Abstract
Bacterial growth occurs in noncarbonated natural mineral waters a few days after filling and storage at room temperature, a phenomenon known for more than 40 years. Using the full-cycle rRNA approach, we monitored the development of the planktonic bacterial community in a noncarbonated natural mineral water after bottling. Seven 16S rRNA gene libraries, comprising 108 clones in total, were constructed from water samples taken at various days after bottling and from two different bottle sizes. Sequence analyses identified 11 operational taxonomic units (OTUs), all but one affiliated with the betaproteobacterial order Burkholderiales (6 OTUs) or the class Alphaproteobacteria (4 OTUs). Fluorescence in situ hybridization (FISH) was applied in combination with DAPI (4′,6′-diamidino-2-phenylindole) staining, viability staining, and microscopic counting to quantitatively monitor changes in bacterial community composition. A growth curve similar to that of a bacterium grown in a batch culture was recorded. In contrast to the current perception that Gammaproteobacteria are the most important bacterial components of natural mineral water in bottles, Betaproteobacteria dominated the growing bacterial community and accounted for 80 to 98% of all bacteria detected by FISH in the late-exponential and stationary-growth phases. Using previously published and newly designed genus-specific probes, members of the betaproteobacterial genera Hydrogenophaga, Aquabacterium, and Polaromonas were found to constitute a significant proportion of the bacterial flora (21 to 86% of all bacteria detected by FISH). For the first time, key genera responsible for bacterial growth in a natural mineral water were identified by applying molecular cultivation-independent techniques.
According to European law (Directives 80/777/EEC and 96/70/EC of the European Parliament and of the Council), natural mineral water is microbiologically unaltered water and thus clearly distinguishable from ordinary drinking water. Furthermore, it is characterized by its constancy of composition concerning certain mineral salts and trace elements. Natural or drilled underground sources of natural mineral water must be protected from pollution to guarantee the original microbiological purity and the chemical composition of essential components of the mineral water. In addition, it is prohibited to subject natural mineral water to any treatment except for (i) the elimination and/or (re)introduction of carbon dioxide and (ii) the decantation and/or filtration of unstable constituents such as iron, manganese, sulfur, or arsenic compounds.
One consequence of these directives is that natural mineral water also contains the indigenous microbial flora present at the source (28). This natural bacterial community appears to be highly preserved throughout the bottling process, as indicated by restriction fragment length polymorphism (RFLP) screening of numerous isolates obtained before and after bottling (47). In 1960, Buttiaux and Boudier (7) were the first to show that within 1 week after bottling and storage at ambient temperatures the natural microbial flora of the water starts to multiply and gives rise to an increase in CFU up to 104 to 105 ml−1. Various research groups confirmed this bacterial growth phenomenon by quantifying the bacteria present in natural mineral waters at the source and at several points in time after bottling and storage at different temperatures (6, 13, 19, 37, 40, 41, 53). These studies focused on quantification of the bacterial community as a whole by determining (i) heterotrophic plate counts, (ii) total cell counts using acridine orange or ethidium bromide, and/or (iii) viable cell counts using 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyl-2H-tetrazolium chloride to quantify the number of actively respiring cells. Regarding the bacterial community composition of bottled mineral waters, only approaches based on isolation and subsequent identification of individual mineral water bacteria have been applied (19, 20, 47). Given that cultivation-dependent community analyses generally suffer from well-recognized quantitative and qualitative biases (45, 50), it is likely that the true bacterial community structure of bottled natural mineral waters remains largely unrecognized to date.
In the present study, the culture-independent full-cycle rRNA approach, involving the establishment of 16S rRNA gene clone libraries and the subsequent design and application of clone-specific probes for quantitative fluorescence in situ hybridization (FISH) (4, 23), was applied to provide a more realistic picture of the bacterial flora growing in a bottled noncarbonated natural mineral water. In contrast to previous reports, the actively growing bacterial community was found to be dominated by members of the betaproteobacterial order Burkholderiales.
MATERIALS AND METHODS
Storage and filtration of natural mineral water samples.
Noncarbonated natural mineral water samples (pH 7.2; mineral content [in mg liter−1]: Na+, 5; K+, 1; Ca2+, 78, Mg2+ 24; Cl−, 4.5; SO42− 10; NO3−, 3.8; HCO3−, 357) were analyzed at 1 to 23 days after bottling. Natural mineral water in 0.5- and 1.5-liter polyethylene terephthalate (PET) bottles was purchased either from a retail outlet in Germany or directly from the manufacturer within 1 day after bottling. Mineral water bottles were stored at room temperature (20 to 22°C) prior to investigation. Bottles were vigorously agitated twice daily to minimize biofilm formation on the bottle walls. For DNA extraction, whole-cell fixation for FISH, determination of viable cell counts, and PCR with whole cells, microbial cells were concentrated from 50 to 3,000 ml of mineral water on white polycarbonate (PC) filters (diameter, 25 mm; pore size, 0.2 μm [GTTP 02500; Millipore, Eschborn, Germany]) by using a stainless steel vacuum filtration unit consisting of six glass filter towers (Sartorius, Göttingen, Germany).
Extraction of genomic DNA.
DNA was extracted from 500 to 3,000 ml of mineral water by using two different methods. Prior to both DNA extractions, planktonic bacteria were enriched on a PC filter by filtration. Subsequently, the PC filter was cut into small pieces with a sterile scalpel. For DNA extraction according to method I, bacteria were resuspended from the PC filter pieces by vortexing with 2 ml of natural mineral water and pelleted by centrifugation (14,000 rpm, 10 min). DNA from pelleted bacteria was extracted by enzymatic cell lysis and chloroform treatment according to a previously established protocol (55). Method II involved the resuspension of PC filter pieces in 400 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). Subsequently, cells were lysed by ultrasonic treatment (Sonorex Super RK 102 H; Bandelin, Berlin, Germany) with sterile glass beads (diameter, 0.10 to 0.11 mm) for 10 min in the presence of 50 μl of 25% sodium dodecyl sulfate and 600 μl of phenol-chloroform-isoamyl alcohol (25:24:1). Lysates were incubated at 65°C for 10 min and subsequently centrifuged (14,000 rpm, 10 min). Nucleic acids were extracted twice with 1 ml of phenol each time. To precipitate nucleic acids from the solution, 0.1 volumes of 3 M sodium acetate (pH 5.2) and 2.5 volumes of ice-cold ethanol were added, followed by incubation for 2 h at −20°C. Precipitated DNA was washed with ice-cold 70% ethanol and resuspended in 50 μl of double-distilled water prior to storage at −20°C.
