Abstract
The cross-feeding of microbial products derived from 14C-labeled nitrifying bacteria to heterotrophic bacteria coexisting in an autotrophic nitrifying biofilm was quantitatively analyzed by using microautoradiography combined with fluorescence in situ hybridization (MAR-FISH). After only nitrifying bacteria were labeled with [14C]bicarbonate, biofilm samples were incubated with and without NH4+ as a sole energy source for 10 days. The transfer of 14C originally incorporated into nitrifying bacterial cells to heterotrophic bacteria was monitored with time by using MAR-FISH. The MAR-FISH analysis revealed that most phylogenetic groups of heterotrophic bacteria except the β-Proteobacteria showed significant uptake of 14C-labeled microbial products. In particular, the members of the Chloroflexi were strongly MAR positive in the culture without NH4+ addition, in which nitrifying bacteria tended to decay. This indicated that the members of the Chloroflexi preferentially utilized microbial products derived from mainly biomass decay. On the other hand, the members of the Cytophaga-Flavobacterium cluster gradually utilized 14C-labeled products in the culture with NH4+ addition in which nitrifying bacteria grew. This result suggested that these bacteria preferentially utilized substrate utilization-associated products of nitrifying bacteria and/or secondary metabolites of 14C-labeled structural cell components. Our results clearly demonstrated that the coexisting heterotrophic bacteria efficiently degraded and utilized dead biomass and metabolites of nitrifying bacteria, which consequently prevented accumulation of organic waste products in the biofilm.
Most bacteria in the natural environment and engineering systems are present in the form of complex multispecies biofilms attached to solid surfaces rather than as planktonic (free-swimming) isolated cells in the bulk water phase (10, 27, 37, 38, 41). Microbial life in such biofilms, which cannot be seen in a planktonic state, is mostly characterized by multiplicity (many species together), nutrient limitation, competition for oxygen, substrates, and/or space (beneficial or inhibitory interactions among microbial species), and a structured distribution of the microbial species. Therefore, several studies have been performed on wastewater treatment biofilms by combining fluorescence in situ hybridization (FISH) and microsensor technology to link the spatial organization of microbial communities and in situ functions at the community level (10, 26, 27, 38).
The coexistence of a high level of heterotrophic bacteria with nitrifying bacteria has often been found in autotrophic nitrifying biofilms cultured without an external organic carbon supply by applying a 16S rRNA approach (16, 27, 28). It has been hypothesized that heterotrophic bacteria scavenge organic matter derived from biomass decay and substrate metabolism of nitrifying bacteria. However, the in situ ecophysiology of heterotrophic bacteria in such autotrophic biofilms is still largely unknown because most of the heterotrophic bacteria are uncultured microorganisms and the use of only the 16S rRNA approach does not allow a direct link between identity and the in situ catabolic activity in the biofilm.
A combination of microautoradiography (MAR) and the FISH approach has recently been used to study the in situ ecophysiology of various cultivable or uncultivable bacteria in activated sludge (8, 23, 24, 25), marine samples (6, 30, 31, 33), freshwater sediments (11, 12), sewer biofilms (14), and autotrophic nitrifying biofilms (16). These studies have demonstrated that the MAR-FISH technique has significant potential for providing a direct link between rRNA-based phylogenetic identification and in situ substrate uptake patterns (metabolic capability) without a requirement for cultivation.
We previously analyzed the phylogenetic identities, spatial organization, and substrate uptake patterns of coexisting heterotrophic bacteria in autotrophic nitrifying biofilms by using MAR-FISH with externally supplied 14C-labeled synthetic organic substrates, including acetate, amino acids, and N-acetyl-d-glucosamine (NAG) (16). However, the radiolabeled substrates added were foreign to the microbial community and might not represent the actual food sources of heterotrophic bacteria in the biofilm. It is, therefore, too early to form a clear picture of the in situ ecophysiological interactions between nitrifying bacteria and heterotrophic bacteria in a biofilm.
The objective of the present study was to determine which phylogenetic groups of heterotrophic bacteria could directly utilize microbial products (e.g., structural cell components and metabolites) derived from nitrifying bacteria in an autotrophic nitrifying biofilm. To achieve this objective, biofilm samples were first incubated with [14C]bicarbonate to radiolabel only nitrifying bacteria, and after this the fate (transfer) of the radiolabeled 14C atoms incorporated into nitrifying bacteria was traced and visualized by using the MAR-FISH approach. In situ ecophysiological interactions between nitrifying bacteria and heterotrophic bacteria in the biofilm were examined at different phylogenetic group levels.
MATERIALS AND METHODS
Biofilm samples.
Autotrophic nitrifying biofilms were cultured with synthetic medium in a partially submerged rotating disk reactor (RDR) consisting of five polymethyl methacrylate disks with four removable slides. The autotrophic nitrifying biofilms were seeded with primary settling tank effluent from the Soseigawa municipal wastewater treatment plant (Sapporo, Japan) and then cultured with synthetic nutrient medium that contained no organic substrate, which led to development of a simple model biofilm ecosystem. The synthetic nutrient medium was composed of 3.6 mM NH4Cl, 17.8 mM NaHCO3, 0.4 mM K2HPO4, 0.41 mM MgSO4 · 7H2O, and 1.25 mM NaCl, and the pH was 7.8 ± 0.2. Distilled water, which contained no detectable dissolved organic carbon, was used to dilute the medium. The RDR was operated as previously described by Kindaichi et al. (16), and biofilm samples were obtained from the RDR after about 2 months.
Incubation with [14C]bicarbonate.
