (869)HITDTNNK(876) in Manduca sexta Bt-R(1) receptor with loop 2 of domain II of Bacillus thuringiensis Cry1A toxins. J. Biol. Chem. 277:30137-30143. [DOI] [PubMed] [Google Scholar]
- 24.Goodman, N. S., R. J. Gottfried, and M. H. Rogoff. 1967. Biphasic system for separation of spores and crystals of Bacillus thuringiensis. J. Bacteriol. 94:485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grochulski, P., L. Masson, S. Borisova, M. Pusztai-Carey, J. L. Schwartz, R. Brousseau, and M. Cygler. 1995. Bacillus thuringiensis CryIA(a) insecticidal toxin: crystal structure and channel formation. J. Mol. Biol. 254:447-464. [DOI] [PubMed] [Google Scholar]
- 26.Hara, H., S. Atsumi, K. Yaoi, K. Nakanishi, S. Higurashi, N. Miura, H. Tabunoki, and R. Sato. 2003. A cadherin-like protein functions as a receptor for Bacillus thuringiensis Cry1Aa and Cry1Ac toxins on midgut epithelial cells of Bombyx mori larvae. FEBS Lett. 538:29-34. [DOI] [PubMed] [Google Scholar]
- 27.Hendrickx, K., A. de Loof, and H. van Mellaert. 1990. Effects of Bacillus thuringiensis δ-endotoxin on the permeability of brush border membrane vesicles from tobacco hornworm (Manduca sexta) midgut. Comp. Biochem. Physiol. Part C 95:241-245. [DOI] [PubMed] [Google Scholar]
- 28.Hofmann, C., P. Lüthy, R. Hütter, and V. Pliska. 1988. Binding of the δ-endotoxin from Bacillus thuringiensis to brush-border membrane vesicles of the cabbage butterfly (Pieris brassicae). Eur. J. Biochem. 173:85-91. [DOI] [PubMed] [Google Scholar]
- 29.Höfte, H., and H. R. Whiteley. 1989. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol. Rev. 53:242-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenkins, J. L., M. K. Lee, A. P. Valaitis, A. Curtiss, and D. H. Dean. 2000. Bivalent sequential binding model of a Bacillus thuringiensis toxin to gypsy moth aminopeptidase N receptor. J. Biol. Chem. 275:14423-14431. [DOI] [PubMed] [Google Scholar]
- 31.Jenkins, J. L., M. K. Lee, S. Sangadala, M. J. Adang, and D. H. Dean. 1999. Binding of Bacillus thuringiensis Cry1Ac toxin to Manduca sexta aminopeptidase-N receptor is not directly related to toxicity. FEBS Lett. 462:373-376. [DOI] [PubMed] [Google Scholar]
- 32.Kleanthous, C. 2000. Protein-protein recognition, p. 20-21. Oxford University Press, Oxford, England.
- 33.Knight, P. J. K., B. H. Knowles, and D. J. Ellar. 1995. Molecular cloning of an insect aminopeptidase N that serves as a receptor for Bacillus thuringiensis CryIA(c) toxin. J. Biol. Chem. 270:17765-17770. [DOI] [PubMed] [Google Scholar]
- 34.Knight, P. J. K., N. Crickmore, and D. J. Ellar. 1994. The receptor for Bacillus thuringiensis CryIAc δ-endotoxin in the brush border membrane of the lepidopteran Manduca sexta is aminopeptidase N. Mol. Microbiol. 11:429-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knowles, B. H., and D. J. Ellar. 1987. Colloid-osmotic lysis is a general feature of the mechanisms of action of Bacillus thuringiensis δ-endotoxins with different insect specificities. Biochim. Biophys. Acta 924:509-518. [Google Scholar]
- 36.Knowles, B. H., W. E. Thomas, and D. J. Ellar. 1984. Lectin-like binding of Bacillus thuringiensis var. kurstaki lepidopteran-specific toxin is an initial step in insecticidal action. FEBS Lett. 168:197-202. [DOI] [PubMed] [Google Scholar]
- 37.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 38.Lee, M. K., B. A. Young, and D. H. Dean. 1995. Domain III exchanges of Bacillus thuringiensis Cry1A toxins affect binding to different gypsy moth midgut receptors. Biochem. Biophys. Res. Commun. 216:306-312. [DOI] [PubMed] [Google Scholar]
