Abstract
The effects of changes in the gut environment upon the human colonic microbiota are poorly understood. The response of human fecal microbial communities from two donors to alterations in pH (5.5 or 6.5) and peptides (0.6 or 0.1%) was studied here in anaerobic continuous cultures supplied with a mixed carbohydrate source. Final butyrate concentrations were markedly higher at pH 5.5 (0.6% peptide mean, 24.9 mM; 0.1% peptide mean, 13.8 mM) than at pH 6.5 (0.6% peptide mean, 5.3 mM; 0.1% peptide mean, 7.6 mM). At pH 5.5 and 0.6% peptide input, a high butyrate production coincided with decreasing acetate concentrations. The highest propionate concentrations (mean, 20.6 mM) occurred at pH 6.5 and 0.6% peptide input. In parallel, major bacterial groups were monitored by using fluorescence in situ hybridization with a panel of specific 16S rRNA probes. Bacteroides levels increased from ca. 20 to 75% of total eubacteria after a shift from pH 5.5 to 6.5, at 0.6% peptide, coinciding with high propionate formation. Conversely, populations of the butyrate-producing Roseburia group were highest (11 to 19%) at pH 5.5 but fell at pH 6.5, a finding that correlates with butyrate formation. When tested in batch culture, three Bacteroides species grew well at pH 6.7 but poorly at pH 5.5, which is consistent with the behavior observed for the mixed community. Two Roseburia isolates grew equally well at pH 6.7 and 5.5. These findings suggest that a lowering of pH resulting from substrate fermentation in the colon may boost butyrate production and populations of butyrate-producing bacteria, while at the same time curtailing the growth of Bacteroides spp.
Microbial metabolism in the colon has an important impact on health and is strongly influenced by the amount and type of dietary components that survive small intestinal digestion. Short-chain fatty acids (SCFA) arising from microbial fermentation provide energy sources for the colonic epithelium, and butyrate in particular exerts important effects on cell differentiation and gut health (2, 6, 30, 44, 48, 50). Products of microbial fermentation, however, can also be toxic or carcinogenic (21). Shifts in microbial community structure caused by diet (32) also have the potential to influence interactions between gut microbes, gut epithelial cells, and the immune system (10, 28, 40).
Conditions for bacterial growth and metabolism in the human large intestine vary with diet and with location in the colon (8, 9, 25, 38, 52). We have little reliable information, however, on the likely impact of dietary and environmental factors on the microbial community of the human colon. The pH of the gut lumen is likely to be a key factor. Several reports indicate that a slightly acidic pH can occur in the proximal colon, increasing distally (4, 38, 46). A major factor tending to reduce colonic pH is the production of SCFA by microbial fermentation of dietary carbohydrate energy sources, including prebiotics, that are digestible by gut microorganisms but not by host enzymes (4, 18, 20). Another key factor that must influence microbial activity and competition is the relative availability of carbohydrate energy sources and nitrogen sources. Less fermentable carbohydrate reaches the distal compared to the proximal colon, leading to differences in the amount of digestible carbohydrate relative to endogenous protein along the colon (36). Protein-rich diets may increase the amount of dietary protein reaching the large intestine (41).
In vitro continuous flow simulations of the colonic lumen allow carefully controlled manipulations that are impossible to create or monitor in human subjects in vivo. Here we investigate the effect of a one-unit shift in pH and of a sixfold change in peptide supply upon microbial fermentation in a single-stage anaerobic continuous flow fermentor inoculated with mixed human fecal bacteria. Culture-independent molecular methods based on a panel of 16S rRNA-targeted fluorescent probes (19) are used to monitor the composition of the whole gut microbial community. This provides the ability for the first time to correlate metabolic responses with shifts in microbial community structure.
MATERIALS AND METHODS
Collection and preparation of fecal samples.
Fresh fecal samples were provided by two adult volunteers, one consuming a vegetarian diet (donor 1) and the other an omnivorous diet (donor 2). The volunteers did not take any antibiotics or other drugs known to influence the fecal flora for 6 months before the study commenced.
Simulated human colonic fermentor studies.
Single-stage fermentor systems were operated as described previously (17) using a medium based on that of Macfarlane et al. (37). The carbon sources present in the mixed substrate medium were potato starch (0.5% [wt/vol]) and xylan, pectin, amylopectin, and arabinogalactan at 0.06% (wt/vol) each. The total peptide concentrations (comprising equal amounts of casein hydrolysate and peptone water) were either 0.6% as described previously or decreased to 0.1%. Bile salts were added at 0.005%, and the medium was buffered by the addition of 0.32% NaHCO3 and reduced by the addition of 0.05% cysteine HCl. The fermentor growth medium was maintained under a stream of CO2. The volume of the medium in the fermentor vessel was kept constant at 250 ml, with a flow rate of fresh medium equating to one turnover per day, giving a dilution rate of 0.042 h−1. SCFA were added initially to give 33 mM acetate, 9 mM propionate, 5 mM butyrate, and 1 mM each of isobutyrate, isovalerate, and valerate but were not included in the supplied medium. Both the sterile medium feed flask and fermentor flasks were mixed by internal stirrer bars powered by external stirring units. A pH controller delivered sterile solutions of either 0.5 M HCl or 0.5 M NaOH to maintain the pH at either 5.5 ± 0.1 or 6.5 ± 0.1 and to gradually shift the pH between the two values. The temperature was monitored by using a temperature probe, and the fermentor was maintained at 37°C using a thermal jacket. Fecal suspensions were prepared by suspending freshly voided feces (5 g) in 20 ml of 50 mM phosphate buffer (pH 6.5) under O2-free CO2 containing 0.05% cysteine. Aliquots of a given fecal suspension were used to inoculate two parallel fermentor vessels through ports in the top, giving a fecal inoculum of 2% (wt/vol) in the vessel.
Bacterial strains.