Enrichment of microbial cells from mineral water for direct PCR.
Bacteria from 500 ml of bottled natural mineral water were enriched on a PC filter by filtration and subsequently resuspended from the filter surface with 2 ml of mineral water by vortexing. After centrifugation (10,000 rpm, 10 min) and disposal of the supernatant, the cell pellet was resuspended in 20 μl of mineral water and stored at −20°C.
PCR amplification.
Oligonucleotide primers (616V-630R) targeting the 16S rRNA genes of almost all bacteria were used for PCR to obtain almost full-length bacterial 16S rRNA gene fragments (∼1.5 kb) as described previously (24). Reaction mixtures containing 25 pmol of each primer were prepared in a total volume of 50 μl by using 10× REDTaq PCR buffer and 3 U of REDTaq DNA polymerase (Sigma-Aldrich, Taufkirchen, Germany). Thermal cycling was carried out by an initial denaturation step at 94°C for 1 min, followed by 30 cycles of denaturation at 94°C for 40 s, annealing at 52°C for 40 s, and elongation at 72°C for 1 min 30 s. Cycling was completed by a final elongation step at 72°C for 10 min. The presence and size of the amplification products were determined by 1% agarose gel electrophoresis. Ethidium bromide-stained bands were digitally recorded with a video documentation system (Cybertech, Hamburg, Germany).
16S rRNA sequence analysis.
Amplified 16S rRNA gene sequences were cloned, sequenced, and phylogenetically analyzed according to previously described procedures (32). All 16S rRNA gene-containing clones were screened by RFLP analysis by using the four-base-specific restriction endonucleases CfoI and MspI in 1× SuRE/Cut buffer L (Roche, Mannheim, Germany) (22). The new 16S rRNA gene sequences were added to an alignment of ca. 50.000 full small-subunit rRNA sequences (http://arb-db-central.swiki.net/1) by using the alignment tool ARB_EDIT of the ARB program package (34). Alignments were refined by visual inspection. Chimeric sequences were identified by independently subjecting base positions 1 to 513, 514 to 1026, and 1027 to 1539 (Escherichia coli numbering) of the 16S rRNA sequence to phylogenetic analysis. Inconsistent affiliation of the gene fragments in the phylogenetic trees was interpreted as being caused by a chimeric sequence. Phylogenetic analyses were performed by applying distance matrix, maximum-parsimony, and maximum-likelihood methods. Only alignment positions that were conserved in ≥50% of either bacterial or proteobacterial sequences were analyzed. Phylogenetic consensus trees were prepared as recommended previously (33). Names of bacterial taxa were used in accordance with the prokaryotic nomenclature proposed in the taxonomic outline of the second edition of Bergey's Manual of Systematic Bacteriology (http://dx.doi.org/10.1007/bergeysoutline200210) (15).
Fixation of microbial cells on PC filters for FISH.
After filtration of microbial cells on white PC filters, all fixation and washing steps were performed in the vacuum filtration unit by successively applying and removing a vacuum. All fixation solutions were prepared as described previously (10). The PC filter was covered with fresh fixation solution (4% paraformaldehyde in 1× phosphate-buffered saline) for 20 min. Fixation solution was removed by applying a vacuum. Subsequently, PC filters were successively washed three times with 1× phosphate-buffered saline and double-distilled water, air dried, and stored in the dark prior to FISH.
Oligonucleotide probes and FISH.
Probes used in the present study are listed in Table 1. Newly developed probes were additionally deposited at probeBase (http://www.microbial-ecology.net/probebase/) (31). Oligonucleotides labeled with the hydrophilic sulfoindocyanine dye Cy3 were purchased from Hybaid-Interaktiva (Ulm, Germany). Optimal hybridization conditions were determined for newly designed probes (23) by using previously established hybridization and washing buffers (36). In situ hybridization of paraformaldehyde-fixed bacteria on a PC filter was performed according to a published protocol (17). Accordingly, the PC filter with the paraformaldehyde-fixed bacteria was cut into four sections. Each filter section was placed on a microscopic slide and covered with 30 μl of hybridization solution. Hybridization was performed in an equilibrated chamber at 46°C. Subsequently, filter sections were stringently washed for 15 min at 48°C and dried on Whatman 3M paper (Whatman International, Ltd., Maidstone, United Kingdom).
TABLE 1.
rRNA-targeted oligonucleotide probe sequences, specificity, and formamide concentration in the hybridization buffer required to ensure optimal hybridization stringency
| Probea | Target rRNA | Probe sequence (5′ to 3′) | Formamide (%) | Specificityb | Source or reference |
|---|---|---|---|---|---|
| NON338 | ACT CCT ACG GGA GGC AGC | Nonbinding control (complementary to EUB338) | 52 | ||
| EUB338 | 16S | GCT GCC TCC CGT AGG AGT | -c | Together targeting most Bacteria | 3 |
| EUB338-II | 16S | GCA GCC ACC CGT AGG TGT | -c | Together targeting most Bacteria | 9 |
| EUB338-III | 16S | GCT GCC ACC CGT AGG TGT | -c | Together targeting most Bacteria | 9 |
| ALF968 | 16S | GGT AAG GTT CTG CGC GTT | 20 | Many Alphaproteobacteria including OTUs 7, 8, and 9 | 38 |
| BET42a | 23S | GCC TTC CCA CTT CGT TT | 35d | Betaproteobacteria | 36 |
| HYD208 | 16S | AAT CGC GCG AGG CCT TAC | 30 | Hydrogenophaga spp., including OTU 1 | 2 |
| HYD56b | 23S | TGG GAC CTC GTT CAG TTA | 50 | Hydrogenophaga spp. | 46 |
| S-*-Aqua-0828-a-A-18 | 16S | GCA AGC CGT CCA ACA ACC | 20 | Aquabacterium spp., including OTU 3 | This study |
| S-*-Pomo-0828-a-A-18 | 16S | CTA ATG CAC CCA ACA ACC | 20 | Polaromonas spp., including OTU 2 | This study |
| GAM42a | 23S | GCC TTC CCA CAT CGT TT | 35d | Gammaproteobacteria | 36 |
| CF319a | 16S | TGG TCC GTG TCT CAG TAC | 35 | Cytophaga-Flavobacterium cluster of the Bacteroidetes | 35 |
| HGC69a | 23S | TAT AGT TAC CAC CGC CGT | 35 | Actinobacteria | 39 |
Probes developed in this study were named according to an established protocol (1).