- 39.Lee, M. K., and D. H. Dean. 1996. Inconsistencies in determining Bacillus thuringiensis toxin binding sites relationship by comparing competition assays with ligand blotting. Biochem. Biophys. Res. Commun. 220:575-580. [DOI] [PubMed] [Google Scholar]
- 40.Lee, M. K., T. H. You, F. L. Gould, and D. H. Dean. 1999. Identification of residues in domain III of Bacillus thuringiensis Cry1Ac toxin that affect binding and toxicity. Appl. Environ. Microbiol. 65:4513-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee, M. K., T. H. You, B. A. Young, J. A. Cotrill, A. P. Valaitis, and D. H. Dean. 1996. Aminopeptidase N purified from gypsy moth brush border membrane vesicles is a specific receptor for Bacillus thuringiensis CryIAc toxin. Appl. Environ. Microbiol. 62:2845-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, M. K., R. E. Milne, A. Z. Ge, and D. H. Dean. 1992. Location of a Bombyx mori receptor binding region on a Bacillus thuringiensis δ-endotoxin. J. Biol. Chem. 267:3115-3121. [PubMed] [Google Scholar]
- 43.Li, J., J. Carroll, and D. J. Ellar. 1991. Crystal structure of an insecticidal protein. The δ-endotoxin from Bacillus thuringiensis subsp. tenebrionis at 2.5 Å resolution. Nature (London) 353:815-821. [DOI] [PubMed] [Google Scholar]
- 44.Liang, Y., S. S. Patel, and D. H. Dean. 1995. Irreversible binding kinetics of Bacillus thuringiensis CryIA δ-endotoxins to gypsy moth brush border membrane vesicles is directly correlated to toxicity. J. Biol. Chem. 270:24719-24724. [DOI] [PubMed] [Google Scholar]
- 45.Lorence, A., A. Darszon, and A. Bravo. 1997. Aminopeptidase dependent pore formation of Bacillus thuringiensis Cry1Ac toxin on Trichoplusia ni membranes. FEBS Lett. 414:303-307. [DOI] [PubMed] [Google Scholar]
- 46.Lu, H., F. Rajamohan, and D. H. Dean. 1994. Identification of amino acid residues of Bacillus thuringiensis δ-endotoxin CryIAa associated with membrane binding and toxicity to Bombyx mori. J. Bacteriol. 176:5554-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reference deleted.
- 48.Luo, K., S. Sangadala, L. Masson, A. Mazza, R. Brousseau, and M. J. Adang. 1997. The Heliothis virescens 170 kDa aminopeptidase functions as ‘receptor A’ by mediating specific Bacillus thuringiensis Cry1A δ-endotoxin binding and pore formation. Insect Biochem. Mol. Biol. 27:735-743. [DOI] [PubMed] [Google Scholar]
- 49.Masson, L., A. Mazza, L. Gringorten, D. Baines, V. Aneliunas, and R. Brousseau. 1994. Specificity domain localization of Bacillus thuringiensis insecticidal toxins is highly dependent on the bioassay system. Mol. Microbiol. 14:851-860. [DOI] [PubMed] [Google Scholar]
- 50.Masson, L., Y. J. Lu, A. Mazza, R. Brousseau, and M. J. Adang. 1995. The CryIA(c) receptor purified from Manduca sexta displays multiple specificities. J. Biol. Chem. 270:20309-20315. [DOI] [PubMed] [Google Scholar]
- 51.Morse, R. J., T. Yamamoto, and R. M. Stroud. 2001. Structure of Cry2Aa suggests an unexpected receptor binding epitope. Structure (Cambridge) 9:409-417. [DOI] [PubMed] [Google Scholar]
- 52.Nagamatsu, Y., S. Toda, F. Yamaguchi, M. Ogo, M. Kogure, M. Nakamura, Y. Shibata, and T. Katsumoto. 1998. Identification of Bombyx mori midgut receptor for Bacillus thuringiensis insecticidal CryIA(a) toxin. Biosci. Biotechnol. Biochem. 62:718-726. [DOI] [PubMed] [Google Scholar]
