Abstract
The availability of cloned luciferase genes from fireflies (luc) and from bacteria (luxAB) has led to the widespread use of bioluminescence as a reporter to measure cell viability and gene expression. The most commonly occurring bioluminescence system in nature is the deep-sea imidazolopyrazine bioluminescence system. Coelenterazine is an imidazolopyrazine derivative which, when oxidized by an appropriate luciferase enzyme, produces carbon dioxide, coelenteramide, and light. The luciferase from the marine copepod Gaussia princeps (Gluc) has recently been cloned. We expressed the Gluc gene in Mycobacterium smegmatis using a shuttle vector and compared its performance with that of an existing luxAB reporter. In contrast to luxAB, the Gluc luciferase retained its luminescence output in the stationary phase of growth and exhibited enhanced stability during exposure to low pH, hydrogen peroxide, and high temperature. The work presented here demonstrated the utility of the copepod luciferase bioluminescent reporter as an alternative to bacterial luciferase, particularly for monitoring responses to environmental stress stimuli.
Bioluminescence is widely distributed in nature, occurring in a remarkably diverse set of organisms, including bacteria, dinoflagellates, fungi, fish, insects, shrimp, and squid (31, 39, 40, 50). Bioluminescence arises from oxidation of a substrate (a luciferin) by an enzyme (a luciferase), usually in the presence of molecular oxygen. Luciferin and luciferase are generic terms as none of the major classes exhibit sequence homology. While phylogenetic analyses suggest that bioluminescence has had more than 30 independent origins, there are five basic luciferin-luciferase systems. The most widely studied bioluminescence systems are those belonging to luminous beetles in the family Lampyridae, more commonly known as fireflies (such as Photinus pyralis) (18, 72), and the luminous bacteria (Vibrio sp., Photobacterium sp., and Photorhabdus luminescens) (2, 3, 14, 22, 23, 26, 64). The firefly luminescence reaction is catalyzed by a monomeric ca. 62-kDa luciferase encoded by a single gene (luc) and involves the oxidation of a benzothiazoyl-thiazole luciferin and ATP, resulting in the production of oxyluciferin, AMP, CO2, and light (18, 47, 51, 72). In contrast, the bacterial (lux) luminescence reaction involves the oxidation of a long-chain aldehyde (RCHO) and reduced flavin mononucleotide, resulting in the production of oxidized flavin mononucleotide and a long-chain fatty acid (RCOOH), along with the emission of blue-green light at 490 nm (10, 32, 73). The reaction is catalyzed by bacterial luciferase, a heterodimeric 77-kDa enzyme composed of an alpha subunit and a beta subunit encoded by the luxA and luxB genes, respectively (22, 25). Bioluminescence is an excellent reporter system (recently reviewed in reference 56), a sensitive marker for microbial detection (13, 15, 27, 28, 49, 57, 58, 70), a real-time, noninvasive reporter for measuring gene expression (11, 12, 19, 33, 36, 48, 51, 53, 54, 55, 69), and a way to measure intracellular biochemical function (cell viability) (1, 4, 5, 16, 17, 27, 28, 29, 30, 34).
The rapid growth of applications of bioluminescence has stimulated research into investigation and exploitation of new bioluminescent systems (44). The most commonly occurring natural bioluminescence system is the deep-sea imidazolopyrazine bioluminescence system that has been found in seven phyla and approximately 90 genera, including copepods, ostracods, cephalopods, and amphipods (66). Coelenterazine is an imidazolopyrazine derivative that acts as the luciferin which, when oxidized by the appropriate luciferase, produces carbon dioxide, coelenteramide, and light (59, 60). One of the most widely studied coelenterazine-catalyzing luciferases is Ruc produced by Renilla reniformans, a sea pansy that displays bioluminescence upon mechanical stimulation. Ruc was first purified and characterized by Matthews et al. (45), and the cDNA was later isolated and expressed in Escherichia coli (42), transgenic plant tissues (46), and mammalian cells (41) and is now commercially available as an assay system (Promega Corporation).
Gaussia princeps is a bioluminescent marine copepod with a 10-mm-long body that lives at depths between 350 and 1,000 m. It emits bioluminescence as a secretion from 30 glands located in the antennas, cephalothorax, thorax, and abdomen in response to mechanical, electrical, or light stimuli (6-8, 38). The release of a luminous bolus from G. princeps is accompanied by rapid swimming that propels the copepod away from the bolus. In this manner, bioluminescence most likely serves as a defense mechanism that startles and blinds dark-adapted predators, providing a glowing decoy to hold the predator's attention while the copepod escapes. The luciferase (Gluc) gene from G. princeps has recently been cloned and shown to oxidize coelenterazine to produce light (9). The Gluc luciferase was subsequently used as a bioluminescent reporter of DNA hybridization and shows promise as a detection reagent in immunoassays (68) and in mammalian cells (65).
The present study was designed to investigate the possible use of the Gluc luciferase as a reporter system in mycobacteria. An estimated 2 billion people are latently infected with Mycobacterium tuberculosis, the causative agent of tuberculosis (21). There are about 8 million new infections per year and 2 million deaths, and many of them are in patients already infected with human immunodeficiency virus. New antituberculosis drugs and better vaccines are urgently needed, and research in these areas is a high priority (63). M. tuberculosis research requires containment level 3 facilities, which coupled with a slow doubling time (nearly 24 h) makes studies using conventional microbiological techniques challenging. In our laboratory (35, 62, 67) and other laboratories (4, 5, 16, 17, 20, 34) extensive use has been made of the bacterial and beetle luciferases as reporter genes in mycobacteria to determine cell numbers and viability. We have shown that the Gluc luciferase is expressed in the fast-growing organism Mycobacterium smegmatis and have characterized its performance under different stress conditions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. smegmatis mc2 155 (61) and E. coli DH5α were used in this study. Liquid cultures of bacteria were grown with shaking at 200 rpm at 37°C in Luria-Bertani (LB) medium supplemented with hygromycin (50 μg ml−1) or kanamycin (100 μg ml−1) as appropriate.