Up-to-date specificity can be checked via probeBase (http://www.microbial-ecology.net/probebase/) (31).
Usable at any formamide concentrations.
Used with unlabeled competitor oligonucleotide (36).
DAPI and ChemChrome staining.
PC filter sections with oligonucleotide probe-stained bacteria were covered with ice-cold 1.5 μM DAPI (4′,6′-diamidino-2-phenylindole) solution (21) and stained for 10 min on ice in the dark. Thereafter, the filter sections were washed with ice-cold double-distilled water followed by 50% ethanol, dried on Whatman 3M paper, and mounted on glass slides with Citifluor AF1 (Citifluor, Ltd., Canterbury, United Kingdom).
Viability staining of microbial cells was performed immediately after filtration by covering the PC filter with the fluorogenic esterase substrate ChemChrome (Chemunex, Maisson-Alfort, France) according to the manufacturer's recommendations.
Total, probe-dependent, and viable cell counts.
An epifluorescence microscope equipped with a mercury lamp (Axioplan HBO 50; Carl Zeiss, Göttingen, Germany) and appropriate fluorescence filter sets for counting of probe (Cy3 filter set HQCy3; excitation, BP535/50 nm; dichroic mirror Q565 LP; emission, BP610/75 nm [Carl Zeiss])-, DAPI (filter set 01; excitation, 365/12; dichroic mirror 397; emission LP397 [Carl Zeiss])-, and ChemChrome (filter set 09, excitation, BP470/40 nm; dichroic mirror, 510 nm; emission, LP520 nm [Carl Zeiss])-stained microbial cells was used. The total and viable cell numbers were determined by counting DAPI- and ChemChrome-stained cells, respectively, in at least 50 randomly chosen fields of view. For analyzing the relative cell numbers of individual bacterial populations, FISH was combined with DAPI staining. For each hybridization experiment, probe- and DAPI-stained cells in 20 randomly chosen fields of view were counted at a magnification of 400 or 1,000. In each microscopic field probe-positive cell counts were determined first. Bleaching of Cy3-labeled cells by UV light prior to their recording could thereby be avoided. All probe-dependent counts were corrected by subtracting the counts obtained with the negative control probe NON338 (Table 1). The ratio of the number of cells labeled by the rRNA-targeted oligonucleotide probe to the total number of cells stained by DAPI was calculated for each field of view.
Nucleotide sequence accession numbers.
The sequences obtained in the present study are available in GenBank under accession numbers AF522997 to AF523070.
RESULTS
The composition of the microbial community approximately 3 weeks after bottling.
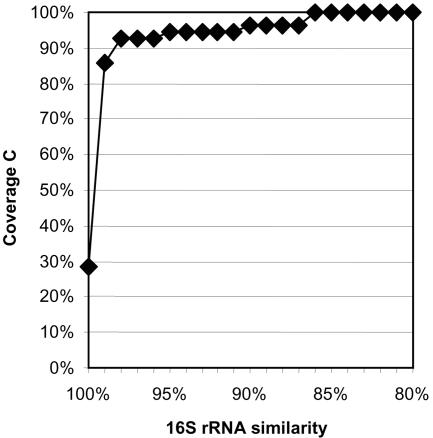
In order to get a first impression of the richness and relative abundance of bacteria in a commercially available natural mineral water, noncarbonated water taken 23 days after filling in 1.5-liter PET bottles (all water bottles derived from the same lot [expiry date 4 January 2001]) was analyzed. In the first step, three 16S rRNA gene libraries (B, C, and D) were established from separate 500-ml samples of mineral water (Table 2). In all, 58 clones were randomly picked and assigned to 10 operational taxonomic units (OTUs) by RFLP analysis (data not shown). Furthermore, all clones were either completely (44 clones) or partially (14 clones) sequenced and phylogenetically analyzed. Two clones, each representing a single-sequence OTU, were identified as chimeras and excluded from further analyses. Based on sequence similarity (97% threshold), the remaining 16S rRNA clone sequences could be grouped in the same OTUs as suggested by RFLP analysis (Table 3) (23, 30). The homologous coverage curve indicated that the 56 clones covered most of the expected richness in the pooled libraries B, C, and D if a sequence similarity threshold of up to 99% is applied (Fig. 1) (16, 42).
TABLE 2.
Characteristics of 16S rRNA gene clone libraries from a natural mineral water analyzed at various days after bottling in PET bottles of different sizes
| Library designation | No. of clonesa | DNA extractionb | Vol of PET bottles (liters) | Days after bottling |
|---|---|---|---|---|
| B | 18 | I | 1.5 | 23 |
| C | 23 | II | 1.5 | 23 |
| D | 17 | -c | 1.5 | 23 |
| G | 3 | II | 1.5 | 7 |
| H | 20 | II | 1.5 | 15 |
| I | 13 | II | 0.5 | 7 |
| J | 14 | II | 0.5 | 15 |
16S rRNA gene clones with insert of the right length (ca. 1.5 kb).
See Materials and Methods.
PCR was performed with enriched whole microbial cells.
TABLE 3.
Affiliation of 16S rRNA gene clones sequenced in this study
| Phylogenetic affiliation | OTUa | Sequenced clones | No. of clones | Bacterium or clone with highest 16S rRNA sequence similarity
|
||
|---|---|---|---|---|---|---|
| Name or designation | Accession no. | % Similarityb | ||||
| Betaproteobacteria | 1 | B-3, B-6, B-9, B-10, B-11, B-15, B-18, B-19, B-20, B-22, C-1, C-3, C-5, C-16, D-4, D-9, D-10, D-13, D-17, D-18, D-20, G-1, H-3, H-9, H-12, J-11 | 26 | Hydrogenophaga palleronii | AF078769 | 98.0-99.3 |
| 2 | B-1, B-2, B-4, B-16, B-21, C-2, C-4, C-6, C-8, C-13, C-14, C-15, C-18, C-22, C-25, D-7, H-2, J-13 | 18 | CDCE-degrading bacterium JS666 | AF408397 | 98.3-99.4 | |
| Polaromonas vacuolata | U14585 | 97.2-98.1 | ||||
| 3 | B-14, C-7, C-9, C-11, C-12, C-19, C-20, C-23, D-2, D-5, D-11, D-12, D-24, H-8, I-17, I-18, J-21 | 17 | Aquabacterium commune | AF035054 | 97.2-99.8 | |
| 4 | D-16, G-2, G-5, H-5 | 4 | Rhodoferax ferrireducens | AF435948 | 97.7-98.3 | |
| 5 | B-5, B-23 | 2 | Wastewater clone T47 | Z93977 | 99.1-99.2 | |
| Hydrogenophaga palleronii | AF078769 | 96.5-96.6 | ||||
| 6 | D-15, I-1 | 2 | Limnobacter thiooxidans | AJ289885 | 99.8-99.9 | |
| 69 | ||||||
| Alphaproteobacteria | 7 | C-26 | 1 | Oligotrophic microbial fuel cell clone oc55 | AY491596 | 98.7 |
| Parvibaculum lavamentivorans | AY387398 | 93.9 | ||||
| 8 | D-1 | 1 | Caulobacter fusiformis | AJ227759 | 97.3 | |
| 9 | I-14 | 1 | Caulobacter henricii | AJ227758 | 99.6 | |
| 10 | I-19 | 1 | Bradyrhizobium liaoningense | AF208513 | 98.8 | |
| 4 | ||||||
| Unknown | 11 | J-1 | 1 | Hot spring clone OPB92 | AF027030 | 73.1 |
| Chimeras | C-24, D-14, J-15 | 3 | ||||
Clones with a 16S rRNA sequence similarity of >97% with each other were grouped into an OTU.