- 53.Nagamatsu, Y., S. Toda, T. Koike, Y. Miyoshi, S. Shigematsu, and M. Kogure. 1998. Cloning, sequencing, and expression of the Bombyx mori receptor for Bacillus thuringiensis insecticidal Cry1Aa toxin. Biosci. Biotechnol. Biochem. 62:727-734. [DOI] [PubMed] [Google Scholar]
- 54.Nagamatsu, Y., T. Koike, K. Sasaki, A. Yoshimoto, and Y. Furukawa. 1999. The cadherin-like protein is essential to specificity determination and cytotoxic action of the Bacillus thuringiensis insecticidal CryIAa toxin. FEBS Lett. 460:385-390. [DOI] [PubMed] [Google Scholar]
- 55.Nagamatsu, Y., Y. Itai, C. Hatanaka, G. Funatsu, and K. Hayashi. 1984. A toxic fragment from the entomocidal crystal protein of Bacillus thuringiensis. Agric. Biol. Chem. 48:611-619. [Google Scholar]
- 56.Nakane, P. K., and G. B. Pierce, Jr. 1967. Enzyme-labeled antibodies for the light and electron microscopic localization of tissue antigens. J. Cell Biol. 33:307-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakanishi, K., K. Yaoi, N. Shimada, T. Kadotani, and R. Sato. 1999. Bacillus thuringiensis insecticidal Cry1Aa toxin binds to a highly conserved region of aminopeptidase N in the host insect leading to its evolutionary success. Biochim. Biophys. Acta 1432:57-63. [DOI] [PubMed] [Google Scholar]
- 58.Nakanishi, K., K. Yaoi, Y. Nagino, H. Hara, M. Kitami, S. Atsumi, N. Miura, and R. Sato. 2002. Aminopeptidase N isoforms from the midgut of Bombyx mori and Plutella xylostella: their classification and the factors that determine their binding specificity to Bacillus thuringiensis Cry1A toxin. FEBS Lett. 519:215-220. [DOI] [PubMed] [Google Scholar]
- 59.Oeda, K., K. Oshie, M. Shimizu, K. Nakamura, H. Yamamoto, I. Nakayama, and H. Ohkawa. 1987. Nucleotide sequence of the insecticidal protein gene of Bacillus thuringiensis strain aizawai IPL7 and its high-level expression in Escherichia coli. Gene 53:113-119. [DOI] [PubMed] [Google Scholar]
- 60.Rajagopal, R., S. Sivakumar, N. Agrawal, P. Malhotra, and R. K. Bhatnagar. 2002. Silencing of midgut aminopeptidase N of Spodoptera litura by dsRNA establishes its role as Bt toxin receptor. J. Biol. Chem. 277:46849-46851. [DOI] [PubMed] [Google Scholar]
- 61.Rajamohan, F., E. Alcantara, M. K. Lee, X. J. Chen, A. Curtiss, and D. H. Dean. 1995. Single amino acid changes in domain II of Bacillus thuringiensis CryIAb δ-endotoxin affect irreversible binding to Manduca sexta midgut membrane vesicles. J. Bacteriol. 177:2276-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rajamohan, F., O. Alzate, J. A. Cotrill, A. Curtiss, and D. H. Dean. 1996. Protein engineering of Bacillus thuringiensis δ-endotoxin: mutations at domain II of Cry1Ab enhance receptor affinity and toxicity towards gypsy moth larvae. Proc. Natl. Acad. Sci. USA 93:14338-14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rajamohan, F., J. A. Cotrill, F. Gould, and D. H. Dean. 1996. Role of domain II, loop 2 residues of Bacillus thuringiensis CryIAb δ-endotoxin in reversible and irreversible binding to Manduca sexta and Heliothis virescens. J. Biol. Chem. 271:2390-2397. [DOI] [PubMed] [Google Scholar]
- 64.Rajamohan, F., S. R. A. Hussain, J. A. Cotrill, F. Gould, and D. H. Dean. 1996. Mutations at domain II, loop 3, of Bacillus thuringiensis CryIAa and CryIAb δ-endotoxins suggest loop 3 is involved in initial binding to lepidopteran midguts. J. Biol. Chem. 271:25220-25226. [DOI] [PubMed] [Google Scholar]