Unless otherwise stated, the bacterial strains referred to in Table 1 and Fig. 4 are from Barcenilla et al. (3). Roseburia intestinalis L1-82 (DSM 14610) (12) and Roseburia sp. strain A2-183 (DSM 16839) are available from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ). Bacteroides ovatus V975 was from T. R. Whitehead (Peoria, Ill.), B. thetaiotaomicron (B5482) DSM 2079, B. vulgatus DSM 1447, B. distasonis DSM 20701, Collinsella aerofaciens DSM 3979, and Bifidobacterium infantis DSM 20088 were obtained from the DSMZ. B. longum NCIMB 8809 was obtained from the National Collection of Industrial and Marine Bacteria, Ltd. (Aberdeen, United Kingdom). B. adolescentis L2-32 is described in Duncan et al. (16). This and Ruminococcus bromii L2-63 were non-butyrate-producing isolates from the study of Barcenilla et al. (3). Art12/1 is a recent isolate of Megamonas hypermegale (S. H. Duncan, unpublished). R. flavefaciens 17 and R. albus SY3 are Rowett Research Institute isolates from the rumen. Lactobacillus acidophilus A274 was from V. Bottazzi, Italy. Enterococcus faecalis JH2-2 was from A. A. Salyers (University of Illinois). Megasphaera elsdenii (LC1) ATCC 25940 is from the American Type Culture Collection and Escherichia coli JM109 was supplied by Promega.
TABLE 1.
Probes and conditions used to estimate abundance of bacterial groups in fermentor samples
| Probe | Sequence (5′-3′) | Probe target | Validationa | Hybridization temp (°C) | Formamide concn (%) | Source or reference |
|---|---|---|---|---|---|---|
| Eub338 | GCTGCCTCCCGTAGGAGT | Domain Bacteria | All strains listed here | 50 | 0 | 1 |
| Erec482 | GCTTCTTAGTCAGGTACCG | Clostridial clusters XIVa+b | E. hallii L2-7, Coprococcus sp. strain L2-50; E. rectale A1-86, R. intestinalis L1-82 | 47 | 0 | 16 |
| Bac303 | CCAATGTGGGGGACCTT | Bacteroides-Prevotella group | B. vulgatus BV1447, B. thetaiotaomicron B5482, B. distasonis DSM 20701 | 47 | 0 | 40 |
| Fprau645 | CCTCTGCACTACTCAAGAAAAAC | F. prausnitzii group | F. prausnitzii A2-165 | 50 | 0 | 50 |
| Bif164 | CATCCGGCATTACCACCC | Bifidobacterium genus | B. adoloscentis L2-32, B. longum NCIMB 8809, B. infantis DSM 20088 | 50 | 0 | 30 |
| Rfla729 | AAAGCCCAGTAAGCCGCC | R. flavefaciens subcluster | R. albus SY3, R. flavefaciens 17 R. bromii L2-63 | 50 | 20 | 20 |
| Rbro730 | TAAAGCCCAG(C/T)AGGCCGC | R. bromii subcluster | 50 | 20 | 20 | |
| Rrec584 | TCAGACTTGCCG(C/T)ACCGC | Roseburia genus | E. rectale A1-86, R. intestinalis L1-82 | 50 | 0 | This study |
| Lab158 | GGTATTAGCA(C/T)CTGTTTCCA | Lactobacillus-Enterococcus group | L. acidophilus A2-74, E. faecalis JH2-2 | 47 | 20 | 19 |
| Ato291 | GGTCGGTCTCTCAACCC | Atopobium cluster | C. aerofaciens DSM 3979 | 50 | 0 | 21 |
| Prop853 | ATTGCGTTAACTCCGGCAC | Clostridial cluster IX | M. elsdenii LC1 | 50 | 0 | This study |
| Enterobact D | TGCTCTCGCGAGGTCGCTTCTCTT | Enterobacteriaceae | E. coli MG1655 | 50 | 0 | 44 |
That is, a positive reaction was seen.
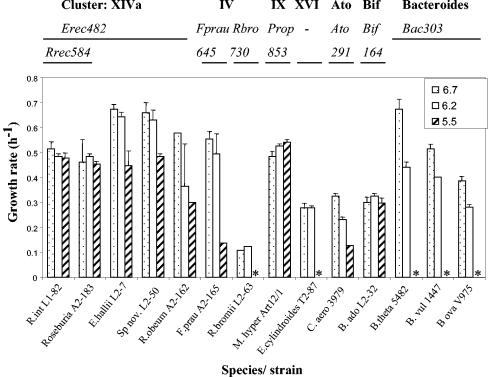
FIG. 4.
Comparison of growth rates (h−1) for 14 representative pure strains of human fecal bacteria grown in batch culture on YCFAG medium at initial pH values of 6.7, 6.2, and 5.5 (see Materials and Methods). Where growth occurred, final pH values were up to 0.8 U lower than the initial pH (lowest value 5.1). ✽, Lack of growth at pH 5.5. The phylogenetic group or cluster to which each strain belongs (clostridial clusters XIVa, IV, IX, and XVI, Atopobium [Ato]; Bifidodobacterium [Bif]; and Bacteroides) and the probes that recognize them are shown above the histogram.
Enumeration of bacteria in the fecal and fermentor samples by FISH analysis.
Immediately after inoculation of the fermentors, and at key points thereafter, a sample was removed for fluorescence in situ hybridization (FISH) analysis and fixed by mixing 1:3 in 4% (wt/vol) paraformaldehyde at 4°C for 16 h and stored at −20°C. FISH analysis was performed as described by Harmsen et al. (23). Diluted cell suspensions were applied to gelatin-coated slides. A total of 10 μl of a 50-ng μl−1 concentration of the oligonucleotide probes plus 100 μl of hybridization buffer was added, and the slides were hybridized overnight. To prevent fading of fluorescence, 50 μl of Vectashield (Vector Laboratories, Burlingame, Calif.) was added to each slide. Cells were counted automatically using image analysis software (17) with a Leica DMRXA epifluorescence microscope, except when the number of cells was less than 10 per field of view, in which case the cells were counted manually. Depending on the number of fluorescent cells, 25 to 50 microscopic fields were counted. The samples were all assessed with the following probes; Bac303, Erec482, Fprau645, Bif164, Lab158, Enterobact D, Ato291, combined Rbro730/Rfla729 (16, 19, 20, 21, 30, 49, 50) and the newly designed Rrec584 and Prop853 probes (Table 1). Total bacterial numbers were estimated by using the universal probe Eub338 (1).
Fermentation product analysis.
SCFA production was determined on samples in duplicate from each fermentor at each sampled time point by capillary gas chromatography after conversion to t-butyldimethylsilyl derivatives (49). The lower limit for reliable detection of SCFA changes was taken as 0.2 mM. Ammonia concentrations in fermentor samples were analyzed by reacting the samples with sodium phenate and sodium hypochlorite. The product, indophenol blue, was measured by determining the optical density at 625 nm (56).
Batch culture incubations at different initial pH values.