Similarities were calculated with completely sequenced clones without application of a conservation filter and after exclusion of columns with insertions or deletions.
FIG. 1.
Homologous coverage curve for the 16S rRNA gene library generated from a natural mineral water approximately 3 weeks after bottling. The homologous coverage was determined according to the formula C = [1 − (n1 × N−1)] × 100%, with n1 being the number of OTUs containing only one sequence and N being the total number of 16S rRNA gene clones analyzed. Different 16S rRNA sequence similarity thresholds were used for OTU definition (x axis) and the respective homologous coverage values are plotted on the y axis. Prior to this analysis the 16S rRNA gene clone sequences obtained from the three libraries (B, C, and D) were combined in a single data set.
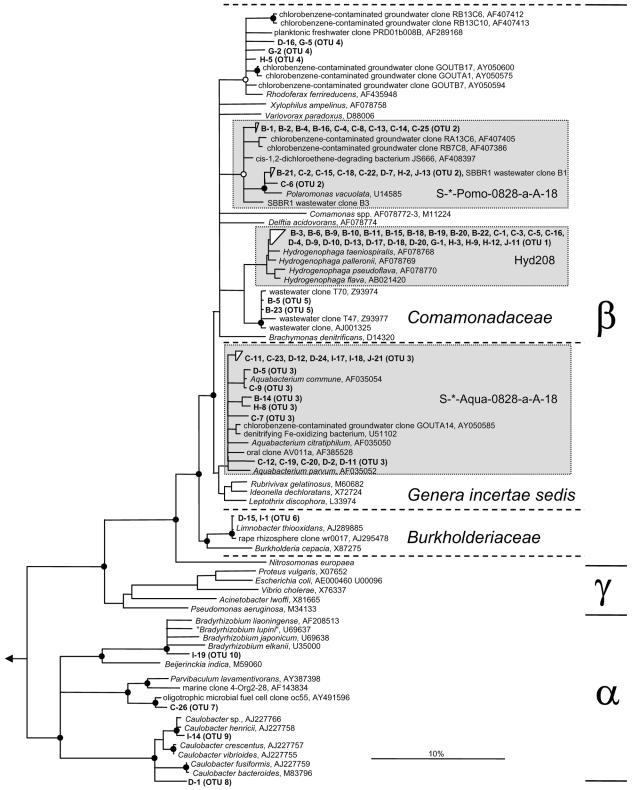
Phylogeny inference showed that the eight OTUs were all affiliated with the phylum Proteobacteria. The majority of clones (54), representing OTUs 1 to 6, belonged to the order Burkholderiales within the class Betaproteobacteria (Table 3). In the phylogenetic 16S rRNA-based tree, OTUs 1, 2, 3, and 4 branched off within the betaproteobacterial genera Hydrogenophaga, Polaromonas, Aquabacterium, and Rhodoferax, respectively (Fig. 2). Two clones, representing OTU 5, formed a monophyletic branch, together with clones derived from wastewater treatment plants. The only betaproteobacterial clone sequence that did belong to the family Burkholderiaceae represented OTU 6 and was almost identical to Limnobacter thiooxidans, a bacterium recently isolated from freshwater lake sediment (43). The remaining two single-sequence OTUs 7 and 8 were affiliated with the class Alphaproteobacteria (Table 3). OTU 8 could be assigned to the genus Caulobacter, whereas OTU 7 clustered with a yet-uncultured alphaproteobacterium from an oligotrophic microbial fuel cell (Fig. 2).
FIG. 2.
Phylogenetic 16S rRNA consensus tree of natural mineral water clones (in boldface) affiliated with the phylum Proteobacteria. Tree topology was determined based on sequences longer than 1,300 nucleotides by neighbor-joining analysis with a 50% proteobacterial conservation filter. An extensive set of reference sequences representing all recognized bacterial and archeal phyla was used as an outgroup. The consensus tree was drawn as recommended previously (33). Filled and open circles on tree knots represent parsimony bootstrap support (100 resamplings) of >90% and 75 to 90%, respectively. Bootstrap values of <75% were omitted. Multifurcations indicate that the branching order could not be unambiguously determined when different treeing methods and conservation filters were applied. The scale bar represents 10% estimated sequence divergence. The α, β, and γ classes of Proteobacteria are delimited by horizontal lines. Different families within the order Burkholderiales are delimited by dashed horizontal lines. Boxes shaded in gray show the coverage of the genus-specific 16S rRNA-targeted oligonucleotide probes used for FISH.
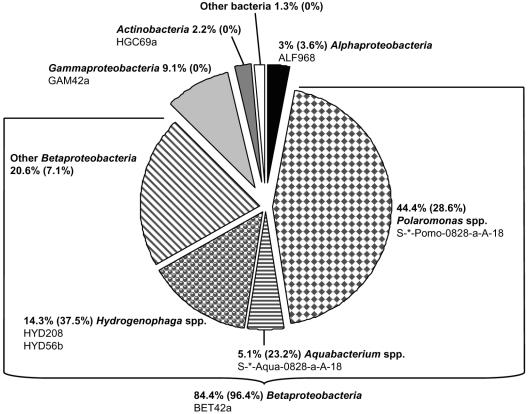
In the second step, the community structure of planktonic bacteria in this mineral water sample was quantitatively analyzed by using DAPI staining and group-specific rRNA-targeted oligonucleotide probes for FISH (Table 1). Microscopy revealed that bacteria were evenly distributed on the surface of the PC filters and appeared as rod-shaped single cells of different length. The total cell count in this mineral water sample was 1.7 × 105 cells ml−1 (standard deviation of 38%). A total of 63% of all cells were detected by the Bacteria-specific probe set EUB338, EUB338-II, and EUB338-III (EUBmix) (3, 9). In accordance with the results of the gene library survey, members of the Betaproteobacteria accounted for 84.4% of bacterial cells stained positive with the EUBmix (Fig. 3). Members of the Gammaproteobacteria, Alphaproteobacteria, and Actinobacteria were detected at substantially lower in situ abundances of 9.1, 3, and 2.2%, respectively (Fig. 3). Only 1.3% of all detectable bacteria in this mineral water sample were not detected with the group-specific probe set used in the present study.