Construction of bioluminescent reporter plasmids.
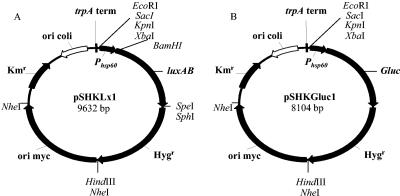
The plasmids and primers used in this study are shown in Tables 1 and 2. Plasmid pTKmx is a mycobacterium-E. coli shuttle vector that harbors a promoterless xylE reporter gene downstream of a transcriptional terminator (37). The xylE gene was excised from the vector by digestion with the restriction enzymes KpnI and SphI. The luxAB genes derived from Vibrio harveyi were obtained by PCR from plasmid pSMT1 (62) using primers P1 and P2 and directionally cloned using the KpnI and SphI sites in pTKmx to obtain the promoterless reporter plasmid pSKLx. Primers P1 and P2 introduced XbaI and BamHI sites upstream of the luxAB genes and an SpeI site downstream of the luxAB genes. Kanamycin is not the antibiotic of choice when working with mycobacteria as spontaneous mutants frequently arise. For this reason the gene for resistance to the antibiotic hygromycin was obtained by PCR from plasmid pSMT1 (62) using primers P3 and P4 and was directionally cloned using the SphI and HindIII sites in pSKLx to obtain plasmid pSHKLx. In order to obtain high levels of light expression, the 600-bp promoter for the heat shock gene hsp60 was obtained by PCR from M. tuberculosis H37Rv DNA using primers P5 and P6 and was directionally cloned using the XbaI and BamHI sites in pSHKLx to obtain the promoted construct pSHKLx1 (Fig. 1A). The luxAB genes were removed from pSHKLx1 as a BamHI and SphI fragment, and the vector was blunt ended using the Klenow fragment (New England Biolabs, United Kingdom) and dephosphorylated. The 540-bp Gluc luciferase gene from G. princes was excised from plasmid pUC19Gluc (Nanolight Technology, Prolume Ltd., Pinetop, AZ) using EcoRI and XbaI and was blunt ended using the Klenow fragment (Fig. 1B) before ligation into the pSHKLx1 plasmid with luxAB deleted to create pSHKGluc1.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pTKmx | Mycobacterium-E. coli shuttle vector encoding a promoterless xylE reporter gene, Kmr | 37 |
| pSMT1 | Mycobacterium-E. coli shuttle vector encoding the luxAB cassette from V. harveyi, Hygr | 62 |
| pSKLx | Mycobacterium-E. coli shuttle vector encoding the promoterless luxAB (V. harveyi) cassette, Kmr | This study |
| pSHKLx | Mycobacterium-E. coli shuttle vector encoding the promoterless luxAB (V. harveyi) cassette, Kmr Hygr | This study |
| pSHKLx1 | Mycobacterium-E. coli shuttle vector encoding a promoted luxAB (V. harveyi) cassette, Kmr Hygr | This study |
| pUC19Gluc | Cloning vector containing the 540-bp luciferase gene (Gluc) from G. princeps | Nanolight Technology, Prolume Ltd. |
| pSHKGluc1 | Mycobacterium-E. coli shuttle vector containing the promoted Gluc gene from G. princeps, Kmr Hygr | This study |
TABLE 2.
Primers used in this studya
| Primer | Sequence |
|---|---|
| P1 | GGTCTAGAGGATCCGGAGGAATGTTATGAAATTTGG |
| P2 | GGACTAGTTTACGAGTGGTATTTGACGA |
| P3 | GGGCATGCGAATTCCCGGGGATCCGGTGATTG |
| P4 | CCCAAGCTTTCAGGCGCCGGGGGCGGTGTCCGGC |
| P5 | GGTCTAGAGGTGACCACAACGACGCCCCCGCT |
| P6 | GGGGATCCGCAATTGTCTTGGCCAT |
Restriction sites are underlined.
FIG. 1.
Mycobacterium-E. coli shuttle vectors pSHKLx1 (encoding a promoted luxAB [V. harveyi] cassette) (A) and pSHKGluc1 (encoding a promoted Gluc luciferase gene from G. princeps) (B). trpA term, terminator sequence; Hygr, hygromycin resistance gene; Kmr, kanamycin resistance gene; ori myc, mycobacterial origin of replication (pAL5000); ori coli, E. coli origin of replication (pUC); Phsp60, promoter from M. tuberculosis hsp60 gene.
Luminescence assays.
Luminescence measurements were obtained in triplicate at room temperature using a tube luminometer (Berthold Autolumat LB953). Luminescence was measured immediately after addition of the substrate for 10 s using an integration time of 1 s, and the results were expressed in relative light units (RLU). For bacteria expressing Gluc, a 10-mmol liter−1 stock of the substrate coelenterazine (Nanolight Technology, Prolume Ltd., Pinetop, AZ) was prepared in methanol for use at a final concentration of 10 μmol liter−1. All coelenterazine solutions were stored at −20°C, and working solutions were kept on ice in the dark during preparation. For bacteria expressing luxAB a 1% stock of the aldehyde substrate (decanal) was prepared in ethanol.