This study consisted of the following two experiments: a 14C-labeling experiment and a 14C-tracing experiment. First, the 14C-labeling experiment was performed to produce 14C-labeled nitrifying bacterial cells. The biofilms were taken from the removable slides (surface area, 18 cm2) and homogenized. The homogenized biofilms were diluted with the synthetic nutrient medium containing 3.6 mM NH4+ to obtain a final concentration of volatile suspended solids of 1.2 g per liter. Five-milliliter portions of the diluted samples were transferred into 12-ml glass serum vials. The bottles were sealed with gas-tight rubber stoppers. Sodium [14C]bicarbonate (specific activity, 58 mCi mmol−1; Amersham Bioscience, Little Chalfont, United Kingdom), a carbon source for nitrifying bacteria, was injected into the vials to obtain a final radioactivity of 10 μCi mmol−1 and a [14C]bicarbonate concentration of 35 μM. Then the vials were incubated with shaking at 60 rpm for 6 h. Biofilm samples pasteurized at 70°C for 15 min were prepared and incubated with [14C]bicarbonate in the same way as a control to test for possible adsorption phenomena and chemography. After 6 h of incubation, the biomass was harvested by centrifugation (10,000 × g for 8 min) and washed twice with phosphate-buffered saline (PBS) (10 mM sodium phosphate buffer and 130 mM sodium chloride; pH 7.2) to remove excess unincorporated [14C]bicarbonate. At this stage, subsamples were taken, and MAR-FISH was conducted as described below to confirm that only nitrifying bacteria were labeled with 14C in mixed populations. We confirmed that only nitrifying bacteria were strongly MAR positive and that other heterotrophic bacteria were not MAR positive, suggesting that 6 h of incubation was optimum for radiolabeling only nitrifying bacteria without 14C cross-feeding to heterotrophic bacteria.
Second, to investigate which phylogenetic groups of heterotrophic bacteria can utilize microbial products derived from nitrifying bacteria, the washed biomass samples (including 14C-labeled nitrifying bacteria) that were prepared in the 14C-labeling experiment were further incubated in fresh synthetic medium with unlabeled bicarbonate (0.5 mM) and with and without 3.6 mM NH4+ as an energy source for 10 days. Since the NH4+ was depleted during the 10-day incubation, additional NH4+ (7.1 mM and 12.8 mM on day 1 and day 3, respectively) was added. All incubations with and without NH4+ were conducted in duplicate, and the means and standard errors were determined.
Sample fixation.
During the 10 days of incubation, subsamples were taken at 1, 3, 7, and 10 days for MAR-FISH analysis. The samples were fixed for 3 h at 4°C by adding 5 ml of 8% paraformaldehyde, which resulted in a final paraformaldehyde concentration of 4%. Subsequently, the samples were centrifuged at 10,000 × g for 8 min, washed three times with 1 ml of PBS to remove excess soluble radioactive compounds, and stored in 50% ethanol in PBS at −18°C. After the fixation and washing steps, the samples were spotted on gelatin-coated glass coverslips as described elsewhere (14, 18).
Liquid scintillation counting.
The degradation of 14C-labeled microbial products derived from nitrifying bacteria by heterotrophic bacteria was also confirmed by determining the 14C contents in the biomass and culture medium fractions by liquid scintillation counting during incubation. The 14C content in the biomass fraction was determined after preparation as follows. One milliliter of a culture sample was centrifuged at 10,000 × g for 8 min and washed three times with 1 ml of PBS. The harvested biomass was resuspended in 1 ml of tap water, and a 0.1-ml aliquot was then added to 3 ml of scintillation liquid (Ultima Gold XR; Packard BioScience Co., Meriden, Conn.). After the sample was thoroughly mixed and stored at room temperature for 3 h, the radioactivity was determined with an Aloka model LSC-1000 liquid scintillation counter as recommended by the manufacturer. The total 14C in the culture samples (biomass plus culture medium) was directly subjected to the liquid scintillation counting procedure. The 14C content of the culture medium (bulk) was obtained by subtracting the 14C content in the biomass from the total 14C content in the culture samples (biomass plus culture medium).
Oligonucleotide probes and FISH.
The 16S and 23S rRNA-targeted oligonucleotide probes and the hybridization conditions used in this study are shown in Table 1. The probes were labeled with fluorescein isothiocyanate (FITC) and tetramethylrhodamine 5-isothiocyanate (TRITC). Dehydration and FISH were performed by using the procedure described by Amann (2) and Okabe et al. (27). Simultaneous hybridizations with probes that required different stringency conditions were performed by using a successive hybridization procedure; hybridization with the probe requiring a higher stringency was performed first, and then hybridization with the probe requiring a lower stringency was performed (42). Some samples were simultaneously stained with 4′,6′-diamidino-2-phenylindole (DAPI) (1 μg ml−1) for 10 min in the dark to determine total cell numbers. The slides were then rinsed briefly with double-distilled H2O, allowed to air dry, and mounted in antifading solution (Slow fade light; Molecular Probes, Eugene, Oreg.).
TABLE 1.