FIG. 3.
Pie chart showing the microbial community composition of a natural mineral water approximately 3 weeks after bottling as determined by FISH and microscopic counting. The probes used for assignment to the different groups are listed. If more than one probe is listed for a single group almost identical results (±1%) were obtained with each of the probes. The percentage refers to the proportion of group- or genus-specific probe-labeled cells of all DAPI-positive cells divided by the proportion of EUBmix-labeled cells of all DAPI-positive cells. The percentage in parentheses indicates the abundance of 16S rRNA gene sequences of the respective bacterial group in the pooled 16S rRNA gene libraries B, C, and D.
Subsequently, the community composition of the numerically dominating Betaproteobacteria was investigated at a higher resolution by using two previously published (HYD208 and HYD56b) and two newly designed genus-specific rRNA-targeted probes (S-*-Pomo-0828-a-A-18, S-*-Aqua-0828-a-A-18) for FISH (Table 1). Polaromonas species were identified as the numerically most important taxon, accounting for 44.4% of all bacteria (52.6% of Betaproteobacteria) detectable by FISH (Fig. 3). Members of the genera Hydrogenophaga and Aquabacterium represented 14.3% and 5.1%, respectively, of the EUBmix-positive cells. In summary, >75% of all Betaproteobacteria present in the mineral water sample were identified with this genus-specific probe set (Fig. 3).
Development of the microbial community within 2 weeks after bottling.
Changes in the microbial structure of the natural mineral water were investigated over 2 weeks after bottling in 1.5- and 0.5-liter PET bottles (all water bottles derived from the same lot [expiry date 15 March 2001]) by applying the aforementioned combined 16S rRNA gene library survey and quantitative FISH approach.
At three different times after bottling (days 1, 7, and 15) genomic DNA was extracted from 3,000 ml of natural mineral water filled in 1.5- and 0.5-liter PET bottles and used for establishment of bacterial 16S rRNA gene libraries (Table 2). However, no 16S rRNA gene amplificate could be obtained from DNA extracted at the first day after bottling. In total, 50 clones were screened by RFLP analysis allowing the identification of four new RFLP banding types (data not shown). Subsequently, 19 clones representing all different RFLP types in the four libraries G, H, I, and J were completely sequenced and phylogenetically analyzed. Clone J-15, which showed one of the new RFLP banding patterns, was identified as a chimera and excluded from further analyses. Two of the three novel OTUs (OTUs 9 and 10) belonged to the genera Caulobacter and Bradyrhizobium, respectively, within the class Alphaproteobacteria (Table 3 and Fig. 2). The third OTU (OTU 11), only represented by clone J-1, showed similarities of <74% to all 16S rRNA sequences deposited in public databases and could not be unambiguously affiliated with any recognized phylum (Table 3).
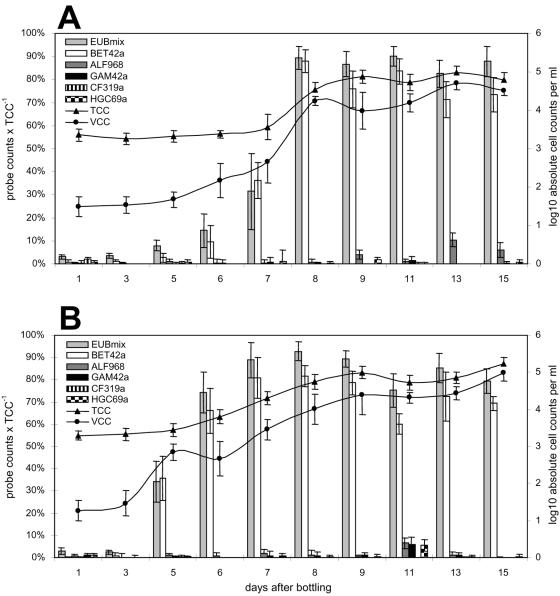
To document the bacterial growth phenomenon, including quantification of those groups responsible for growth, total, viable, and probe-specific cell counts were determined on natural mineral water samples from days 1, 3, 5, 6, 7, 8, 9, 11, 13, and 15 after bottling. In both bottle sizes, total cell counts remained constant at ca. 2 × 103 cells ml−1 between days 1 and 5, the majority of these DAPI-positive cells being of small coccoid shape. The total cell counts increased to ca. 105 cells ml−1 between days 5 and 9 and remained at this level until the end of the analyzed period (Fig. 4). Fewer than 50 viable cells ml−1 could be identified by ChemChrome staining within the first 3 days after bottling. Thereafter, between days 3 and 8, viable cell counts increased to over 104 cells ml−1 and remained in the range of 2 to 10 × 104 viable cells ml−1 until day 15. Comparably, the number of FISH-detectable cells increased dramatically from a maximal level of 3% at day 3 after bottling to 75 to 93% from day 8 onward (Fig. 4). All cells that were visualized by FISH or ChemChrome staining were rods of different length that appeared either as single cells or in divisional states. These observations corroborated previous findings that the microbial flora in natural mineral waters experiences a rapid transition from predominantly inactive resting cells to actively multiplying stages a few days after bottling.
FIG. 4.
The microbial community composition of natural mineral water filled in 1.5-liter (A) and 0.5-liter (B) PET bottles monitored for 2 weeks after bottling. The fraction of all DAPI-positive cells detected by domain- and group-specific oligonucleotide probes is indicated by bars (primary y axis). Absolute cell numbers of all cells (TCC) and viable cells (VCC) are depicted as log10 cells per ml on the secondary y axis. Error bars show the standard deviations of different counts on the same sample.