The YCFAG medium (which contains yeast extract, Casitone, vitamins, SCFA, and 10 mM glucose) used to assess influence of initial pH values on bacterial growth rates is described fully in reference 12. The pH of the medium was adjusted to give values of 5.5, 6.2, and 6.7 and then dispensed into Hungate tubes that were flushed with CO2; the tubes were heat sterilized. Heat-labile vitamins and glucose were added after the medium was autoclaved. Each test bacterial strain was inoculated into medium at the three different initial pH values in triplicate, and growth was measured spectrophotometrically as the absorbance at 650 nm. Growth rates were calculated in exponential phase.
Statistical analysis.
To determine the abundance of bacteria and bacterial groups in fermentor and fecal samples by FISH, between 25 to 50 fields were counted to give coefficient of variance values of <10. Statistical analyses, including analysis of variance, were performed by using Genstat Release 7.2 (Lawes Agricultural Trust, Rothamsted, United Kingdom).
RESULTS
Investigation of the effects of peptide supply and pH on microbial communities in continuous culture.
The fermentor medium used here and in previous studies (17) is based on that of Macfarlane et al. (37) with amylopectin starch as the main component of a mixture of polysaccharide energy sources (see Materials and Methods). The main sources of amino acid nitrogen in this medium are peptides in the form of casein hydrolysate and peptone water. Two types of experiment, each repeated with fecal inocula from two different donors, were conducted here. In the first type (illustrated for one donor in Fig. 1) two fermentors that received the same inoculum were run in parallel at pH 6.5 with the normal peptide concentration of 0.6% (high) or a reduced peptide concentration of 0.1% (low). In the second type, parallel fermentors were again run with 0.6 or 0.1% peptide inputs. In this case, however, the initial pH was 5.5, and this pH was maintained for a period of 6 to 9 days before a shift to pH 6.5 (illustrated in Fig. 2 for repeat experiments at the high peptide input with the two donors). Metabolite concentrations at the end of each distinct pH regime in these experiments are summarized in Table 2.
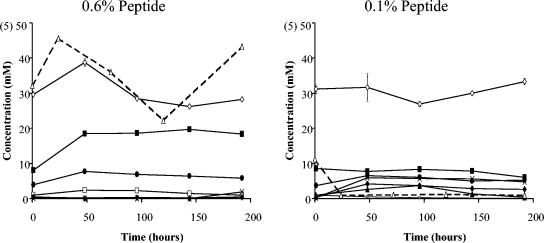
FIG. 1.
Time course of SCFA and ammonia formation in continuous fermentor cultures maintained at pH 6.5 on high (0.6% peptide) and low (0.1%) peptide, following inoculation with a fecal sample from donor 2. Concentrations of products are indicated by symbols as follows: formate (▴), acetate (⋄), propionate (▪), butyrate (•), valerate (□), lactate (⧫), and succinate (×). Ammonium concentrations (▵) are shown on a 10-fold expanded scale, indicated in parentheses. Each value is the average of three independent analyses; error bars indicate the standard deviations.
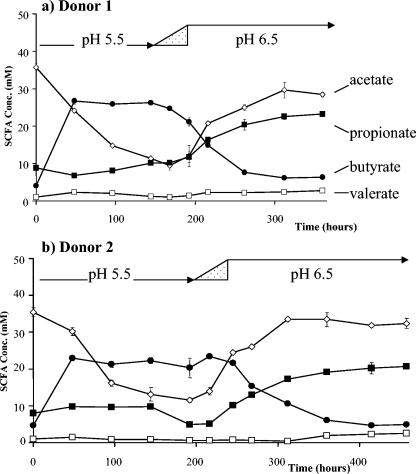
FIG. 2.
Time course of SCFA formation in continuous fermentor cultures initiated at pH 5.5 and 0.6% peptide input after inoculation with a fecal sample from donor 1 (a) and donor 2 (b). The pH was shifted progressively to pH 6.5 within the interval indicated by the triangle. The outcome of parallel experiments (not shown) at 0.1% peptide input is summarized in Table 2. Concentrations of products are indicated as follows: acetate (⋄), propionate (▪), butyrate (•), and valerate (□). Concentrations of formate, lactate, succinate, and caproate (not shown) did not exceed 2 mM. Each value is the average of three independent analyses; error bars represent the standard deviations.
TABLE 2.
Effect of pH and peptide input on final metabolite concentrations for fecal microbial communities in continuous culture
| Metabolite | Concn (mM)a upon incubation at:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| pH 6.5b
|
pH 5.5c
|
pH 6.5c (shifted from pH 5.5)
|
Meand
|
|||||
| Donor 1 | Donor 2 | Donor 1 | Donor 2 | Donor 1 | Donor 2 | pH 6.5 | pH 5.5 | |
| Medium + 0.6% peptide | ||||||||
| Formate | 0.5 | 0.5 | 0.3 | 0.1 | 0.3 | 0.3 | 0.4 | 0.2 |
| Acetate | 32.3 | 28.2 | 11.4 | 14.0 | 28.4 | 32.3 | 30.3 | 12.7 |
| Propionate | 20.1 | 18.3 | 10.1 | 5.1 | 23.2 | 20.6 | 20.6 | 7.6 |
| Butyrate | 4.3 | 5.8 | 26.3 | 23.5 | 6.3 | 4.9 | 5.3 | 24.9 |
| Valerate | 2.4 | 0.2 | 1.2 | 0.5 | 2.8 | 2.5 | 2.0 | 0.9 |
| Succinate | 1.3 | 1.9 | 0 | 0 | 0 | 0 | 0.8 | 0 |
| dl-Lactate | 1.4 | 0.3 | 0 | 0 | 0 | 0.4 | 0.5 | 0 |
| Ammonia | 7.7 | 4.3 | 2.3 | 1.5 | 0.9 | 1.0 | 3.5 | 1.9 |
| Medium + 0.1% peptide | ||||||||
| Formate | 7.8 | 0.3 | 0.3 | 5.6 | 6.4 | 0.6 | 3.8 | 3.0 |
| Acetate | 26.4 | 33.3 | 42.2 | 34.1 | 45.3 | 32.1 | 34.3 | 38.2 |
| Propionate | 12.0 | 6.1 | 4.2 | 0.9 | 6.6 | 5.6 | 7.6 | 2.6 |
| Butyrate | 7.3 | 5.2 | 16.1 | 11.4 | 5.3 | 12.7 | 7.6 | 13.8 |
| Valerate | 1.2 | 0.1 | 0.5 | 0.5 | 0.5 | 0.9 | 0.7 | 0.5 |
| Succinate | 0 | 4.7 | 0 | 3.6 | 1.7 | 2.5 | 2.2 | 1.8 |
| dl-Lactate | 3.7 | 2.7 | 0 | 1.2 | 1.9 | 0.6 | 2.2 | 0.6 |
| Ammonia | 0.07 | 0.09 | 0.2 | 0.03 | 0.08 | 0.02 | 0.07 | 0.12 |
From the experiment shown in Fig. 1 (but including data for donor 1). Data refer to the final (192-h) time point.