16S and 23S rRNA-targeted oligonucleotide probes and hybridization conditions
| Probe | Sequence (5′ to 3′) | FA (%)a | Specificity | Reference |
|---|---|---|---|---|
| EUB338 | GCTGCCTCCCGTAGGAGT | —b | Most Bacteria | 1 |
| EUB338 II | GCAGCCACCCGTAGGTGT | —b | Planctomycetales | 7 |
| EUB338 III | GCTGCCACCCGTAGGTGT | —b | Verrucomicrobiales | 7 |
| ALF968 | GGTAAGGTTCTGCGCGTT | 20 | α-Proteobacteria, except Rickettsiales | 22 |
| BET42ac | GCCTTCCCACTTCGTTT | 35 | β-Proteobacteria | 19 |
| GAM42ad | GCCTTCCCACATCGTTT | 35 | γ-Proteobacteria | 19 |
| GNSB-941 | AAACCACACGCTCCGCT | 35 | Phylum Chloroflexi | 9 |
| CFX1223 | CCATTGTAGCGTGTGTGTMG | 35 | Phylum Chloroflexi | 4 |
| CF319a/b | TGGTCCGTRTCTCAGTAC | 35 | CF cluster of the Bacteroidetes | 20 |
| Nso190 | CGATCCCCTGCTTTTCTCC | 35e | Ammonia oxidizers | 21 |
| Ntspa662 | GGAATTCCGCGCTCCTCT | 35 | Genus Nitrospira | 8 |
| Comp Ntspa662f | GGAATTCCGCTCTCCTCT | Competitor for Ntspa662 | 8 | |
| NIT3 | CCTGTGCTCCATGCTCCG | 40 | Nitrobacter spp. | 43 |
| Comp NIT3g | CCTGTGCTCCAGGCTCCG | Competitor for NIT3 | 43 |
FA, formamide concentration in the hybridization buffer.
The probe can be used at any formamide concentration.
Unlabeled probe GAM42a was used as a competitor to enhance specificity.
Unlabeled probe BET42a was used as a competitor to enhance specificity.
Although the original description of the use of Nso190 indicated that 55% formamide should be used, we experimentally confirmed that 35% formamide was sufficient to discriminate ammonia-oxidizing β-Proteobacteria in our autotrophic nitrifying biofilm, which is consistent with other descriptions of the use of Nso190 at much lower stringencies than those originally described (32).
Unlabeled probe Ntspa662 was used as a competitor to enhance specificity.
Unlabeled probe NIT3 was used as a competitor to enhance specificity.
For quantitative determination of the microbial composition in the autotrophic nitrifying biofilm, the surface fractions of the specific probe-hybridized cell area and the mixed EUB338 (EUB338, EUB338 II, and EUB338 III) probe-hybridized cells (total biomass) were determined after simultaneous in situ hybridizations with various probe sets. The average surface fraction was determined from at least 10 representative laser scanning microscopy projection images obtained from each of duplicate biofilm cultures by using image analysis software provided by Zeiss (16, 27).
Autoradiographic procedure.
Microautoradiography was performed directly on a coverslip as described by Lee et al. (18). After the FISH procedure, an autoradiographic liquid film emulsion (LM-1; Amersham Biosciences, Little Chalfont, United Kingdom) was used. The optimal exposure time was previously determined to be 2 days.
Microscopy and enumeration by MAR-FISH.
A model LSM510 confocal laser scanning microscope (Carl Zeiss, Oberkochen, Germany) equipped with an Ar ion laser (458 and 488 nm) and two HeNe ion lasers (543 nm) was used. The formation of silver grains in the autoradiographic film was observed by using the transmission mode of the system. All images were combined and processed with the standard software package provided with the LSM510 and were printed by using Photoshop 5.0 (Adobe Systems Inc., Mountain View, Calif.).
A MAR-positive cell was defined as a cell covered with more than five silver grains in this study. The numbers of MAR-positive cells and total probe-hybridized cells were determined by directly counting at least 500 silver grain-covered cells in randomly chosen microscopic fields of a few slides prepared from each sample.
Analytical measurements.
Concentrations of NH4+, NO2−, and NO3− were determined by ion-exchange chromatography using a DX-100 (DIONEX, Osaka, Japan) with an IonPac CS3 cation column and an IonPac AS9 anion column after filtration with 0.2-μm-pore-size membranes.
RESULTS
Community composition in the biofilm.
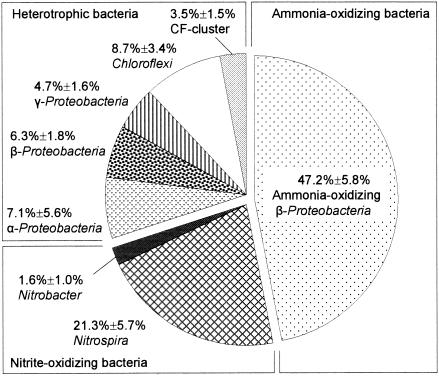
The microbial community composition in the autotrophic nitrifying biofilm was first analyzed by FISH with various sets of oligonucleotide probes (Table 1 and Fig. 1). The total bacteria detected with the mixed EUB338 probes accounted for 77.0% ± 7.0% of the total DAPI counts. Members of the ammonia-oxidizing bacteria (AOB) (detected with probe Nso190) and the nitrite-oxidizing bacteria (NOB) (the Nitrospira phylum plus the genus Nitrobacter belonging to the α-Proteobacteria) accounted for 70.0% of the total bacteria detected with the mixed EUB338 probes. The ratio of total nitrifiers (AOB plus NOB) to all heterotrophs was approximately 2:1 in the biofilm.
FIG. 1.
Microbial community composition of the autotrophic nitrifying biofilm as quantitatively determined by FISH. The α-Proteobacteria is the bacterial group that hybridized with probe ALF968, excluding the genus Nitrobacter that hybridized with probe NIT3. The β-Proteobacteria is the bacterial group that hybridized with probe BET42a, excluding the ammonia-oxidizing bacteria that hybridized with probe Nso190. The number of cells that hybridized with a given probe in each microscopic field was expressed as a percentage of the total surface area of bacteria that hybridized with mixed probes EUB338, EUB338 II, and EUB338 III. The values are the means ± standard errors of duplicate samples.