Effects of growth phase on the bioluminescence responses of M. smegmatis.
Liquid cultures of bacteria were grown with shaking at 200 rpm at 37°C in LB medium supplemented with hygromycin. Samples were analyzed during the exponential phase (optical density at 600 nm [OD600], 0.5) and during the stationary phase (OD600, 1.2 and 2.5) to determine the effects of the growth phase on bioluminescence. Assays were performed with three independent replicate cultures.
Bioluminescence responses of M. smegmatis to pH.
Assays were performed with exponential-phase cultures (OD600, 0.5). In the wells of a 24-well microtiter plate, 500-μl aliquots of bacterial cells were added to 500-μl aliquots of LB medium acidified using 2 M HCl as previously described (52). Cultures were incubated statically at 37°C and assayed for bioluminescence (as described above) and cell viability (by plating serial dilutions onto Middlebrook 7H11 agar plates supplemented with 10% oleic acid-albumin-dextrose-catalase and hygromycin as appropriate) at 0 and 1 h. Assays were performed twice with three independent replicate cultures.
Bioluminescence responses of M. smegmatis to hydrogen peroxide.
Assays were performed with exponential-phase cultures (OD600, 0.5). In the wells of a 24-well microtiter plate, 500-μl aliquots of bacterial cells were added to 500-μl aliquots of LB medium containing hydrogen peroxide that resulted in final concentrations in the range from 0 to 32 mM. Cultures were incubated statically at 37°C and were assayed for bioluminescence and cell viability at 0 and 90 min. Assays were performed twice with three independent replicate cultures.
Bioluminescence responses of M. smegmatis to heat shock.
Assays were performed with exponential-phase cultures (OD600, 0.5). Bacterial cultures were subjected to 30 min of heat shock at 45°C and 48°C and then incubated at 37°C to recover. Cultures were assayed for bioluminescence prior to heat shock and at regular intervals during recovery. Assays were performed twice with three independent replicate cultures.
RESULTS
Expression of a novel bioluminescence gene in M. smegmatis.
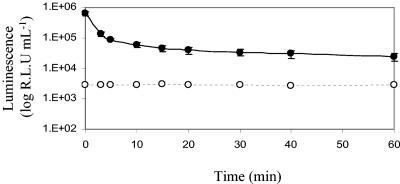
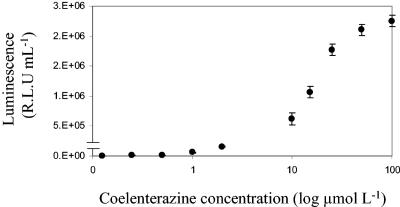
The gene encoding the luciferase enzyme from G. princeps (Gluc) was cloned downstream from an hsp60 promoter in the mycobacterial shuttle vector pSHKGluc1 (Fig. 1) and introduced into M. smegmatis by electroporation. The novel bioluminescence reporter gene was stably expressed in both M. smegmatis and E. coli and catalyzed a flash reaction in which luminescence decreased approximately 10-fold in the first 5 min (Fig. 2). After this initial decrease the rate of decreased slowed, and luminescence remained detectable above the background level after 60 min. The amount of luminescence was dependent on the concentration of substrate added and followed a sigmoid curve. At very low coelenterazine concentrations (below 0.5 μmol liter−1) the flash reaction proceeded extremely fast and the luminescence rapidly decayed to background levels, while at high concentrations the substrate became saturating. However, at final concentrations between 0.1 and 10 μmol liter−1 there was a linear relationship between light output and coelenterazine concentration (Fig. 3). A final concentration of coelenterazine of 10 μmol liter−1 was selected for all further assays. At this concentration, coelenterazine exhibited background chemiluminescence of approximately 100 RLU ml−1. The background signal was higher in the Middlebrook 7H9 medium commonly used for mycobacterial growth, and in order to minimize this, LB medium was used throughout the present study. This is in contrast to the lower background signal of the aldehyde substrate used by the bacterial luciferase, which is around 50 RLU ml−1 and is not affected by medium composition.
FIG. 2.
M. smegmatis is capable of expressing the Gluc bioluminescence reporter gene, and the luminescence reaction that it catalyzes is a flash reaction. The luminescence is shown for a culture of M. smegmatis pSHKGluc1 (OD600, 0.5) after addition of 10 μmol liter−1 coelenterazine (•). The luminescence of a comparable culture of M. smegmatis without the Gluc gene, demonstrating the background chemiluminescent signal of the coelenterazine substrate when it was incubated with bacterial cells, is also shown (○). The error bars indicate standard deviations.
FIG. 3.
Relationship between luminescence and coelenterazine concentration in M. smegmatis expressing Gluc. The results are corrected for the background. The error bars indicate standard deviations.
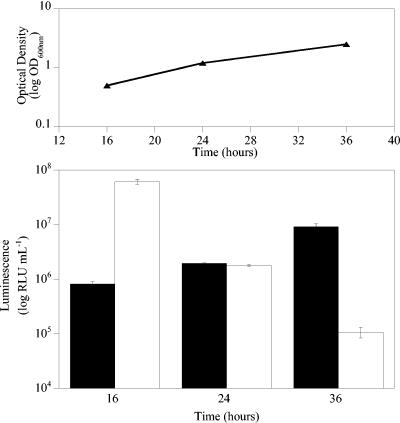
Gluc bioluminescence correlates with bacterial cell number throughout the exponential and stationary phases of growth in vitro.