From the experiment shown in Fig. 2 (but including data for the low-peptide condition). Data refer to the final time point at the relevant pH (donor 1, 144 h, pH 5.5, and 360 h, pH 6.5; donor 2, 216 h, pH 5.5, and 458 h, pH 6.5).
Overall means of preceding four values at pH 6.5, two at 5.5; the statistical analysis (analysis of variance) is described in the text.
As can be seen from Fig. 1 and Table 2, ammonia was almost undetectable in the low-peptide conditions, reflecting assimilation into bacterial protein. The higher peptide input resulted in significant ammonia concentrations and must be considered more relevant to conditions in vivo (9).
Impact of pH and peptide supply on SCFA formation.
A one-unit shift in pH between 5.5 and 6.5 at the higher peptide input gave the most striking impact upon SCFA production. In particular, pH 5.5 resulted in a highly butyrogenic fermentation, and in a progressive net decrease in acetate concentration (Fig. 2). Butyrate reached concentrations of 24 to 28 mM and in fact exceeded both acetate and propionate concentrations after 150 h. Switching to pH 6.5 resulted in SCFA concentrations very similar to those seen in the separate experiments that were maintained at pH 6.5 throughout (Fig. 1), with acetate becoming the predominant SCFA accompanied by high propionate (>18 mM) and low butyrate (4 to 7 mM) concentrations (Fig. 2 and Table 2). At the lower peptide input, the metabolic effects of a one-unit pH shift were less dramatic (time course not shown). Net acetate consumption was not evident at pH 5.5, and butyrate concentrations did not exceed 20 mM. Nevertheless, butyrate comprised a higher final percentage of total SCFA at pH 5.5 (mean, 23%) than at pH 6.5 (mean, 10.8%) for the low-peptide input (from Table 2). The corresponding percentages for butyrate at the higher peptide input were 53.9% at pH 5.5 and 9% at pH 6.5. Statistical analysis of the data shown in Table 2 confirm that the effect of pH upon butyrate concentration was significant at the 0.001% level.
Propionate concentrations showed a marked response to the peptide supply, which was significant at the 0.001% level (Fig. 1; Table 2). Final propionate concentrations were at least 1.7-fold and up to 5.6-fold higher in all of the high peptide conditions compared to the parallel low-peptide treatment (Table 2). Propionate concentrations were lower in the pH 5.5 incubations compared to pH 6.5 incubations at the same peptide input (P < 0.001).
Changes in microbial populations monitored by FISH.
In order to determine whether these metabolic changes correlated with the populations of any of the major bacterial groups, 12 different 16S rRNA-based FISH probes were used that target predominant groups of human fecal bacteria: Eub338 (total eubacteria), Bac303 (Bacteroides/Prevotella), Erec482 (clostridial clusters XIVa and b), Rrec584 Roseburia/E. rectale (a component of cluster XIVa), Fprau645 (a component of clostridial cluster IV), Rfla729/Rbro730 (cluster IV ruminococci), Bif164 (bifidobacteria), Prop853 (clostridial cluster IX), Ato291(Atopobium cluster), Enterobact D (Enterobacteriaceae), and Lab158 (Lactobacillus/Enterococcus). Probes were first validated against a panel of representative human gut bacteria to ensure their specificity (Table 1). The results from the enumeration of fermentor samples are summarized in Table 3 and Fig. 3.
TABLE 3.
Counts for major bacterial groups in fermentor samples estimated by FISHa
| Probeb | Counts (108 cells ml−1)c
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor 1
|
Donor 2
|
|||||||||||||||
| Type 1 expt
|
Type 2 exptd
|
Type 1 expte
|
Type 2 exptd
|
|||||||||||||
| 0.5 h | 0.6%, 6.5, 192 h | 0.1%, 6.5, 192 h | 0.5 h | 0.6%
|
0.1%
|
0.5 h | 0.6%, 6.5, 192 h | 0.1%, 6.5, 192 h | 0.5 h | 0.6%
|
0.1%
|
|||||
| 5.5, 144 h | 6.5, 360 h | 5.5, 144 h | 6.5, 360 h | 5.5, 216 h | 6.5, 458 h | 5.5, 216 h | 6.5 458 h | |||||||||
| Eub338 | 5.45 | 14.1 | 10.1 | 5.04 | 12.8 | 26.4 | 2.86 | 4.55 | 2.73 | 19.3 | 4.57 | 4.82 | 24.3 | 34.1 | 2.73 | 8.50 |
| Bac303 | 0.15 | 8.54 | 2.81 | 0.21 | 4.10 | 22.1 | 0.38* | 1.54 | 0.47 | 14.8 | 0.74 | 0.62 | 2.52 | 27.6 | 0.40* | 0.36 |
| Erec482 | 1.89 | 0.59 | 3.73 | 1.22 | 6.05 | 1.42 | 1.03 | 1.81 | 0.35 | 1.82 | 1.33 | 2.02 | 8.32 | 3.79 | 1.10 | 7.31 |
| Rrec584 | 0.18 | ND | 0.04* | 0.25 | 2.41 | ND | 0.49 | ND | 0.34 | ND | 0.18* | 0.70 | 3.41 | ND | 0.32 | ND |
| Fprau645 | 0.15 | ND | 0.14 | 0.22 | 1.00 | 0.32 | 0.10* | 0.09 | 0.03 | 0.02 | ND | 0.57 | 0.82 | 0.61 | 0.25 | 0.02* |
| Prop853 | 0.20 | 0.03* | 0.10 | 0.13 | 0.18 | 1.30 | 0.16 | 0.58 | 0.39 | 0.09 | 0.01* | 0.37 | 0.56 | 0.63 | 0.17 | ND |
| Rfla729+Rbro730 | ND | ND | ND | 0.19 | ND | 0.45 | ND | ND | ND | ND | ND | 0.14* | ND | ND | ND | ND |
| Bif164 | ND | ND | 0.20 | ND | ND | ND | 0.29 | 0.41 | 0.43 | 0.08* | 0.14* | 0.19 | ND | ND | 0.38 | 0.15 |
| Enterobact D | ND | 0.03* | 0.03* | ND | 0.01* | 0.05* | ND | 0.10* | ND | ND | ND | ND | ND | ND | ND | 0.02 |
| Lab158 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Ato291 | 0.11 | ND | ND | 0.05 | 0.15* | 0.03* | 0.05* | ND | 0.12* | ND | ND | 0.04* | ND | ND | ND | ND |
Sampling points correspond to those for the metabolic data shown for the same experiments in Table 2, with the addition of initial (0.5-h) counts.