- 65.Sacchi, V. F., P. Parenti, G. M. Hanozet, B. Giordana, P. Lüthy, and M. G. Wolfersberger. 1986. Bacillus thuringiensis toxin inhibits K+-gradient-dependent amino acid transport across the brush border membrane of Pieris brassicae midgut cells. FEBS Lett. 204:213-218. [Google Scholar]
- 66.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schnepf, H. E., H. C. Wong, and H. R. Whiteley. 1985. The amino acid sequence of a crystal protein from Bacillus thuringiensis deduced from the DNA base sequence. J. Biol. Chem. 260:6264-6272. [PubMed] [Google Scholar]
- 68.Schwartz, J. L., L. Garneau, D. Savaria, L. Masson, R. Brousseau, and E. Rousseau. 1993. Lepidopteran-specific crystal toxins from Bacillus thuringiensis form cation- and anion-selective channels in planar lipid bilayers. J. Membr. Biol. 132:53-62. [DOI] [PubMed] [Google Scholar]
- 69.Schwartz, J. L., L. Potvin, X. J. Chen, R. Brousseau, R. Laprade, and D. H. Dean. 1997. Single-site mutations in the conserved alternating-arginine region affect ionic channels formed by CryIAa, a Bacillus thuringiensis toxin. Appl. Environ. Microbiol. 63:3978-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwartz, J. L., Y. J. Lu., P. Sohnlein, R. Brousseau, R. Laprade, L. Masson, and M. J. Adang. 1997. Ion channels formed in planar lipid bilayers by Bacillus thuringiensis toxins in the presence of Manduca sexta midgut receptors. FEBS Lett. 412:270-276. [DOI] [PubMed] [Google Scholar]
- 71.Shinkawa, A., K. Yaoi, T. Kadotani, M. Imamura, N. Koizumi, H. Iwahana, and R. Sato. 1999. Binding of phylogenetically distant Bacillus thuringiensis Cry toxins to a Bombyx mori aminopeptidase N suggests importance of Cry toxin's conserved structure in receptor binding. Curr. Microbiol. 39:14-20. [DOI] [PubMed] [Google Scholar]
- 72.Smedley, D. P., and D. J. Ellar. 1996. Mutagenesis of three surface-exposed loops of a Bacillus thuringiensis insecticidal toxin reveals residues important for toxicity, receptor recognition and possibly membrane insertion. Microbiology 142:1617-1624. [DOI] [PubMed] [Google Scholar]
- 73.Vadlamudi, R. K., E. Weber, I. H. Ji, T. H. Ji, and L. A. Bulla. 1995. Cloning and expression of a receptor for an insecticidal toxin of Bacillus thuringiensis. J. Biol. Chem. 270:5490-5494. [DOI] [PubMed] [Google Scholar]
- 74.Valaitis, A. P., M. K. Lee, F. Rajamohan, and D. H. Dean. 1995. Brush border membrane aminopeptidase-N in the midgut of the gypsy moth serves as the receptor for the CryIA(c) δ-endotoxin of Bacillus thuringiensis. Insect Biochem. Mol. Biol. 25:1143-1151. [DOI] [PubMed] [Google Scholar]
- 75.Wolfersberger, M. G., X. J. Chen, and D. H. Dean. 1996. Site-directed mutations in the third domain of Bacillus thuringiensis δ-endotoxin CryIAa affect its ability to increase the permeability of Bombyx mori midgut brush border membrane vesicles. Appl. Environ. Microbiol. 62:279-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu, S. J., and D. H. Dean. 1996. Functional significance of loops in the receptor binding domain of Bacillus thuringiensis CryIIIA δ-endotoxin. J. Mol. Biol. 255:628-640. [DOI] [PubMed] [Google Scholar]
- 77.Yaoi, K., T. Kadotani, H. Kuwana, A. Shinkawa, T. Takahashi, H. Iwahana, and R. Sato. 1997. Aminopeptidase N from Bombyx mori as a candidate for the receptor of Bacillus thuringiensis Cry1Aa toxin. Eur. J. Biochem. 246:652-657. [DOI] [PubMed] [Google Scholar]
- 78.Yaoi, K., K. Nakanishi, T. Kadotani, M. Imamura, N. Koizumi, H. Iwahana, and R. Sato. 1999. Bacillus thuringiensis Cry1Aa toxin-binding region of Bombyx mori aminopeptidase N. FEBS Lett. 463:221-224. [DOI] [PubMed] [Google Scholar]