14C labeling of nitrifying bacteria.
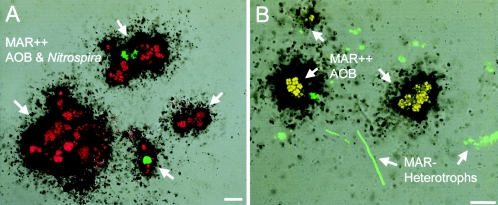
In order to radiolabel only nitrifying bacteria in the biofilm community, the homogenized biofilm samples were incubated in the synthetic nutrient medium with [14C]bicarbonate as a carbon source for 6 h. In preliminary experiments, we confirmed that the combination of 6 h of incubation and a 2-day exposure time was the optimum conditions for detection of enough radioactivity incorporated into nitrifying bacterial cells without substrate cross-feeding from nitrifying bacteria to heterotrophic bacteria. MAR-FISH analysis revealed that almost 100% of the AOB and NOB were strongly MAR positive (Fig. 2A), but no MAR-positive heterotrophic bacteria were detected (Fig. 2B). Approximately 1 mM NH4+ was consumed, and an equivalent amount of NO2− and NO3− was produced during 6 h of incubation, indicating that AOB and NOB were both active. Liquid scintillation counting revealed that the nitrifying bacteria took up ca. 35% of the [14C]bicarbonate added for 6 h of incubation. The control experiments with pasteurized biomass did not show detectable uptake of radioactivity and MAR-positive cells (data not shown). This result indicated that [14C]bicarbonate was truly incorporated into nitrifying bacteria rather than adsorbed on the cell surface.
FIG. 2.
Combined MAR and FISH images of homogenized autotrophic nitrifying biofilms. Only nitrifying bacteria were successfully radiolabeled by incubation with [14C]bicarbonate for 6 h (A). Heterotrophic bacteria were all MAR negative under these incubation conditions (B). In situ hybridizations were performed with a combination of FITC-labeled Ntspa662 and TRITC-labeled Nso190 (red) (A) and with a combination of FITC-labeled mixed EUB338 probes (green) and TRITC-labeled Nso190 (red) (B). Bars = 10 μm. MAR++, strongly MAR positive; MAR−, MAR negative.
Fate of 14C incorporated into nitrifying bacteria.
The total number of cells detected by DAPI direct counting decreased slightly from 4.8 × 108 ± 1.1 × 108 cells ml−1 at day 0 to 3.1 × 108 ± 0.6 × 108 cells ml−1 at day 10 in the culture without NH4+ addition. In the culture with NH4+ addition, the total number of cells was relatively constant (4.8 × 108 ± 1.1 × 108 cells ml−1 to 5.3 × 108 ± 1.0 × 108 cells ml−1). In addition, the percentages of domain Bacteria cells detected with the mixed EUB probes (EUB338, EUB338 II, and EUB338 III) relative to the number of DAPI-stained cells after 10 days of incubation were about 85.6% ± 7.8% and 70.4% ± 6.5% in the cultures with and without NH4+ addition, respectively. This indicated that more active cells were present in the culture to which NH4+ was added. However, no significant change in the community composition of the heterotrophic bacteria was observed in both cultures during the 10-day incubation (data not shown). On the other hand, the populations of AOB and NOB decreased with time in the culture without NH4+ addition due to the absence of the energy source, whereas both populations were slightly increased in the culture with NH4+ addition (Table 2). Furthermore, the concentration of NH4+ decreased with time, and the equivalent amount of NO3− was simultaneously produced with no accumulation of NO2−, indicating that both types of nitrifying bacteria (AOB and NOB) actively oxidized NH4+ and NO2− and grew in the culture to which NH4+ was added during the 10-day incubation.
TABLE 2.
Changes in populations of nitrifying bacteria (AOB and NOB) during 10 days of incubation as determined by FISH
| Incubation time (days) | AOB (107 cells ml−1)
|
NOB (107 cells ml−1)
|
||
|---|---|---|---|---|
| With NH4+ addition | Without NH4+ addition | With NH4+ addition | Without NH4+ addition | |
| 0 | 2.2 ± 0.4a | 2.1 ± 0.3 | 1.0 ± 0.3 | 1.1 ± 0.3 |
| 3 | 2.1 ± 0.4 | 1.6 ± 0.2 | 0.9 ± 0.1 | 0.5 ± 0.1 |
| 10 | 2.6 ± 0.3 | 1.5 ± 0.3 | 1.6 ± 0.4 | 0.4 ± 0.3 |
The values are means ± standard errors.
14C contents in culture samples.
The 14C contents in samples (biomass plus culture medium) from cultures with and without NH4+ addition at day 0 and day 10 were determined by liquid scintillation counting and compared. The majority (91 and 88%, respectively) of the 14C was associated with the biomass fraction in both culture samples, and only about 10% of the 14C-labeled material was released into the bulk fraction at day 0. After 10 days of incubation, the 14C contents in the biomass (14C-labeled nitrifying bacteria plus heterotrophic bacteria) decreased from 88% to 51% in the culture without NH4+ addition and from 91% to 78% in the culture with NH4+ addition.
Uptake of 14C-labeled microbial products derived from nitrifying bacteria by heterotrophic bacteria.