See Table 1.
Values for each donor indicate the percent peptide concentration, the pH, and the time point. ND, not detected (counts of <106 cells ml−1); *, coefficient of variance >10, <15.
Time courses shown in Fig. 2 (for 0.6% peptide).
Time course shown in Fig. 1.
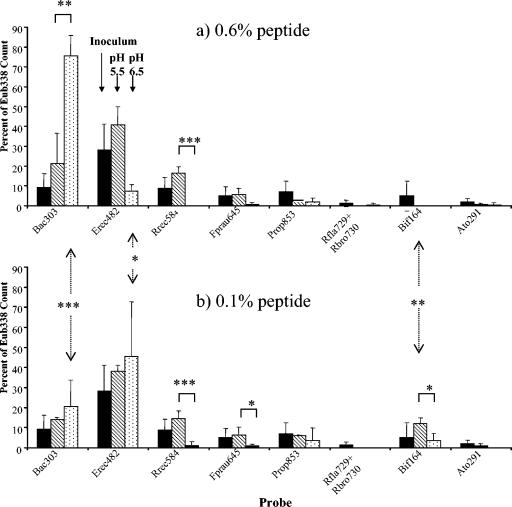
FIG. 3.
Effect of pH upon the abundance of major bacterial groups, expressed as a proportion of the total (Eub338) counts, in continuous fermentor cultures with high (0.6%) (a) and low (0.1%) (b) peptide input. Mean proportions are calculated from the FISH counts given in Table 3 and refer to all of the experiments involving the two donors that are summarized in Table 2 and illustrated in Fig. 1 and 2. Means are given for the initial 0.5-h time point (▪), after incubation at pH 5.5 (▧), and after incubation at pH 6.5 (□). Statistical significance for the effects of different pH and peptide regimes is indicated as follows: ✽, P = <0.05; ✽✽, P = <0.01; ✽✽✽, P = <0.001.
Analysis of an early time point (0.5 h) from each experiment indicated marked differences in the composition of the fecal inoculum between the two donors and between different experiments with the same donor (Table 3). Inocula from donor 2 (consuming an omnivorous diet), for example, yielded higher proportions of Bacteroides (13 to 17% of total eubacteria) and Bifidobacterium (4 to 16%) organisms than donor 1 (3 to 4% and undetectable, respectively). It is already known that individuals show wide variation with respect to bacterial strains and species found in the fecal microbiota (39). It was therefore of interest to discover whether consistent responses might be revealed in bacterial populations using FISH probes in spite of these differences in the inoculum.
At the end of all four incubations conducted at pH 6.5 and high peptide input the Bacteroides group dominated, accounting for between 61 and 84% (mean, 78%) of the total eubacterial counts (Fig. 3). In contrast, Bacteroides accounted for between 4.3 and 34% (mean, 18.5%) of total eubacteria after incubation at pH 5.5 or at pH 6.5 and low peptide input. The most abundant group in the fecal inocula was clostridial cluster XIVab, detected by the Erec482 probe. This group accounted for 86% of total eubacteria at the end of one experiment (donor 2, low peptide [pH 6.5]) but showed wide variation (4 to 86%). A component of this cluster, the butyrate-producing E. rectale-Roseburia group, however, showed more consistent behavior, being low or undetectable at pH 6.5 but comprising 12 to 19% of the total at pH 5.5 (Fig. 3). Meanwhile a second important group of butyrate producers, Faecalibacterium prausnitzii, also showed its highest populations (3 to 9% of the total) at pH 5.5. Bifidobacterial populations were only significant (up to 10%) under conditions of low peptide supply.
Cluster IX representatives (detected with Prop853) were present (up to 13% of total eubacteria) in most of the final samples. Ruminococcal numbers (those species belonging to clostridial cluster IV detected by the Rfla729/Rbro730 probes) were low in the fermentors, although this group can be significant components of fecal microbiota (19, 45, 53).
Effect of pH on the growth and metabolism of representative human colonic anaerobes in pure culture.
In view of these findings a panel of 14 bacteria representative of the dominant groups found in human feces was selected in order to assess their ability to grow in YCFAG medium buffered to different initial pH values (see Materials and Methods). These included three Bacteroides species, five representatives from clostridial cluster XIVa, two from cluster IV, one (Eubacterium cylindroides) from cluster XVI, one (M. hypermegale) from cluster IX, one Bifidobacterium strain, and one strain (C. aerofaciens) belonging to the Atopobium group (Fig. 4). Also shown in Fig. 4 are the oligonucleotide probes that were used to detect relatives of each of these strains in the mixed ecosystem. All of the cluster XIVa strains tested—which, with the exception of the R. obeum strain, were all butyrate producers—grew at pH 5.5 and showed at least 50% of the growth rate seen at pH 6.7. B. adolescentis and M. hypermegale grew as well at pH 5.5 as at 6.7. In contrast, none of the three Bacteroides strains grew at pH 5.5 (Fig. 4). The single E. cylindroides and R. bromii strains tested also failed to grow at pH 5.5. The F. prausnitzii and C. aerofaciens strains were intermediate in their tolerance of pH 5.5, showing 25 and 38%, respectively, of their growth rates at pH 6.7.
DISCUSSION
Impact of a one unit pH change upon fermentation.
The most important finding from the present study is the profound effect of a pH shift of 1 U between 5.5 and 6.5 on fermentation patterns. The lower pH strongly favored butyrate production, which was ∼4-fold higher at pH 5.5 than at pH 6.5 under nitrogen-sufficient conditions. High levels of butyrate formation at pH 5.5 and 0.6% peptide input were accompanied by decreasing acetate concentrations that may reflect net acetate consumption (15). Conversely, the switch to pH 6.5 at the same peptide input led to decreased butyrate and a twofold increase in propionate formation. These changes in relative SCFA production rates appear more extreme than those normally detected for SCFA ratios in fecal samples. It should be noted, however, that fecal SCFA ratios do not equate to production rates because of the rapid absorption of most of the SCFA formed in vivo by the colonic mucosa.