In addition, FISH with the betaproteobacterial probe BET42a demonstrated that, almost exclusively, members of this proteobacterial class were responsible for the microbial growth in the mineral water. At days 7 to 8, when bacterial growth passed into the stationary phase, 80 to 98% of all FISH-detectable cells were Betaproteobacteria (Fig. 4). In contrast, members of the Alphaproteobacteria, Gammaproteobacteria, and Actinobacteria showed much lower in situ abundances, varying temporally from <1 to 9% (Fig. 4).
The genus-specific probes HYD208, S-*-Pomo-0828-a-A-18, and S-*-Aqua-0828-a-A-18 were used to identify the key betaproteobacterial genera involved in the growth phenomenon in this natural mineral water (Tables 1 and 4). Independent of bottle size, Hydrogenophaga species occurred at in situ abundances of 4 to 12% of all BET42a-positive cells. Aquabacterium and Polaromonas species were always present in comparable proportions. In addition, these two betaproteobacterial genera numerically dominated the mineral water sample in the smaller 0.5-liter PET bottles at day 7, with 34% (Aquabacterium) and 39% (Polaromonas) of all detectable bacteria (corresponding to 42 and 49% of all Betaproteobacteria, respectively). However, they were less dominant in the other samples analyzed, comprising 4 to 11% of all EUBmix-positive cells (corresponding to 4 to 15% of all Betaproteobacteria) (Table 4).
TABLE 4.
Relative abundances of selected betaproteobacterial genera in natural mineral water samples at different times after bottling as determined by counting and comparison of FISH- positive and DAPI-positive cells
| 16S rRNA-targeted oligonucleotide probe | Mean % fraction of total cell counts (SD) at various times after bottling in:
|
|||
|---|---|---|---|---|
| 1.5-liter PET bottles
|
0.5-liter PET bottles
|
|||
| 8 days | 15 days | 7 days | 15 days | |
| HYD208 | 10.3 (4.1) | 6.6 (3.4) | 3.0 (2.3) | 3.2 (1.8) |
| S-*-Pomo-0828-a-A-18 | 6.5 (2.0) | 10.8 (3.3) | 39.3 (8.5) | 7.9 (2.0) |
| S-*-Aqua-0828-a-A-18 | 3.5 (1.9) | 8.9 (4.2) | 34.1 (8.1) | 5.5 (1.5) |
DISCUSSION
Although the factors responsible for the growth of bacteria in natural mineral waters after bottling have been extensively discussed previously (29), knowledge of the actual microbial diversity is still limited. Community structure data for natural mineral waters have thus far been obtained solely by cultivation-based methods. The most frequently isolated microorganisms from bottled natural mineral water were aerobic heterotrophs belonging mainly to the Gamma but also to the Alpha and Beta classes of Proteobacteria (for reviews, see references 28 and 29). However, because of their inherent qualitative and quantitative biases, cultivation-based diversity surveys are rather unlikely to reflect the true microbial community structure present in situ (12, 27, 45, 50). The present study provides for the first time quantitative in situ data on the bacterial community composition in a noncarbonated natural mineral water at different points in time after filling. Using cultivation-independent 16S rRNA gene-based molecular techniques we could (i) identify most of the bacterial groups responsible for the bacterial growth, and (ii) reveal their relative contributions to overall cell numbers.
16S rRNA gene library surveys allow the determination of species richness (measured as number of OTUs) in any given habitat. In contrast to eutrophic wastewater systems (51), the oligotrophic natural mineral water investigated in the present study showed a much lower species richness (11 OTUs) (Table 3). The observed low species number probably reflects the limited availability of dissolved organic carbon in this highly oligotrophic habitat (28). Surprisingly, eight of the 11 OTUs identified had high 16S rRNA gene similarities (above 97%) to described species, and phylogenetic analyses unambiguously placed these in the betaproteobacterial genera Hydrogenophaga, Aquabacterium, Polaromonas, Rhodoferax, and Limnobacter (all in the order Burkholderiales), and in the alphaproteobacterial genera Caulobacter (two OTUs) and Bradyrhizobium (Fig. 2 and Table 3). In accordance with previous reports on the lifestyle of bacteria in bottled mineral waters (28, 29), most described members of these genera are heterotrophs, preferably using oxygen as an electron acceptor (14, 26, 43, 54). Recently, 34 clusters of typical planktonic bacteria in lakes and rivers were delineated by phylogenetic analysis of available 16S rRNA gene sequences (56). However, none of the clones from our study belong to these surface freshwater clusters. In contrast, some of our OTUs are affiliated with sequences retrieved from groundwater habitats (Fig. 2), supporting the hypothesis that bacteria growing in bottled mineral waters mainly originate from the underground source (47). Biofilms growing in the bottling plant could represent an additional origin for bacteria in the bottled mineral water; e.g., Aquabacterium and Caulobacter species are known to inhabit freshwater biofilms (26, 44).
For an encompassing assessment of microbial diversity, knowledge of species richness has to be supplemented with quantitative data about the relative proportions of the individual bacterial groups (species evenness). By using quantitative FISH, we could demonstrate that Betaproteobacteria dominated the growing bacterial consortium in the mineral water analyzed. This observation is consistent with the general importance of this class in diverse oligotrophic freshwater habitats including lake waters (18) and drinking water distribution systems (25). Although bacteria of the genera Hydrogenophaga, Aquabacterium, and Polaromonas (order Burkholderiales) contributed substantially to overall bacterial growth, the proportion of Betaproteobacteria that could not be identified varied depending on bottle size and sampling time. Given that the relative numbers of Betaproteobacteria (class level) remained essentially constant after the late logarithmic growth phase (Fig. 4), these observations might best be explained by shifts in community composition among betaproteobacterial genera or species. Thus, bacteria belonging to the other three identified betaproteobacterial OTUs (also members of the order Burkholderiales), for which no specific probes were designed and applied, might have been numerically more abundant in situ than suggested by the number of clones representing these organisms in the gene libraries.
Many novel fluorescent and nonfluorescent Pseudomonas species have been isolated from natural mineral waters (5, 8, 11, 48, 49). Although these Pseudomonas species have been described as being by far the most important members of the natural mineral water flora (28, 29), it should be noted that (i) none of the 16S rRNA gene clones retrieved in our study were of gammaproteobacterial origin and (ii) only a maximum of 9% of all FISH-positive cells were identified as Gammaproteobacteria by using oligonucleotide probe GAM42a. We additionally analyzed bottled natural mineral waters of five other brands by FISH (data not shown) and the proportion of Gammaproteobacteria never exceeded 6% of the bacterial cells. Although only a limited number of different mineral water brands was analyzed, our data are not consistent with the current perception that gammaproteobacterial species, such as Pseudomonas, are key players in bottled natural mineral waters. It is thus tempting to speculate that members of the Betaproteobacteria, more precisely of the order Burkholderiales, may also be the dominant bacteria in some other natural mineral waters. The genus-specific probe set developed in the present study could be used in future diversity surveys to explore this possibility.