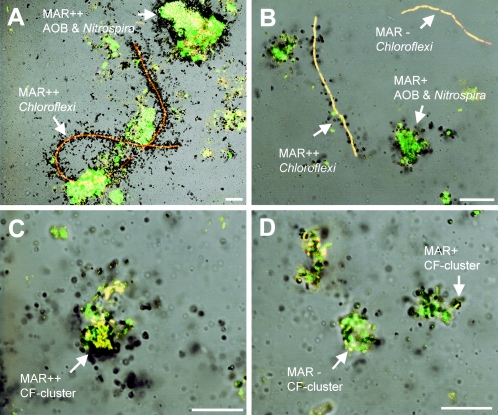
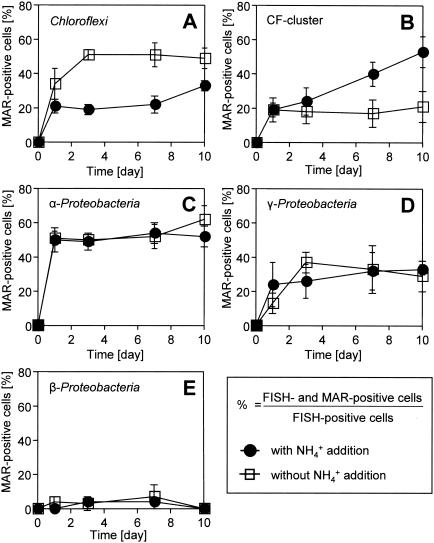
The coexisting heterotrophic bacteria that could incorporate 14C-labeled microbial products derived from nitrifying bacteria were directly visualized by MAR-FISH (Fig. 3) and quantitatively analyzed (Fig. 4). On day 0 of incubation, approximately 100% of the AOB and NOB (Nitrobacter and Nitrospira) cells were strongly MAR positive in both cultures (Fig. 2A), but no MAR-positive heterotrophic bacteria were detected (Fig. 2B). For the member of the Chloroflexi that hybridized with the GNSB-941 and CFX1223 probes, the percentage of probe-hybridized cells that simultaneously took up 14C-labeled microbial products derived from nitrifying bacteria significantly increased to more than 50% within the first 3 days and remained at this level in the culture without NH4+ addition (Fig. 3A and 4A). On the other hand, only 20% of the Chloroflexi cells were weakly MAR positive in the culture with NH4+ addition (Fig. 3B and 4A). The percentage of probe CF319a/b-hybridized cells (members of the Cytophaga-Flavobacterium [CF] cluster) that simultaneously were strongly MAR positive gradually increased to 55% at day 10 in the culture with NH4+ addition (Fig. 3C and 4B), whereas the fraction was unchanged (around 20%) and the MAR signal was weak in the culture without NH4+ addition (Fig. 3D and 4B). More than 50% of the α-Proteobacteria cells that hybridized with probe ALF968 (excluding the genus Nitrobacter cells that hybridized with probe NIT3) became MAR positive within 1 day in both cultures (Fig. 4C), and thereafter this fraction was unchanged. About 30% of the γ-Proteobacteria cells that hybridized with probe GAM42a also incorporated 14C-labeled microbial products derived from nitrifying bacteria in both cultures (Fig. 4D). In contrast, less than 7% of the β-Proteobacteria cells that hybridized with probe BET42a were weakly MAR positive in both cultures (Fig. 4E). These results clearly demonstrated that [14C]bicarbonate originally incorporated into nitrifying bacterial cells could be released and subsequently utilized by the coexisting heterotrophic bacteria.
FIG. 3.
(A and B) Combined MAR and FISH images showing uptake of 14C-labeled microbial products derived from nitrifying bacteria by Chloroflexi in the cultures without NH4+ (A) and with NH4+ (B) at day 3. (C and D) Uptake of 14C-labeled microbial products derived from nitrifying bacteria by the members of CF cluster in the cultures with NH4+ (C) and without NH4+ (D) at day 10. In situ hybridizations were performed with a combination of the FITC-labeled mixed EUB338 probes (green) and TRITC-labeled probes GNSB-941 and CFX1223 (A and B) and with a combination of FITC-labeled mixed EUB338 probes (green) and probe CF319a/b (C and D) (red). Bars = 10 μm. MAR++, strongly MAR positive; MAR+, weakly MAR positive; MAR−, MAR negative.
FIG. 4.
Patterns of uptake of 14C-labeled microbial products derived from nitrifying bacteria by Chloroflexi that hybridized with probes GNSB-941 and CFX1223 (A), by the members of the CF cluster that hybridized with probe CF319a/b (B), by α-Proteobacteria that hybridized with probe ALF968 (excluding the probe NIT3-hybridized genus Nitrobacter) (C), by γ-Proteobacteria that hybridized with probe GAM42a (D), and by β-Proteobacteria that hybridized with probe BET42a (excluding the probe Nso190-hybridized AOB) (E). The fractions of probe-hybridized and MAR-positive cells that incorporated 14C-labeled microbial products derived from nitrifying bacteria were expressed as percentages of the total specific probe-hybridized cells. The error bars indicate the standard errors of duplicate samples.
DISCUSSION
The MAR-FISH technique used in this study was very powerful for tracking the fate of radioactivity originally incorporated into nitrifying bacterial cells. Although various radioactive substrates have been used for microautoradiographic studies (6, 12, 14, 16, 18, 24, 25, 30), we directly labeled nitrifying bacterial cells with [14C]bicarbonate and used it as a substrate for the first time to investigate the substrate cross-feeding from nitrifying bacteria to heterotrophic bacteria in this study. We successfully demonstrated that microbial products derived from nitrifying bacteria were subsequently utilized by different phylogenetic groups of heterotrophic bacteria, which was directly visualized and quantified by MAR-FISH at the single-cell resolution level for the first time.