The pH in the distal colon, where carbohydrate fermentation is generally assumed to be slow, is estimated to be 6.5 or higher, but more acidic pHs have been reported for the proximal colon (4). The present results imply that acidic pHs as low as 5.5 tend to favor butyrate formation compared to a near-neutral pH. Butyrate is the preferred energy source for the colonic epithelium and is rapidly absorbed by the mucosa in vivo. Many carbohydrates that escape digestion in the small intestine, such as resistant starch, and prebiotics, such as fructo-oligosaccharides, are reported to be butyrogenic (11, 31, 34, 55). The present findings therefore suggest that such butyrogenic effects might be explained largely by a lowering of the lumenal pH of the proximal colon consequent upon increased fermentative activity (4). On the other hand, a pH of 6.5, in the presence of a carbohydrate supply and the higher (0.6%) peptide input, was found to favor propionate rather than butyrate production. Such conditions may apply in vivo in the early stages of carbohydrate fermentation, or with slowly degraded substrates, or as a result of effective buffering.
Butyrate production rates were previously shown to be much higher in in vitro batch incubations compared to continuous fermentor incubations held at pH 6.5 (15). It now seems clear that this is likely to reflect the drop in pH that occurs in this type of batch culture experiment, which has been widely used to predict the effects of different substrates on colonic fermentation (7). Decreasing the pH is considered to be an important mechanism of action of dietary carbohydrates, but the pH drop in in vitro batch cultures may differ from that occurring in vivo, where absorption and turnover remove the fermentation products.
Impact of peptide input upon fermentation.
Microbial fermentation in the human colon is considered to be energy rather than nitrogen limited (36). Colonic ammonia concentrations of 10 to 40 mM are reported in vivo, derived from urea and from protein breakdown (38, 39). The lower of the two peptide inputs (0.1%) used in the present study resulted in low concentrations of free ammonia, indicating nitrogen limitation. Furthermore, total bacterial numbers detected by the Eub338 probe (Table 3) were consistently lower for the 0.1% peptide input (mean, 5.5 × 108/ml) than for the 0.6% peptide input (mean, 2.2 × 109). The higher (0.6%) peptide condition therefore provides the more realistic approximation to the in vivo situation. The higher peptide input led to increased propionate formation, suggesting that the relative availability of carbohydrate and protein in different regions of the colon could have significant consequences for SCFA production ratios (36).
Relationship between metabolic changes and microbial populations.
The availability of FISH detection techniques makes it possible to track changes in the colonic community and therefore to attempt to interpret metabolic changes in terms of changes in bacterial populations. The majority of anaerobic bacteria found in the human gut produce acetate, but only a limited number of species produce propionate or butyrate (3, 36).
The most abundant butyrate producers are believed to be the Roseburia-E. rectale and Faecalibacterium groups (3, 15, 27, 48). In these fermentor experiments the Roseburia-E. rectale group accounted for up to 20% of the total bacterial count under high-peptide conditions at pH 5.5, when the highest butyrate concentrations were also seen. Similar changes were observed for the F. prausnitzii group. Interestingly, many of these bacteria are net consumers of acetate in pure culture (3, 12, 13, 14, 15). This, compounded by a probable reduction in net acetate producers, provides a likely explanation for the remarkable decline in acetate that was observed in this condition at pH 5.5 and a high peptide input. Isolated Roseburia-related strains also grew well at pH 5.5 in pure culture (Fig. 4). Conversely, the lowest butyrate concentrations were found here at pH 6.5 when the Roseburia-E. rectale and F. prausnitzii populations were apparently less able to compete in the mixed system. Thus, populations of the known major butyrate-producing bacteria correlated closely with butyrate formation in these experiments. In addition, however, butyrate production by isolated strains of Roseburia and F. prausnitzii also shows a slight stimulation (by up to 30%) at pH 5.5 compared to 6.7 in vitro (S. H. Duncan et al., unpublished results).
Many of the known propionate-producing bacteria (Selenomonas, Mitsuokella, Megamonas, Megasphaera, Veillonella, etc.) are detected with the cluster IX probe used here, and their combined populations accounted for up to 13% of total eubacteria. A second and more numerous group, Bacteroides, is, however, capable of producing succinate and propionate depending on the culture conditions, in particular on the state of N and/or C limitation (36). At the turnover rate used here, continuous pure cultures of B. fragilis growing under a CO2 gas phase were found to produce more propionate than succinate when glucose limited (5), although succinate became the more significant product for B. ovatus when carbon was in excess (35). This is thought to reflect the availability of CO2 for the synthesis of oxaloacetate, the precursor for succinate, from PEP (36). If we assume that there are sufficient populations of other bacteria able to decarboxylate succinate, the dominant populations of Bacteroides provide the most obvious explanation for the high propionate concentrations observed at the high peptide input. Indeed, the SCFA proportions seen in the high peptide condition at pH 6.5 (30 mM acetate, 23 mM propionate) are similar to those observed in a pure culture of B. fragilis under C limitation (5).
Very high Bacteroides populations (up to 80% of total eubacteria) and propionate concentrations were also found previously at pH 6.5 and 0.6% peptide input (17). Other factors that probably favored this group were the supply of readily utilized carbohydrates and a high CO2 concentration in the gas phase (5, 29). In contrast, Bacteroides populations typically range between 5 and 30% of bacterial cells detected in fecal and colonic samples (17, 19, 26, 54) although their populations may be elevated in Crohn's disease patients (42). The present study suggests that a mildly acidic pH might act to limit Bacteroides populations in the proximal colon. The three Bacteroides species tested here in vitro all grew very poorly at pH 5.5, although it seems likely that some Bacteroides-related bacteria must show greater resistance to low pH, since the group was still detectable at pH 5.5 in the fermentor experiments.
These experiments were designed to examine the extent to which two human colonic microbial communities responded to two specific factors (pH, peptide supply) in the gut environment. Many other factors are of course subject to change in vivo: in particular the carbohydrate energy source and transit time were not varied here but can result in significant changes in microbial populations and fermentation (see, for example, references 17, 20, 31, 32, and 51). The present results, however, reveal the potentially dramatic effects of a one-unit pH change, particularly under nitrogen-sufficient conditions, upon microbial community composition and SCFA production ratios. Specifically, a mildly acidic pH (5.5) stimulated butyrate production and the populations of the known butyrate-producing species such as Roseburia spp. and F. prausnitzii. Propionate formation, on the other hand, was maximized at pH 6.5, correlating with greatly increased populations of Bacteroides-related bacteria.