To determine the effects of the growth phase on bioluminescence, M. smegmatis cultures were grown in LB medium, and samples were taken at various stages during the exponential and stationary phases of growth. While luminescence from M. smegmatis expressing luxAB correlated with cell number during the exponential phase of growth, when cells entered the stationary phase, the luminescence decreased (Fig. 4) (62). Indeed, the RLU/CFU ratios ranged from 0.25 during the mid-exponential phase to 5.81 × 10−5 in the late stationary phase. In contrast, while the luminescence of Gluc-expressing cells was lower than that of luxAB-expressing cells during the exponential phase, it correlated with the cell number throughout both the exponential and stationary phases of growth (Fig. 4). Indeed, the RLU/CFU ratios remained constant at ca. 0.004 throughout the experiment.
FIG. 4.
Gluc bioluminescence correlates with bacterial cell number throughout the exponential and stationary phases of growth. The upper panel shows optical density plotted against time, and the lower panel shows a plot of luminescence versus time. Solid bars, luminescence from M. smegmatis expressing Gluc; open bars, luminescence from M. smegmatis expressing luxAB. The error bars indicate standard deviations.
Gluc bioluminescence is not affected by exposure to low pH.
To determine the effects of pH on bioluminescence and viability, M. smegmatis cultures were incubated for 1 h in LB medium acidified with HCl. With the exception of incubation at pH 1.5, which resulted in a 10-fold decrease in CFU, this treatment was found to have no effect on the viability of M. smegmatis, as assessed by the ability of washed cells to form colonies on Middlebrook 7H11 agar (data not shown) (52). The luminescence output from the luxAB reporter decreased significantly under these conditions, dropping 10-fold after 1 h of incubation at pH 2.75 and 100-fold after incubation at pH 1.5 (Table 3). In contrast, this treatment had no effect on the bioluminescence of Gluc-expressing cells (Table 3).
TABLE 3.
Gluc bioluminescence is not affected by exposure to low pHa
| pH | Luminescence (RLU ml−1) from luxAB-expressing cells | Luminescence (RLU ml−1) from Gluc-expressing cells |
|---|---|---|
| 6.80 | 3.95 × 105 (± 0.77 × 105) | 3.13 × 105 (± 0.38 × 105) |
| 4.75 | 4.22 × 105 (± 0.94 × 105) | 7.39 × 105 (± 0.81 × 105) |
| 3.40 | 4.09 × 105 (± 0.10 × 105) | 5.19 × 105 (± 0.32 × 105) |
| 2.75 | 8.07 × 104 (± 3.14 × 104) | 3.73 × 105 (± 0.20 × 105) |
| 1.50 | 3.48 × 102 (± 0.18 × 102) | 3.90 × 105 (± 0.28 × 105) |
The data show luminescence (standard deviations in parentheses) measured after 1 h of exposure to LB medium acidified with HCl.
Gluc bioluminescence is not affected by exposure to hydrogen peroxide.
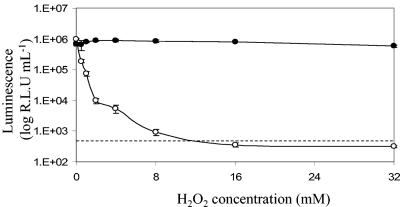
To determine the effects of reactive oxygen species on bioluminescence and viability, M. smegmatis cultures were incubated in LB medium containing various concentrations of hydrogen peroxide. This treatment was found to have no effect on the viability of M. smegmatis (data not shown). In contrast, the luminescence of luxAB-expressing cells was very sensitive to the presence of hydrogen peroxide; there was an almost 10-fold decrease in light output after 90 min of incubation with 0.5 mM hydrogen peroxide, and the number of RLU fell below the limit of detection when the concentration of hydrogen peroxide was greater than 8 mM (Fig. 5). Again, the sustained bioluminescence output of Gluc-expressing cells paralleled the viability as assessed by CFU (Fig. 5).
FIG. 5.
Gluc bioluminescence is not affected by exposure to hydrogen peroxide. Symbols: •, luminescence from M. smegmatis expressing Gluc; ○, luminescence from M. smegmatis expressing luxAB. The limit of detection of the luxAB reaction is indicated by the dashed line. The error bars indicate standard deviations.
Gluc bioluminescence is not affected by heat shock.
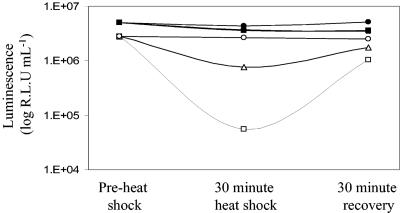
M. smegmatis cultures were subjected to 30 min of heat shock at 45°C and 48°C. Samples were analyzed before the heat shock, immediately after the heat shock, and after 30 min of recovery at 37°C. This treatment was found to have no effect on the viability of M. smegmatis (data not shown). This was reflected by the sustained luminescence of Gluc-expressing cells, in contrast to the 5- and 10-fold reductions in luxAB luminescence after incubation at 45°C and 48°C, respectively (Fig. 6) (24, 43). The luminescence of luxAB-expressing cells was rapidly restored after a 30-min recovery period at 37°C (Fig. 6).
FIG. 6.
Gluc bioluminescence is not affected by heat shock. Solid symbols, luminescence from M. smegmatis expressing Gluc; open symbols, luminescence from M. smegmatis expressing luxAB. M. smegmatis cultures were subjected to 30 min of heat shock at 45°C (triangles) and 48°C (squares). Control samples were incubated at 37°C (circles). The standard deviations are smaller than the symbols.