14C labeling of nitrifying bacteria.
To prepare 14C-labeled nitrifying bacteria, it is important to select the right combination of [14C]bicarbonate concentration and incubation time which provides nitrifying bacteria with enough time to incorporate [14C]bicarbonate but not to cross-feed 14C-labeled microbial products to heterotrophic bacteria. We conducted several sets of preliminary experiments and found that the combination of a specific radioactivity of 10 μCi mmol−1 (corresponding to a [14C]bicarbonate concentration of 35 μM), 6 h of incubation, and 2 days of exposure was the optimum combination for this purpose. However, when the incubation time was increased to more than 24 h (and the concentration of [14C]bicarbonate and the exposure time were the same), we started to observe 14C cross-feeding. It has been shown that most, if not all, heterotrophic bacteria can assimilate CO2 in various carboxylation reactions during biosynthesis (often 5 to 10% of the biomass carbon produced) (36). However, under the incubation conditions for MAR used in this study, heterotrophic bacteria were always MAR negative, indicating that there was no significant uptake of 14CO2. This was probably because we incubated the biofilm samples with low concentrations of [14C]bicarbonate (35 μM) and did not add any external organic carbon substrates for heterotrophic bacteria. Therefore, we could ignore the direct CO2 incorporation by heterotrophic bacteria in this study.
Fate of [14C]bicarbonate incorporated into nitrifying bacteria.
Nitrifying bacteria are known to produce relatively large amounts of soluble microbial products, which consist of utilization-associated products (UAP) and biomass-associated products (BAP) (3, 34, 35). It is also known that nitrifying bacteria produce large amounts of extracellular polymeric substances (EPS). The details of soluble microbial product and EPS compositions and their production kinetics are not well known at present. To differentiate UAP from BAP, we conducted 14C-tracing experiments under two different culture conditions with and without addition of NH4+ (an energy source for nitrifying bacteria). Based on the experimental results for the numbers of AOB and NOB, NH4+ and NO2− consumption, and the 14C contents in the culture samples during the 10-day incubation, we concluded that nitrifying bacteria (AOB and NOB) actively utilized substrates (i.e., NH4+ and NO2−) and grew in the culture with NH4+ addition, which led to formation of mainly substrate utilization (growth)-associated products and EPS. On the other hand, nitrifying bacteria died and decayed in the culture without NH4+ addition, which led to the formation of mainly BAP (i.e., structural cell components and EPS). Although the composition and concentrations of UAP, BAP, and EPS were not measured in this study, this does not negate our conclusions.
Most phylogenetic groups of heterotrophic bacteria except the β-Proteobacteria showed uptake of 14C-labeled microbial products, as shown in Fig. 4. MAR-FISH analysis revealed that more than 50% of the members of the Chloroflexi took up 14C-labeled microbial products within the first 3 days in the culture without NH4+ addition, but they did not show strong uptake in the culture with NH4+ addition (Fig. 4A). To the contrary, the percentage of a member of the MAR-positive bacteria belonging to the CF cluster gradually increased to 55% at day 10 in the culture with NH4+ addition but not in the culture without NH4+ addition (Fig. 4B). Based on these results, we speculated that the member of the Chloroflexi preferentially utilized the decaying nitrifying bacteria cell materials, whereas the member of the CF cluster utilized substrate utilization (growth)-associated products, including EPS or the secondary metabolites of the decayed biomass. It is, however, difficult to differentiate between primary substrate-consuming heterotrophic groups that live on the secretions or lysis products (i.e., structural cell components and EPS) of nitrifying bacteria and the groups that live on the metabolites of these primary consumers. Kindaichi et al. (16) previously demonstrated that despite their low abundance in the biofilm community, the members of the Chloroflexi and the CF cluster were only two heterotrophic groups that could utilize [1-14C]NAG added as a model substrate of structural cell components. Depolymerization of cell wall peptidoglycan (i.e., cell lysis) liberates NAG, which could provide a main pool of organic matter in the autotrophic nitrifying biofilm. The members of the Chloroflexi and possibly the CF cluster could play an important role in the degradation of dead nitrifying and heterotrophic bacterial cells. The members of the Chloroflexi detected in our biofilm were filamentous bacteria and were tightly associated with the nitrifying bacterial clusters. Based on 16S rRNA gene analysis, all our clone sequences were affiliated with Chloroflexi subdivision I (13), which contained the most diverse environmental clones, but only a few isolated cultures (39, 40). The first isolates in subdivision I, Anaerolinea thermophila and Caldilinea aerophila, could preferentially utilize a complex yeast extract-like substrate but not acetate (39, 40). In addition, it has been reported that some members of the CF cluster can preferentially degrade various refractory biomacromolecules, such as cellulose, chitin, and proteins, which should have been abundant in the biofilm (29). All these previous studies support the experimental results obtained in this study. However, the in situ spatial organization of the microbial community in the biofilm was disturbed in this study because homogenization was essential for the quantitative MAR-FISH analysis. It is possible that under nondisturbed in situ biofilm conditions the uptake behavior of the various phylogenetic groups is different from the uptake behavior found in this study. To determine nondisturbed and localized in situ substrate uptake patterns of nitrifying bacteria and heterotrophic bacteria in biofilms at the community level, the MAR-FISH technique should be directly applied to thin sections of the biofilm after cryosectioning in the future.