Acknowledgments
This study was supported by a BBSRC-SEERAD research grant to the Rowett Institute and to U. Dundee.
We thank Grietje Holtrop of BioSS for statistical advice and Petra Louis and Colin Stewart for critical reading of the manuscript. We are indebted to Gail Skene for supplying unpublished information on the Prop853 probe.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, A. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avivi-Green, C., S. Polak-Charcon, Z. Madar, and B. Schwartz. 2000. Apoptosis cascade proteins are regulated in vivo by high intracolonic butyrate concentration: correlation with colon cancer inhibition. Oncol. Res. 12:83-95. [DOI] [PubMed] [Google Scholar]
- 3.Barcenilla, A., S. E. Pryde, J. C. Martin, S. H. Duncan, C. S. Stewart, and H. J. Flint. 2000. Phylogenetic relationships of dominant butyrate producing bacteria from the human gut. Appl. Environ. Microbiol. 66:1654-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowns, R. L., J. A. Gibson, G. E. Sladen, B. Hicks, and A. M. Dawson. 1974. Effects of lactulose and other laxatives on ileal and colonic pH as measured by a radiotelemetry device. Gut 15:999-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caspari, D., and J. M. Macy. 1983. The role of carbon dioxide in glucose metabolism of Bacteroides fragilis. Arch. Microbiol. 135:16-24. [DOI] [PubMed] [Google Scholar]
- 6.Csordas, A. 1996. Butyrate, aspirin and colorectal cancer. Eur. J. Cancer Prevent. 5:221-231. [DOI] [PubMed] [Google Scholar]
- 7.Cummings, J. H. 1995. Short chain fatty acids, p. 101-130. In G. R. Gibson and G. T. Macfarlane (ed.), Human colonic bacteria: role in nutrition, physiology, and pathology. CRC Press, Boca Raton, Fla.
- 8.Cummings, J. H., and H. N. Englyst. 1987. Fermentation in the human large intestine and the available substrates. Am. J. Clin. Nutr. 45:1243-1255. [DOI] [PubMed] [Google Scholar]
- 9.Cummings, J. H., and G. T. Macfarlane. 1991. The control and consequences of bacterial fermentation in the human colon. J. Appl. Bacteriol. 70:443-459. [DOI] [PubMed] [Google Scholar]
- 10.Deplancke, B., and H. R. Gaskins. 2001. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. Am. J. Clin. Nutr. 73:1131S-1141S. [DOI] [PubMed] [Google Scholar]
- 11.Djouzi, Z., and C. Andrieux. 1997. Compared effects of three oligosaccharides on metabolism of intestinal microflora in rats inoculated with a human faecal flora. Br. J. Nutr. 78:313-324. [DOI] [PubMed] [Google Scholar]
- 12.Duncan, S. H., A. Barcenilla, C. S. Stewart, S. E. Pryde, and H. J. Flint. 2002. Acetate utilization and butyryl CoA:acetate CoA transferase in human colonic bacteria. Appl. Environ. Microbiol. 68:5186-5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan, S. H., G. L. Hold, A. Barcenilla, C. S. Stewart, and H. J. Flint. 2002. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int. J. Syst. Evol. Microbiol. 52:1-6. [DOI] [PubMed] [Google Scholar]
- 14.Duncan, S. H., G. L. Hold, H. J. M. Harmsen, C. S. Stewart, and H. J. Flint. 2002. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify the species into a new genus Faecalibacterium gen. nov. Int. J. Syst. Evol. Microbiol. 52:2141-2146. [DOI] [PubMed] [Google Scholar]
- 15.Duncan, S. H., G. Holtrop, G. E. Lobley, G. Calder, C. S. Stewart, and H. J. Flint. 2004. Contribution of acetate to butyrate formation by human faecal bacteria. Br. J. Nutr. 91:915-923. [DOI] [PubMed] [Google Scholar]
- 16.Duncan, S. H., P. Louis, and H. J. Flint. 2004. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 70:5810-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duncan, S. H., K. P. Scott, A. G. Ramsay, H. J. M. Harmsen, G. W. Welling, C. S. Stewart, and H. J. Flint. 2003. Effects of alternative dietary substrates on competition between human colonic bacteria in an anaerobic fermentor. Appl. Environ. Microbiol. 69:1136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fedorak, R. N., and K. L. Madsen. 2004. Probiotics and the management of inflammatory bowel disease. Inflamm. Bowel Dis. 10:286-299. [DOI] [PubMed] [Google Scholar]
- 19.Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces quantified by fluorescent in situ hybridization with group specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 21.Gill, C. I. R., and I. R. Rowland. 2002. Diet and cancer: assessing the risk. Br. J. Nutr. 88(Suppl. 1):S73-S87. [DOI] [PubMed] [Google Scholar]
- 22.Harmsen, H. J. M., P. Elfferich, F. Schut, and G. W. Welling. 1999. A 16S rRNA-targeted probe for detection of lactobacilli and enterococci in faecal samples by fluorescent in situ hybridization. Microb. Ecol. Health Dis. 11:3-12. [Google Scholar]
- 23.Harmsen, H. J. M., G. C. Raangs, T. He, J. E. Degener, and G. W. Welling. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harmsen, H. J. M., A. C. Wildeboer-Veloo, J. Grijpstra, J. Knol, J. E. Degener, and G. W. Welling. 2000. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human faeces from volunteers of different age groups. Appl. Environ. Microbiol. 66:4523-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill, M. J. 1998. Composition and control of ileal contents. Eur. J. Cancer Prevent. 7:S75-S78. [DOI] [PubMed] [Google Scholar]
- 26.Hold, G. L., S. E. Pryde, V. J. Russell, E. Furrie, and H. J. Flint. 2002. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 39:33-39. [DOI] [PubMed] [Google Scholar]
- 27.Hold, G. L., A. Schwiertz, R. I. Aminov, M. Blaut, and H. J. Flint. 2003. Oligonucleotide probes that detect quantitatively significant groups of butyrate producing bacteria in human feces. Appl. Environ. Microbiol. 69:4320-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hooper, L. V. 2004. Bacterial contributions to mammalian gut development. Trends Microbiol. 12:129-134. [DOI] [PubMed] [Google Scholar]
- 29.Howlett, M. R., D. O. Mountford, K.W., Turner, and A. M. Robertson. 1976. Metabolism and growth yields in Bacteroides ruminicola strain B14. Appl. Environ. Microbiol. 32:274-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobasch, G., D. Schmiedl, M. Kruschewski, and K. Schmehl. 1999. Dietary resistant starch and chronic inflammatory bowel diseases. Int. J. Colorectal Dis. 14:201-211. [DOI] [PubMed] [Google Scholar]
- 31.Kleessen, B., L. Hartmann, and M. Blaut. 2001. Oligofructose and long chain inulin: influence on the gut microbial ecology of rats associated with a human faecal flora. Br. J. Nutr. 86:291-300. [DOI] [PubMed] [Google Scholar]
- 32.Kruse, H. P., B. Kleessen, and M. Blaut. 1999. Effects of inulin on faecal bifidobacteria in human subjects. Br. J. Nutr. 82:375-382. [DOI] [PubMed] [Google Scholar]
- 33.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. F. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeBlay, G., C. Michel, H. M. Blottiere, and C. Cherbut. 1999. Prolonged intake of fructo-oligosaccharides induces a short-term elevation of lactic acid producing bacteria and a persistent increase in cecal butyrate in rats. J. Nutr. 129:2231-2235. [DOI] [PubMed] [Google Scholar]
- 35.Macfarlane, G. T., and G. R. Gibson. 1991. Co-utilization of polymerized carbon sources by Bacteroides ovatus grown in a two-stage continuous culture system. Appl. Environ. Microbiol. 57:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macfarlane, G. T., and G. R. Gibson. 1997. Carbohydrate fermentation, energy transduction, and gas metabolism in the human large intestine, p. 269-318. In R. I. Mackie and B. A. White (ed.), Gastrointestinal microbiology. Chapman and Hall, London, England.