DISCUSSION
We demonstrated that the copepod Gluc luciferase can be expressed in mycobacteria. It catalyzes a flash reaction, but, while luminescence decays rapidly after addition of the coelenterazine substrate, it is still detectable above the background level after 60 min. This compares favorably with both the luxAB (bacterial) and luc (firefly) bioluminescence systems (Wiles, unpublished data). A drawback to the Gluc system is that the chemiluminescent background of the coelenterazine substrate is different in different diluent buffers. While a low background signal was observed with 10 mM coelenterazine in LB medium (102 RLU ml−1), the signal was 100-fold greater in the Middlebrook 7H9 medium commonly used for culture of the more fastidious organism M. tuberculosis. Alternative media (Sauton's medium, for example [71]) are required when the Gluc system is used with M. tuberculosis.
Experiments with the luxAB reporter system in mycobacteria have shown that there is a strong dependence on changes in the availability of bacterial cofactors under different growth conditions (62, 67). While this is advantageous in signaling a rapid response to the action of some drugs, a sharp decline in luminescence presents a limitation in studying nondividing bacteria in stationary-phase cultures. This is certainly not unique to mycobacteria; in many bacterial species harboring the lux operon, bioluminescence declines when cells enter the stationary phase during in vitro growth (28, 70), and this is most likely due to a decrease in metabolic activity. In contrast, the bioluminescence of Gluc-expressing cells appears to be independent of cofactors that become limited during the stationary phase. Thus, the Gluc reporter provides a correlate of bioluminescence with bacterial number irrespective of the growth phase.
The Gluc system also offers advantages in experiments involving exposure to environmental stress. The ability to survive exposure to low pH and oxidative stress plays an important role in the intracellular survival and pathogenesis of M. tuberculosis, and the heat shock response has been used extensively as a model for studying mycobacterial gene regulation (63). However, in our hands multicopy genes under the control of the hsp60 promoter do not respond to heat shock, presumably because the link with the heat shock regulatory circuit has been broken. Indeed, expression of the luxAB genes under the hsp60 promoter results in the strongest luminescent signal when this expression is compared to the expression with other mycobacterial promoters (Wiles, unpublished results). Exposure of mycobacteria to stress conditions results in rapid dissociation between viability and the output of luminescence from the luxAB reporter. This dissociation is not observed with the Gluc reporter. Given that the two genes are cloned into and expressed in the same vector backbone under control of the same promoter, it is unlikely that this dissociation is due to increased gene expression or enzyme turnover. This difference may again reflect the relative independence of the Gluc reaction from bacterial metabolism or perhaps greater physiological stability of the Gluc luciferase enzyme. The Gluc system may provide a particularly appropriate reporter for studying responses associated with survival under adverse conditions, both during pathogenesis and in the environment.
Conclusions.
In this paper we report the expression of a novel luciferase gene (the Gluc gene) from the marine copepod G. princeps in M. smegmatis that results in detectable luminescence that correlates with viable counts throughout the exponential and stationary phases of bacterial growth under normal conditions in vitro. The work presented here indicated that this luciferase should also be suitable under conditions in which the use of the more traditional bioluminescence genes is limited (for example, in studying bacterial responses to such stresses as oxidative damage and pH). We are currently constructing dual luxAB-Gluc systems with the aim of exploiting the relative advantages and disadvantages of the different luciferase reporters.
Acknowledgments
This work was supported by the NIH TB Research Unit.
We thank Bruce Bryan (Nanolight Technology, Prolume Ltd., Pinetop, AZ) for the kind gift of plasmid pUC19Gluc and the colenterazine used in this study.
REFERENCES
- 1.Arain, T. M., A. E. Resconi, D. C. Singh, and C. K. Stover. 1996. Reporter gene technology to assess activity of antimycobacterial agents in macrophages. Antimicrob. Agents Chemother. 40:1542-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin, T. O., T. Berends, T. A. Bunch, T. F. Holzman, S. K. Rausch, L. Shamansky, M. L. Treat, and M. M. Ziegler. 1984. Cloning of the luciferase structural genes from Vibrio harveyi and expression of bioluminescence in Escherichia coli. Biochemistry 23:3663-3667. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin, T. O., J. H. Devine, R. C. Heckel, J. W. Lin, and G. S. Shadel. 1989. The complete nucleotide sequence of the lux regulon of Vibrio fischeri and the luxABN region of Photobacterium leiognathi and the mechanism of control of bacterial bioluminescence. J. Biolumin. Chemilumin. 4:326-341. [DOI] [PubMed] [Google Scholar]
- 4.Banaiee, N., M. Bobadilla-del-Valle, P. F. Riska, S. Bardarov, P. M. Small, A. Ponce-de-Leon, W. R. Jacobs, G. F. Hatfull, and J. Sifuentes-Osornio. 2003. Rapid identification and susceptibility testing of Mycobacterium tuberculosis from MGIT cultures with luciferase reporter mycobacteriophages. J. Med. Microbiol. 52:557-561. [DOI] [PubMed] [Google Scholar]
- 5.Bardarov, S., H. Dou, K. Eisenach, N. Banaiee, S. U. Ya, J. Chan, W. R. Jacobs, and P. F. Riska. 2003. Detection and drug-susceptibility testing of M. tuberculosis from sputum samples using luciferase reporter phage: comparison with the mycobacteria growth indicator tube (MGIT) system. Diagn. Microbiol. Infect. Dis. 45:53-61. [DOI] [PubMed] [Google Scholar]
- 6.Barnes, A. T., and J. F. Case. 1972. Bioluminescence in the mesopelagic copepod, Gaussia princeps (T. Scott). J. Exp. Mar. Biol. Ecol. 8:53-71. [Google Scholar]
- 7.Bowlby, M. R., and J. F. Case. 1991. Flash kinetics and spatial patterns of bioluminescence in the copepod Gaussia princeps. Mar. Biol. 110:329-336. [Google Scholar]
- 8.Bowlby, M. R., and J. F. Case. 1991. Ultrastructure and neuronal control of luminous cells in the copepod Gaussia princeps. Biol. Bull. 180:440-446. [DOI] [PubMed] [Google Scholar]
- 9.Bryan, B. J., and C. S. Szent-Gyorgyi. May 2001. U.S. patent 6232107.