Finally, it is important to know whether the autochthonous bacteria are harmful to human health. Although this possibility cannot be ruled out by the applied methods, it is noteworthy that none of the identified bacteria was closely related to any known human pathogen.
Acknowledgments
This study was supported by the European Union (the project FAIR-CT96-1037). A.L. was financed by a Marie Curie Intra-European Fellowship (VENTSULFURMICDIV) within the 6th European Community Framework Programme.
We thank Silvia Weber (GPII-WS 2001/02) for excellent technical assistance and Michael Taylor for critical reading of the manuscript and inspiring discussions.
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R., W. Ludwig, R. Schulze, S. Spring, E. Moore, and K.-H. Schleifer. 1996. rRNA-targeted oligonucleotide probes for the identification of genuine and former pseudomonads. Syst. Appl. Microbiol. 19:501-509. [Google Scholar]
- 3.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baida, N., A. Yazourh, E. Singer, and D. Izard. 2001. Pseudomonas brenneri sp. nov., a new species isolated from natural mineral waters. Res. Microbiol. 152:493-502. [DOI] [PubMed] [Google Scholar]
- 6.Bischofberger, T., S. K. Cha, R. Schmitt, B. Konig, and W. Schmidt-Lorenz. 1990. The bacterial flora of non-carbonated, natural mineral water from the springs to reservoir and glass and plastic bottles. Int. J. Food Microbiol. 11:51-71. [DOI] [PubMed] [Google Scholar]
- 7.Buttiaux, R., and A. Boudier. 1960. Comportement des bactéries autotrophes dans les eaux minérales conservées en récipients hermétiquement clos. Ann. Inst. Pasteur Lille 11:43-52. [Google Scholar]
- 8.Coroler, L., M. Elomari, B. Hoste, M. Gillis, D. Izard, and H. Leclerc. 1996. Pseudomonas rhodesiae sp. nov., a new species isolated from natural mineral waters. Syst. Appl. Microbiol. 19:600-607. [Google Scholar]
- 9.Daims, H., A. Brühl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 10.Daims, H., K. Stoecker, and M. Wagner. Fluorescence in situ hybridization for the detection of prokaryotes. In A. M. Osborn and C. J. Smith (ed.), Advanced methods in molecular microbial ecology, in press. BIOS Scientific Publishers, Abingdon, United Kingdom.
- 11.Elomari, M., L. Coroler, B. Hoste, M. Gillis, D. Izard, and H. Leclerc. 1996. DNA relatedness among Pseudomonas strains isolated from natural mineral waters and proposal of Pseudomonas veronii sp. nov. Int. J. Syst. Bacteriol. 46:1138-1144. [DOI] [PubMed] [Google Scholar]
- 12.Felske, A., A. Wolterink, R. van Lis, W. M. de Vos, and A. D. Akkermans. 1999. Searching for predominant soil bacteria: 16S rDNA cloning versus strain cultivation. FEMS Microbiol. Ecol. 30:137-145. [DOI] [PubMed] [Google Scholar]
- 13.Ferreira, A. C., P. V. Morais, and M. S. da Costa. 1993. Alterations in total bacteria, iodonitrophenyltetrazolium (INT)-positive bacteria, and heterotrophic plate counts of bottled mineral water. Can. J. Microbiol. 40:72-77. [Google Scholar]
- 14.Finneran, K. T., C. V. Johnsen, and D. R. Lovley. 2003. Rhodoferax ferrireducens sp. nov., a psychrotolerant, facultatively anaerobic bacterium that oxidizes acetate with the reduction of Fe(III). Int. J. Syst. Evol. Microbiol. 53:669-673. [DOI] [PubMed] [Google Scholar]
- 15.Garrity, G. M., and J. G. Holt. 2001. The road map to the manual, p. 119-166. In G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, N.Y. [Google Scholar]
- 16.Giovannoni, S. J., T. D. Mullins, and K. G. Field. 1995. Microbial diversity in oceanic systems: rRNA approaches to the study of unculturable microbes, p. 217-248. In J. Joint (ed.), Molecular ecology of aquatic microbes, vol. G38. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 17.Glöckner, F. O., R. Amann, A. Alfreider, J. Pernthaler, R. Psenner, K.-H. Trebesius, and K.-H. Schleifer. 1996. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst. Appl. Microbiol. 19:403-406. [Google Scholar]
- 18.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez, C., C. Gutierrez, and T. Grande. 1987. Bacterial flora in bottled uncarbonated mineral drinking water. Can. J. Microbiol. 33:1120-1125. [DOI] [PubMed] [Google Scholar]
- 20.Guillot, E., and H. Leclerc. 1993. Bacterial flora in natural mineral waters: characterization by ribosomal ribonucleic acid gene restriction patterns. Syst. Appl. Microbiol. 16:483-493. [DOI] [PubMed] [Google Scholar]
- 21.Hicks, R. E., R. I. Amann, and D. A. Stahl. 1992. Dual staining of natural bacterioplankton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl. Environ. Microbiol. 58:2158-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juretschko, S., A. Loy, A. Lehner, and M. Wagner. 2002. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25:84-99. [DOI] [PubMed] [Google Scholar]
- 24.Juretschko, S., G. Timmermann, M. Schmid, K. H. Schleifer, A. Pommerening-Roser, H. P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalmbach, S., W. Manz, and U. Szewzyk. 1997. Isolation of new bacterial species from drinking water biofilms and proof of their in situ dominance with highly specific 16S rRNA probes. Appl. Environ. Microbiol. 63:4164-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalmbach, S., W. Manz, J. Wecke, and U. Szewzyk. 1999. Aquabacterium gen. nov., with description of Aquabacterium citratiphilum sp. nov., Aquabacterium parvum sp. nov., and Aquabacterium commune sp. nov., three in situ dominant bacterial species from the Berlin drinking water system. Int. J. Syst. Bacteriol. 49:769-777. [DOI] [PubMed] [Google Scholar]
- 27.Kämpfer, P., R. Erhart, C. Beimfohr, J. Böhringer, M. Wagner, and R. Amann. 1996. Characterization of bacterial communities from activated sludge: culture-dependent numerical identification versus in situ identification using group- and genus-specific rRNA-targeted oligonucleotide probes. Microb. Ecol. 32:101-121. [DOI] [PubMed] [Google Scholar]
- 28.Leclerc, H., and M. S. Da Costa. 1998. Microbiology of natural mineral waters, p. 223-273. In D. A. G. Senior and P. R. Ashurst (ed.), Technology of bottled water. Sheffield Academic Press, Ltd., Sheffield, England.