- 37.Macfarlane, G. T., S. Hay, and G. R. Gibson. 1989. Influence of mucin on glycosidase, protease and arylamidase activities of human gut bacteria grown in a 3-stage continuous culture system. J. Appl. Bacteriol. 66:407-417. [DOI] [PubMed] [Google Scholar]
- 38.Macfarlane, G. T., G. R. Gibson, and J. H. Cummings. 1992. Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 72:57-64. [DOI] [PubMed] [Google Scholar]
- 39.Macfarlane, S., and G. T. Macfarlane. 1995. Proteolysis and amino acid fermentation, p. 75-100. In G. R. Gibson and G. T. Macfarlane (ed.), Human colonic bacteria: role in nutrition, physiology, and pathology. CRC Press, Boca Raton, Fla.
- 40.Madsen, K. L. 2004. Interactions between probiotic bacteria and the intestinal epithelium. Reprod. Nutr. Dev. 44:S100. [Google Scholar]
- 41.Magee, E. A., C. J. Richardson, R. Hughes, and J. H. Cummings. 2000. Contribution of dietary protein to sulfide production in the large intestine: an in vitro and a controlled feeding study in humans. Am. J. Clin. Nutr. 72:1488-1494. [DOI] [PubMed] [Google Scholar]
- 42.Mangin, I., R. Bonnet, P. Seksik, L. Rigottier-Gois, M. Sutren, Y. Bouhnik, C. Neut, M. D. Collins, J.-F. Colombel, P. Marteau, and J. Dore. 2004. Molecular inventory of faecal microflora in patients with Crohn's disease. FEMS Microbiol. Ecol. 50:25-36. [DOI] [PubMed] [Google Scholar]
- 43.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 44.Mariadason, J. M., G. A. Corner, and L.H. Augenlicht. 2000. Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: comparison with trichostatin A, sulindac, and curcumin and implications for chemoprevention of colon cancer. Cancer Res. 60:4561-4572. [PubMed] [Google Scholar]
- 45.Moore, W. E. C., and L. H. Moore. 1995. Intestinal floras of populations that have a high risk of colon cancer. Appl. Environ. Microbiol. 61:3202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nugent, S. G., D. Kumar, D. S. Rampton, and D. F. Evans. 2001. Intestinal luminal pH in inflammatory bowel disease: possible determinants and implications for therapy with aminosalicylates and other drugs. Gut 48:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ootsubo, M., T. Shimizu, R. Tanaka, T. Sawabe, K. Tajima, M. Yoshimizu, Y. Ezaki, and H. Oyaizu. 2002. Oligonucleotide probe for detecting Enterobacteriaceae by in situ hybridization. J. Appl. Microbiol. 93:60-68. [DOI] [PubMed] [Google Scholar]
- 48.Pryde, S. E., S. H. Duncan, G. L. Hold, C. S. Stewart, and H. J. Flint. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217:133-139. [DOI] [PubMed] [Google Scholar]
- 49.Richardson, A. J., G. C. Calder, C. S. Stewart, and A. Smith. 1989. Simultaneous determination of volatile and nonvolatile fermentation products of anaerobes by capillary gas chromatography. Lett. Appl. Microbiol. 9:5-8. [Google Scholar]
- 50.Scheppach, W., H. Luehrs, and T. Menzel. 2001. Beneficial health effects of low digestible carbohydrate consumption. Br. J. Nutr. 85:S23-S30. [DOI] [PubMed] [Google Scholar]
- 51.Sharp, R., and G. T. Macfarlane. 2000. Chemostat enrichments of human feces with resistant starch are selective for adherent butyrate-producing clostridia at high dilution rates. Appl. Environ. Microbiol. 66:4212-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silvester, K. R., and J. H. Cummings. 1995. Does digestibility of meat protein help explain large bowel cancer risk? Nutr. Cancer 24:279-288. [DOI] [PubMed] [Google Scholar]
- 53.Suau, A., V. Rochet, A. Sghir, G. Gramet, S. Brewaeys, M. Sutren, L. Rigottier-Gois, and J. Dore. 2001. Fusobacterium prausnitzii and related species represent a dominant group within the human faecal flora. Syst. Appl. Microbiol. 24:139-145. [DOI] [PubMed] [Google Scholar]
- 54.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Topping, D. L., and P. M. Clifton. 2001. Short chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81:1031-1064. [DOI] [PubMed] [Google Scholar]
- 56.Whitehead, R., G. H. Cooke, and B. T. Chapman. 1967. Problems associated with the continuous monitoring of ammoniacal nitrogen in a river water. Anal. Chem. 11:337. [Google Scholar]
- 57.Zoetendal, E. G., A. D. L. Akkermans, and W. M. DeVos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]