- 10.Campbell, A. K. 1989. Living light: biochemistry, function and biomedical applications. Essays Biochem. 24:41-81. [PubMed] [Google Scholar]
- 11.Burlage, R. S., G. S. Sayler, and F. Larimer. 1990. Monitoring of naphthalene catabolism by bioluminescence with nah-lux transcriptional fusions. J. Bacteriol. 172:4749-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatterjee, J., and E. A. Meighen. 1995. Biotechnological applications of bacterial bioluminescence (lux) genes. Photochem. Photobiol. 62:641-650. [Google Scholar]
- 13.Chen, J., and M. W. Griffiths. 1996. Salmonella detection in eggs using Lux+ bacteriophages. J. Food Prot. 59:908-914. [DOI] [PubMed] [Google Scholar]
- 14.Colepicolo, P., K. Cho, G. O. Poinar, and J. W. Hastings. 1989. Growth and luminescence of the bacterium Xenorhabdus luminescens from a human wound. Appl. Environ. Microbiol. 55:2601-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Contag, C. H., P. R. Contag, J. I. Mullins, S. D. Spilman, D. K. Stevenson, and D. A. Benaron. 1995. Photonic detection of bacterial pathogens in living hosts. Mol. Microbiol. 18:593-603. [DOI] [PubMed] [Google Scholar]
- 16.Cooksey, R. C., J. T. Crawford, W. R. Jacobs, and T. M. Shinnick. 1993. A rapid method for screening antimicrobial agents for activities against a strain of Mycobacterium tuberculosis expressing firefly luciferase. Antimicrob. Agents Chemother. 37:1348-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooksey, R. C., G. P. Morlock, M. Beggs, and J. T. Crawford. 1995. Bioluminescence method to evaluate antimicrobial agents against Mycobacterium avium. Antimicrob. Agents Chemother. 39:754-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Wet, J. R., K. V. Wood, D. R. Helinski, and M. DeLuca. 1985. Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proc. Natl. Acad. Sci. USA 82:7870-7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorn, J. G., R. J. Frye, and R. M. Maier. 2003. Effect of temperature, pH, and initial cell number on luxCDABE and nah gene expression during naphthalene and salicylate catabolism in the bioreporter organism Pseudomonas putida RB1353. Appl. Environ. Microbiol. 69:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'Souza, S., V. Rosseels, O. Denis, A. Tanghe, N. De Smet, F. Jurion, K. Palfliet, N. Castiglioni, A. Vanonckelen, C. Wheeler, and K. Huygen. 2002. Improved tuberculosis DNA vaccines by formulation in cationic lipids. Infect. Immun. 70:3681-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dye, C., S. Scheele, P. Dolin, V. Pathania, and R. C. Raviglione. 1999. Global burden of tuberculosis—estimated incidence, prevalence, and mortality by country. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 22.Engebrecht, J., K. Nealson, and M. Silverman. 1983. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32:773-781. [DOI] [PubMed] [Google Scholar]
- 23.Engebrecht, J., and M. Silverman. 1984. Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. USA 81:4154-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Escher, A., D. J. O'Kane, J. Lee, and A. A. Szalay. 1989. Bacterial luciferase αβ fusion protein is fully active as a monomer and highly sensitive in vivo to elevated temperature. Proc. Natl. Acad. Sci. USA 86:6528-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foran, D. R., and W. M. Brown. 1988. Nucleotide sequence of the luxA and luxB genes of the bioluminescent marine bacterium Vibrio fischeri. Nucleic Acids Res. 16:777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frackman, S., M. Anhalt, and K. H. Nealson. 1990. Cloning, organization, and expression of the bioluminescence genes of Xenorhabdus luminescens. J. Bacteriol. 172:5767-5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francis, K. P., D. Joh, C. Bellinger-Kawahara, M. J. Hawkinson, T. F. Purchio, and P. R. Contag. 2000. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect. Immun. 68:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Francis, K. P., J. Yu, C. Bellinger-Kawahara, D. Joh, M. J. Hawkinson, G. Xiao, T. F. Purchio, M. G. Caparon, M. Lipsitch, and P. R. Contag. 2001. Visualizing pneumococcal infections in the lungs of live mice using bioluminescent Streptococcus pneumoniae transformed with a novel gram-positive lux transposon. Infect. Immun. 69:3350-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu, M. B., and S. H. Choi. 2001. Monitoring and classification of toxicity using recombinant bioluminescent bacteria. Water Sci. Technol. 43:147-154. [PubMed] [Google Scholar]
- 30.Gu, M. B., and G. C. Gil. 2001. A multi-channel continuous toxicity monitoring system using recombinant bioluminescent bacteria for classification of toxicity. Biosens. Bioelectron. 16:661-666. [PubMed] [Google Scholar]
- 31.Hastings, J. W. 1986. Bioluminescence in bacteria and dinoflagellates, p. 363-398. In A. J. Govindjee, J. Amesz, and D. C. Fork (ed.), Light emission by plants and bacteria. Academic Press, New York, N.Y.