The members of the α-Proteobacteria and γ-Proteobacteria rapidly became MAR positive within the first few days of incubation. This is probably because they utilized easily degradable soluble microbial products (e.g., amino acids and fatty acids) that were released from nitrifying bacterial cells into the bulk liquid during sample preparation (centrifugation, washing, and resuspension). The radioactivity in the bulk liquid (approximately 10% of the total 14C content) rapidly decreased within 3 days in both cultures (data not shown), which corresponded well with the rapid increases in the numbers of MAR-positive cells belonging to the α- and γ-Proteobacteria but not the β-Proteobacteria. This result was also consistent with the previous finding that members of the α-Proteobacteria and γ-Proteobacteria preferentially utilized low-molecular-weight organic matter (i.e., acetic acid and amino acids) but not NAG (16). Furthermore, Cottrell and Kirchman (6) have also reported that the α- and γ-proteobacterial populations in marine environments predominantly utilized amino acids. The members of the α- and γ-Proteobacteria and the CF cluster are in general abundant in low-organic-load environments (e.g., less polluted rivers and marine environments) (15, 17), whereas the β-Proteobacteria is the dominant group in heavily polluted rivers (5) and in activated sludge systems having relatively high organic carbon loads (44). In this study, the β-Proteobacteria accounted for only 6% of the autotrophic nitrifying biofilm community and did not show significant uptake of radioactivity derived from nitrifying bacteria. This might suggest that they were less active and competitive in low-organic-load environments.
In conclusion, the fate of the radioactivity of [14C]bicarbonate originally incorporated into nitrifying bacteria was successfully determined by using the MAR-FISH approach. The uptake patterns for 14C-labeled microbial products derived from nitrifying bacteria differed greatly among the different phylogenetic groups. In complex multispecies biofilms, exchange of substrates among different phylogenetic groups is the most important ecophysiological interaction, which cannot be determined in individual pure-culture studies.
Acknowledgments
We gratefully thank the Central Institute of Isotope Science, Hokkaido University, for providing the facilities for the isotope experiments.
This research was supported in part by grant-in-aid 09750627 for developmental scientific research from the Ministry of Education, Science and Culture of Japan. This study was also carried out as a part of “The Project for Development of Technologies for Analyzing and Controlling the Mechanism of Biodegrading and Processing,” which was entrusted by the New Energy and Industrial Technology Development Organization (NEDO), Japan.
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I. 1995. In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes, p. 1-15. In A. D. L. Akkerman, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 3.Barker, D. J., and D. C. Stuckey. 1999. A review of soluble microbial products (SMP) in wastewater treatment systems. Water Res. 33:3063-3082. [DOI] [PubMed] [Google Scholar]
- 4.Björnsson, L., P. Hugenholtz, G. W. Tyson, and L. L. Blackall. 2002. Filamentous Chloroflexi (green non-sulfur bacteria) are abundant in wastewater treatment processes with biological nutrient removal. Microbiology 148:2309-2318. [DOI] [PubMed] [Google Scholar]
- 5.Brümmer, I. H. M., W. Fehr, and I. Wagner-Döbler. 2000. Biofilm community structure in polluted rivers: abundance of dominant phylogenetic groups over a complete annual cycle. Appl. Environ. Microbiol. 66:3078-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daims, H., A. Brühl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 8.Daims, H., J. L. Nielsen, P. H. Nielsen, K.-H. Schleifer, and M. Wagner. 2001. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67:5273-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gich, F., J. Garcia-Gil, and J. Overmann. 2001. Previously unknown and phylogenetically diverse members of the green nonsulfur bacteria are indigenous to freshwater lakes. Arch. Microbiol. 177:1-10. [DOI] [PubMed] [Google Scholar]
- 10.Gieseke, A., L. Bjerrum, M. Wagner, and R. Amann. 2003. Structure and activity of multiple nitrifying bacterial populations co-existing in a biofilm. Environ. Microbiol. 5:355-369. [DOI] [PubMed] [Google Scholar]
- 11.Gray, N. D., R. Howarth, A. Rowan, R. W. Pickup, J. G. Jones, and I. M. Head. 1999. Natural communities of Achromatium oxaliferum comprise genetically, morphologically, and ecologically distinct subpopulations. Appl. Environ. Microbiol. 65:5089-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray, N. D., R. Howarth, R. W. Pickup, J. G. Jones, and I. M. Head. 2000. Use of combined microautoradiography and fluorescence in situ hybridization to determine carbon metabolism in mixed natural communities of uncultured bacteria from the genus Achromatium. Appl. Environ. Microbiol. 66:4518-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito, T., J. L. Nielsen, S. Okabe, Y. Watanabe, and P. H. Nielsen. 2002. Phylogenetic identification and substrate uptake patterns of sulfate-reducing bacteria inhabiting an oxic-anoxic sewer biofilm determined by combining microautoradiography and fluorescent in situ hybridization. Appl. Environ. Microbiol. 68:356-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenzaka, T., N. Yamaguchi, K. Tani, and M. Nasu. 1998. rRNA-targeted fluorescent in situ hybridization analysis of bacterial community structure in river water. Microbiology 144:2085-2093. [DOI] [PubMed] [Google Scholar]
- 16.Kindaichi, T., T. Ito, and S. Okabe. 2004. Eco-physiological interaction between nitrifying bacteria and heterotrophic bacteria in autotrophic nitrifying biofilms as determined by MAR-FISH. Appl. Environ. Microbiol. 70:1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 18.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K.-H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 20.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K.-H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteriodes in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 21.Mobarry, B. K., M. Wagner, V. Urbain, B. E. Rittmann, and D. A. Stahl. 1996. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 62:2156-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neef, A. 1997. Anwendung der in situ Einzelzell-Identifizierung von Bakterien zur Populationsanalyse in komplexen mikrobiellen Biozönosen. Doctoral thesis. Technische Universität München, Munich, Germany.