- 29.Leclerc, H., and A. Moreau. 2002. Microbiological safety of natural mineral water. FEMS Microbiol. Rev. 26:207-222. [DOI] [PubMed] [Google Scholar]
- 30.Loy, A., H. Daims, and M. Wagner. 2002. Activated sludge: molecular techniques for determining community composition, p. 26-43. In G. Bitton (ed.), The encyclopedia of environmental microbiology. John Wiley & Sons, Inc., New York, N.Y.
- 31.Loy, A., M. Horn, and M. Wagner. 2003. probeBase: an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loy, A., C. Schulz, S. Lücker, A. Schöpfer-Wendels, K. Stoecker, C. Baranyi, A. Lehner, and M. Wagner. 2005.. 16S rRNA gene-based oligonucleotide microarray for environmental monitoring of the betaproteobacterial order “Rhodocyclales.” Appl. Environ. Microbiol. 71:1373-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 34.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K.-H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 36.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 37.Morais, P. V., and M. S. da Costa. 1990. Alterations in the major heterotrophic bacterial populations isolated from a still bottled mineral water. J. Appl. Bacteriol. 69:750-757. [DOI] [PubMed] [Google Scholar]
- 38.Neef, A. 1997. Anwendung der in situ-Einzelzell-Identifizierung von Bakterien zur Populationsanalyse in komplexen mikrobiellen Biozönosen. Ph.D. thesis. Technische Universität München, Munich, Germany.
- 39.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K.-H. Schleifer. 1994. In situ probing of gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology 140:2849-2858. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt-Lorenz, W. 1974. Untersuchungen über den Keimgehalt von unkarbonisiertem, natürlichem Mineralwasser und Überlegungen zum bakteriologisch-hygienischem Beurteilen von unkarbonisiertem Mineralwasser. Teil I. Chem. Technol. Lebensmittel. 4:97-107. [Google Scholar]
- 41.Schwaller, P., and W. Schmidt-Lorenz. 1980. Flore microbienne de quatre eaux minérales non gazéifiées et mises en bouteilles. I. Dénombrement de colonies, compositions grossière de la flore, et charactères du groupe des bactéries Gram négatif pigmentées en jaune. Zentbl. Bakteriol. Hyg. I. Abt. Orig. C 1:330-347. [Google Scholar]
- 42.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spring, S., P. Kämpfer, and K.-H. Schleifer. 2001. Limnobacter thiooxidans gen. nov., sp. nov., a novel thiosulfate-oxidizing bacterium isolated from freshwater lake sediment. Int. J. Syst. Evol. Microbiol. 51:1463-1470. [DOI] [PubMed] [Google Scholar]
- 44.Stahl, D. A., R. Key, B. Flesher, and J. Smit. 1992. The phylogeny of marine and freshwater caulobacters reflects their habitats. J. Bacteriol. 174:2193-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staley, J. T., and A. Konopka. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39:321-346. [DOI] [PubMed] [Google Scholar]
- 46.Stoffels, M. 1998. Identifizierung und Anreicherung von Bakterien durch verschiedene molekulare in situ und in vitro Hybridisierungstechniken. Ph.D. thesis. Technische Universität München, Munich, German.
- 47.Vachée, A., P. Vincent, C. B. Struijk, D. A. A. Mossel, and H. Leclerc. 1997. A study of the fate of the autochtonous bacterial flora of still mineral waters by analysis of restriction fragment length polymorphism of genes coding for rRNA. Syst. Appl. Microbiol. 20:492-503. [Google Scholar]
- 48.Verhille, S., N. Baida, F. Dabboussi, M. Hamze, D. Izard, and H. Leclerc. 1999. Pseudomonas gessardii sp. nov. and Pseudomonas migulae sp. nov., two new species isolated from natural mineral waters. Int. J. Syst. Bacteriol. 49:1559-1572. [DOI] [PubMed] [Google Scholar]
- 49.Verhille, S., N. Baida, F. Dabboussi, D. Izard, and H. Leclerc. 1999. Taxonomic study of bacteria isolated from natural mineral waters: proposal of Pseudomonas jessenii sp. nov. and Pseudomonas mandelii sp. nov. Syst. Appl. Microbiol. 22:45-58. [DOI] [PubMed] [Google Scholar]
- 50.Wagner, M., R. Amann, H. Lemmer, and K.-H. Schleifer. 1993. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl. Environ. Microbiol. 59:1520-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner, M., and A. Loy. 2002. Bacterial community composition and function in sewage treatment systems. Curr. Opin. Biotechnol. 13:218-227. [DOI] [PubMed] [Google Scholar]
- 52.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 53.Warburton, D. W., P. I. Peterkin, K. F. Weiss, and M. A. Johnston. 1986. Microbiological quality of bottled water sold in Canada. Can. J. Microbiol. 32:891-893. [DOI] [PubMed] [Google Scholar]
- 54.Willems, A., J. Busse, J. Goor, B. Pot, E. Falsen, E. Jantzen, B. Hoste, M. Gillis, K. Kersters, G. Auling, and J. De Ley. 1989. Hydrogenophaga, a new genus of hydrogen-oxidizing bacteria that includes Hydrogenophaga flava comb. nov. (formerly Pseudomonas flava), Hydrogenophaga palleronii (formerly Pseudomonas palleronii), Hydrogenophaga pseudoflava (formerly Pseudomonas pseudoflava and “Pseudomonas carboxydoflava”), and Hydrogenophaga taeniospiralis (formerly Pseudomonas taeniospiralis). Int. J. Syst. Bacteriol. 39:319-333. [Google Scholar]
- 55.Wisotzkey, J. D., P. Jurtshuk, Jr., and G. E. Fox. 1990. PCR amplification of 16S rDNA from lyophilized cell cultures facilitates studies in molecular systematics. Curr. Microbiol. 21:325-327. [DOI] [PubMed] [Google Scholar]
- 56.Zwart, G., M. P. Kamst-van Agterveld, F. Hagen, S.-K. Han, and B. C. Crump. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquatic Microb. Ecol. 28:141-155. [Google Scholar]