- 32.Hastings, J. W., T. O. Baldwin, and M. Z. Nicoli. 1978. Bacterial luciferase: assay purification and properties. Methods Enzymol. 57:135-152. [Google Scholar]
- 33.Heitzer, A., B. Applegate, S. Kehrmeyer, H. Pinkart, O. F. Webb, T. J. Phelps, D. C. White, and G. S. Sayler. 1998. Physiological considerations of environmental applications of lux reporter fusions. J. Microbiol. Methods 33:45-57. [Google Scholar]
- 34.Hickey, M. J., T. M. Arain, R. M. Shawar, D. J. Humble, M. H. Langhorne, J. N. Morgenroth, and C. K. Stover. 1996. Luciferase in vivo expression technology: use of recombinant mycobacterial reporter strains to evaluate antimycobacterial activity in mice. Antimicrob. Agents Chemother. 40:400-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kampmann, B., P. O. Gaora, V. A. Snewin, M. P. Gares, D. B. Young, and M. Levin. 2000. Evaluation of human antimycobacterial immunity using recombinant reporter mycobacteria. J. Infect. Dis. 182:895-901. [DOI] [PubMed] [Google Scholar]
- 36.Katayama, M., N. F. Tsinoremas, T. Kondo, and S. S. Golden. 1999. cpmA, a gene involved in an output pathway of the cyanobacterial circadian system. J. Bacteriol. 181:3516-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenney, T. J., and G. Churchward. 1996. Genetic analysis of the Mycobacterium smegmatis rpsL promoter. J. Bacteriol. 178:3564-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Latz, M. I., M. R. Bowlby, and J. F. Case. 1990. Recovery and stimulation of copepod bioluminescence. J. Exp. Mar. Biol. Ecol. 136:1-22. [Google Scholar]
- 39.Lee, J. 1989. Bioluminescence, p. 391-417. In K. C. Smith (ed.), The science of photobiology. Plenum Publishing Corp., New York, N.Y.
- 40.Lloyd, J. E. 1971. Bioluminescent communication in insects. Annu. Rev. Entomol. 16:97-122. [Google Scholar]
- 41.Lorenz, W. W., M. J. Cormier, D. J. O'Kane, D. Hua, A. A. Escher, and A. A. Szalay. 1996. Expression of the Renilla reniformans luciferase gene in mammalian cells. J. Biolumin. Chemilumin. 11:31-37. [DOI] [PubMed] [Google Scholar]
- 42.Lorenz, W. W., R. O. McCann, M. Longiaru, and M. J. Cormier. 1991. Isolation and expression of a cDNA encoding Renilla reniformans luciferase. Proc. Natl. Acad. Sci. USA 88:4438-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mackey, B. M., D. Cross, and S. F. Park. 1994. Thermostability of bacterial luciferase expressed in different microbes. J. Appl. Bacteriol. 77:149-154. [DOI] [PubMed] [Google Scholar]
- 44.Markova, S. V., S. Golz, L. A. Frank, B. Kalthof, and E. S. Vysotski. 2004. Cloning and expression of cDNA for a luciferase from the marine copepod Metridia longa: a novel secreted bioluminescent reporter enzyme. J. Biol. Chem. 279:3212-3217. [DOI] [PubMed] [Google Scholar]
- 45.Matthews, J. C., K. Hori, and M. J. Cormier. 1977. Purification and properties of Renilla reniformans luciferase. Biochemistry 16:85-95. [DOI] [PubMed] [Google Scholar]
- 46.Mayerhofer, R., W. H. R. Langridge, M. J. Cormier, and A. A. Szalay. 1995. Expression of recombinant Renilla luciferase in transgenic plants results in high levels of light emission. Plant J. 7:1031-1038. [Google Scholar]
- 47.McElroy, W. D. 1951. Properties of the reaction utilising adenosine triphosphate for bioluminescence. J. Biol. Chem. 191:547-557. [PubMed] [Google Scholar]
- 48.Mihalcescu, M., W. Hsing, and S. Leibler. 2004. Resilient circadian oscillator revealed in individual cyanobacteria. Nature 430:81-85. [DOI] [PubMed] [Google Scholar]
- 49.Moaz, A., R. Mayr, G. Bresolin, K. Neuhaus, K. P. Francis, and S. Schere. 2002. Sensitive in situ monitoring of a recombinant bioluminescent Yersinia enterocolitica reporter mutant in real time on Camembert cheese. Appl. Environ. Microbiol. 68:5737-5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nealson, K. H., and J. W. Hastings. 1992. The luminous bacteria, p. 625-639. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification and applications, vol. 1. Springer-Verlag, New York, N.Y. [Google Scholar]
- 51.Neilson, J. W., S. A. Pierce, and R. M. Maier. 1999. Factors influencing expression of luxCDABE and nah genes in Pseudomonas putida RB1353 (NAH7, pUTK9) in dynamic systems. Appl. Environ. Microbiol. 65:3473-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Brien, L. M., S. V. Gordon, I. S. Roberts, and P. W. Andrew. 1996. Response of Mycobacterium smegmatis to acid stress. FEMS Microbiol. Lett. 139:11-17. [DOI] [PubMed] [Google Scholar]
- 53.O'Connell-Rodwell, C. E., S. M. Burns, M. H. Bachmann, and C. H. Contag. 2002. Bioluminescent indicators for in vivo measurements of gene expression. Trends Biotechnol. 20:S19-S23.12570155 [Google Scholar]