- 23.Nielsen, J. L., D. Christensen, M. Kloppenborg, and P. H. Nielsen. 2003. Quantification of cell-specific substrate uptake by probe-defined bacteria under in situ conditions by microautoradiography and fluorescence in situ hybridization. Environ. Microbiol. 5:202-211. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen, P. H., M. Aquino de Muro, and J. L. Nielsen. 2000. Studies on the in situ physiology of Thiothrix spp. present in activated sludge. Environ. Microbiol. 2:389-398. [DOI] [PubMed] [Google Scholar]
- 25.Nielsen, P. H., P. Roslev, T. E. Dueholm, and J. L. Nielsen. 2002. Microthrix parvicella, a specialized lipid consumer in anaerobic-aerobic activated sludge plants. Water Sci. Technol. 46:73-80. [PubMed] [Google Scholar]
- 26.Okabe, S., T. Itoh, H. Satoh, and Y. Watanabe. 1999. Analyses of spatial distributions of sulfate-reducing bacteria and their activity in aerobic wastewater biofilms. Appl. Environ. Microbiol. 65:5107-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okabe, S., H. Satoh, and Y. Watanabe. 1999. In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 65:3182-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okabe, S., H. Naitoh, H. Satoh, and Y. Watanabe. 2002. Structure and function of nitrifying biofilms as determined by molecular techniques and the use of microelectrodes. Water Sci. Technol. 46:233-241. [PubMed] [Google Scholar]
- 29.O'Sullivan, L. A., A. J. Weightman, and J. C. Fry. 2002. New degenerate Cytophaga-Flexibacter-Bacteroides-specific 16S ribosomal DNA-targeted oligonucleotide probes reveal high bacterial diversity in River Taff epilithon. Appl. Environ. Microbiol. 68:201-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ouverney, C. C., and J. A. Fuhrman. 2000. Marine planktonic archaea take up amino acids. Appl. Environ. Microbiol. 66:4829-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pynaert, K., B. F. Smets, S. Wyffels, D. Beheydt, S. D. Siciliano, and W. Verstraete. 2003. Characterization of an autotrophic nitrogen-removing biofilm from a highly loaded lab-scale rotating biological contactor. Appl. Environ. Microbiol. 69:3626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riemann, L., and F. Azam. 2002. Widespread N-acetyl-d-glucosamine uptake among pelagic marine bacteria and its ecological implications. Appl. Environ. Microbiol. 68:5554-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rittmann, B. E., J. M. Regan, and D. A. Stahl. 1994. Nitrification as a source of soluble organic substrate in biological treatment. Water Sci. Technol. 30:1-8. [Google Scholar]
- 35.Rittmann, B. E., and P. L. McCarty. 2001. Microbial kinetics: soluble microbial products, p. 176-178. In B. E. Rittmann and P. L. McCarty (ed.), Environmental biotechnology: principles and applications. McGraw Hill, New York, N.Y.
- 36.Roslev, P., M. B. Larsen, D. Jørgensen, and M. Hesselsoe. 2004. Use of heterotrophic CO2 assimilation as a measure of metabolic activity in planktonic and sessile bacteria. J. Microbiol. Methods 59:381-393. [DOI] [PubMed] [Google Scholar]
- 37.Schembri, M. A., K. Kjærgaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253-267. [DOI] [PubMed] [Google Scholar]
- 38.Schramm, A., L. H. Larsen, N. P. Revsbech, N. B. Ramsing, R. Amann, and K.-H. Schleifer. 1996. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 62:4641-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekiguchi, Y., Y. Kamagata, K. Nakamura, A. Ohashi, and H. Harada. 1999. Fluorescence in situ hybridization using 16S rRNA-targeted oligonucleotides reveals localization of methanogens and selected uncultured bacteria in mesophilic and thermophilic sludge granules. Appl. Environ. Microbiol. 65:1280-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sekiguchi, Y., T. Yamada, S. Hanada, A. Ohashi, H. Harada, and Y. Kamagata. 2003. Anaerolinea thermophila gen. nov., sp. nov. and Caldilinea aerophila gen. nov., sp. nov., novel filamentous thermophiles that represent a previously uncultured lineage of the domain Bacteria at the subphylum level. Int. J. Syst. Evol. Microbiol. 53:1843-1851. [DOI] [PubMed] [Google Scholar]
- 41.Tolker-Nielsen, T., and S. Molin. 2000. Spatial organization of microbial biofilm communities. Microb. Ecol. 40:75-84. [DOI] [PubMed] [Google Scholar]
- 42.Wagner, M., R. I. Amann, P. Kampfer, B. Assmus, A. Hartmann, P. Hutzler, N. Springer, and K.-H. Schleifer. 1994. Identification and in situ detection of gram-negative filamentous bacteria in activated sludge. Syst. Appl. Microbiol. 17:405-417. [Google Scholar]
- 43.Wagner, M., G. Rath, H. P. Koops, J. Flood, and R. Amann. 1996. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci. Technol. 34:237-244. [Google Scholar]
- 44.Wagner, M., and A. Loy. 2002. Bacterial community composition and function in sewage treatment systems. Curr. Opin. Biotechnol. 13:218-227. [DOI] [PubMed] [Google Scholar]