- 54.Park, S. F., G. S. A. B. Stewart, and R. G. Kroll. 1992. The use of bacterial luciferase for monitoring the environmental regulation of expression of genes encoding virulence factors in Listeria monocytogenes. J. Gen. Microbiol. 138:2619-2627. [DOI] [PubMed] [Google Scholar]
- 55.Qazi, S. N., E. Counil, J. Morrissey, C. E. Rees, A. Cockayne, K. Winzer, W. C. Chan, P. Williams, and P. J. Hill. 2001. agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect. Immun. 69:7074-7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roda, A., P. Pasini, M. Mirasoli, E. Michelini, and M. Guardigli. 2004. Biotechnological applications of bioluminescence and chemiluminescence. Trends Biotechnol. 22:295-303. [DOI] [PubMed] [Google Scholar]
- 57.Shaw, J. J., F. Dane, D. Gieger, and J. W. Kloepper. 1992. Use of bioluminescence for detection of genetically engineered microorganisms released into the environment. Appl. Environ. Microbiol. 58:267-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaw, J. J., and C. I. Kado. 1986. Development of a Vibrio bioluminescence gene-set to monitor phytopathogenic bacteria during the ongoing disease process in a non-disruptive manner. Bio/Technology 4:560-564. [Google Scholar]
- 59.Shimomura, O., T. Masugi, F. H. Johnson, and Y. Haneda. 1978. Properties and reaction mechanism of the bioluminescence system of the deep-sea shrimp Oplophorus gracilorostris. Biochemistry 17:994-998. [DOI] [PubMed] [Google Scholar]
- 60.Shimomura, O., and K. Teranishi. 2000. Light-emitters involved in the luminescence of coelenterazine. Luminescence 15:51-58. [DOI] [PubMed] [Google Scholar]
- 61.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 62.Snewin, V. A., M.-P. Gares, P. O'Gaora, Z. Hasan, I. N. Brown, and D. B. Young. 1999. Assessment of immunity to mycobacterial infection with luciferase reporter constructs. Infect. Immun. 67:4586-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stewart, G. R., B. D. Robertson, and D. B. Young. 2003. Tuberculosis: a problem with persistence. Nat. Rev. Microbiol. 1:97-105. [DOI] [PubMed] [Google Scholar]
- 64.Szittner, R., and E. A. Meighen. 1990. Nucleotide sequence, expression and properties of luciferase coded by lux genes from a terrestrial bacterium. J. Biol. Chem. 265:16581-16587. [PubMed] [Google Scholar]
- 65.Tannous, B. A., D.-E. Kim, J. L. Fernandez, R. Weissleder, and X. O. Breakefield. 2005. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol. Ther. 11:435-443. [DOI] [PubMed]
- 66.Thomson, C. M., P. J. Herring, and A. K. Campbell. 1997. The widespread occurrence and tissue distribution of the imidazolopyrazine luciferins. J. Biolumin. Chemilumin. 12:87-91. [DOI] [PubMed] [Google Scholar]
- 67.Turner, D. J., S. L. Hoyle, V. A. Snewin, M.-P. Gares, I. N. Brown, and D. B. Young. 2002. An ex vivo culture model for screening drug activity against in vivo phenotypes of Mycobacterium tuberculosis. Microbiology 148:2929-2936. [DOI] [PubMed] [Google Scholar]
- 68.Verhaegen, M., and T. K. Christopoulos. 2002. Recombinant Gaussia luciferase. Overexpression, purification, and analytical application of a bioluminescent reporter for DNA hybridization. Anal. Chem. 74:4378-4385. [DOI] [PubMed] [Google Scholar]
- 69.Vollmer, A. C., S. Belkin, D. R. Smulski, T. K. Van Dyk, and R. A. LaRossa. 1997. Detection of DNA damage by use of Escherichia coli carrying recA′::lux, uvrA′::lux, or alkA′::lux reporter plasmids. Appl. Environ. Microbiol. 63:2566-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiles, S., S. Clare, J. Harker, A. Huett, D. B. Young, G. Dougan, and G. Frankel. 2005. Organ specificity, colonization and clearance dynamics in vivo following oral challenges with the murine pathogen Citrobacter rodentium. Cell. Microbiol. 6:963-972. [DOI] [PubMed] [Google Scholar]
- 71.Wiles, S., K. Ferguson, B. D. Robertson, and D. B. Young. 2005. Optimisation of conditions for the use of a novel bioluminescent reporter system in Mycobacterium spp., p. 543-545. In A. Tsuji et al. (ed.), Bioluminescence and chemiluminescence: progress and perspectives. World Scientific, Singapore, Republic of Singapore.
- 72.Wood, K. V., Y. A. Lam, H. H. Seliger, and W. D. McElroy. 1989. Complementary DNAs encoding click beetle luciferases can elicit bioluminescence of different colors. Science 244:700-702. [DOI] [PubMed] [Google Scholar]
- 73.Ziegler, M. M., and T. O. Baldwin. 1981. Biochemistry of bacterial bioluminescence. Curr. Top. Bioenerg. 12:65-113. [Google Scholar]