Abstract
Campylobacter fetus is a gram-negative bacterial pathogen of both humans and animals. Two subspecies have been identified, Campylobacter fetus subsp. fetus and Campylobacter fetus subsp. venerealis, and there are two serotypes, A and B. To further investigate the genetic diversity among C. fetus strains of different origins, subspecies, and serotypes, we performed multiple genetic analyses by utilizing random amplification of polymorphic DNA (RAPD), pulsed-field gel electrophoresis (PFGE), and DNA-DNA hybridization. All 10 primers used for the RAPD analyses can distinguish C. fetus strains of reptile and mammal origin, five can differentiate between C. fetus subsp. fetus and C. fetus subsp. venerealis strains, and four showed differences between type A and type B isolates from mammals. PFGE with SmaI and SalI digestion showed varied genome patterns among different C. fetus strains, but for mammalian C. fetus isolates, genome size was well conserved (mean, 1.52 ± 0.06 Mb for SmaI and 1.52 ± 0.05 Mb for SalI). DNA-DNA hybridization demonstrated substantial genomic-homology differences between strains of mammal and reptile origin. In total, these data suggest that C. fetus subsp fetus strains of reptile and mammal origin have genetic divergence more extensive than that between the two subspecies and that between the type A and type B strains. Combining these studies with sequence data, we conclude that there has been substantial genetic divergence between Campylobacter fetus of reptile and mammal origin. Diagnostic tools have been developed to differentiate among C. fetus isolates for taxonomic and epidemiologic uses.
Campylobacter species are gram-negative, slender, spiral, curved rods. Sixteen valid Campylobacter species have been described (9, 18, 20, 28, 34), of which Campylobacter fetus is the type species (26). C. fetus includes two subspecies, fetus and venerealis (2, 28). Campylobacter fetus subsp. fetus causes abortion in sheep and sporadic abortion in cattle, and Campylobacter fetus subsp. venerealis causes abortion and infertility in cattle (11, 13, 24, 27). Either of the subspecies can be a human pathogen, causing enteritis, abortion, bacteremia, septicemia, endocarditis, or meningitis (3, 4, 27, 30, 37).
Based on the lipopolysaccharide structure and surface layer protein (SLP) composition, C. fetus strains may be designated either type A or type B (7, 19, 23); C. fetus subsp. venerealis strains are always type A, whereas C. fetus subsp. fetus strains can be either type A or type B, or rarely type AB (19, 23, 29). Sequence analyses of the housekeeping gene recA and the C. fetus-specific gene sapD suggested that C. fetus type A and type B strains diverged before C. fetus subspecies fetus and C. fetus subsp. venerealis diverged from one another (29). In contrast, C. fetus strains of reptile origin showed very high divergence from C. fetus strains of mammalian origin. The 16S rRNA sequence analysis also suggested that reptile C. fetus isolates form a distinct phylogenotype between C. fetus and Campylobacter hyointestinalis (29).
Analyses using specific gene sequences can provide only limited insights into differences between C. fetus strains at the genomic level, and the entire C. fetus genomic sequence has not been determined. In contrast, random amplification of polymorphic DNA (RAPD), pulsed-field gel electrophoresis (PFGE), and whole-genome DNA-DNA hybridization can be employed to aid in understanding genetic divergence among C. fetus strains at a more global level.
In the present study, we used such methods to further address questions regarding genetic divergence and evolutionary relationships for C. fetus strains of different subspecies, serotypes, and host origins.
MATERIALS AND METHODS
Bacterial strains and growth media.
A total of 19 C. fetus strains were analyzed in this study, including 13 C. fetus subsp. fetus, 2 C. fetus subsp. venerealis, and 4 C. fetus strains of reptile origin. Of these 19 strains, 12 are type A, 6 are type B, and 1 is type A/B (Table 1). Details of the characterization of these strains have been reported elsewhere (29, 31, 32, 33). The strains were routinely grown on brucella broth (BBL Microbiology Systems, Cockeysville, Md.) or on Trypticase soy agar plates at 37°C in a 5% CO2 incubator.
TABLE 1.
Characteristics and genomic sizes of the C. fetus strain studied
| Strain no. | Strain designation | Origin | Type | Subspecies | SmaI
|
SalI
|
||
|---|---|---|---|---|---|---|---|---|
| Genomic size (Mb) | Type (no. of bands) | Genomic size (Mb) | Type (no. of bands) | |||||
| 1 | 80-109 | Mammal | A | fetus | 1.55 | A (15) | 1.55 | A (15) |
| 2 | 82-40 | Mammal | A | fetus | 1.42 | B (14) | 1.47 | B (14) |
| 3 | 83-94 | Mammal | A | fetus | 1.42 | C (14) | 1.47 | B (14) |
| 4 | 84-32 (23D) | Mammal | A | fetus | 1.60 | D (15) | 1.47 | B (14) |
| 5 | 84-86 | Mammal | A | fetus | 1.54 | E (15) | 1.47 | B (14) |
| 6 | 84-92 | Mammal | A | fetus | 1.54 | E (15) | 1.47 | B (14) |
| 7 | 99-256 | Mammal | A | fetusa | 1.56 | A (15) | 1.54 | C (15) |
| 8 | 84-87 | Mammal | B | fetus | 1.55 | F (16) | 1.49 | D (16) |
| 9 | 84-90 | Mammal | B | fetus | 1.42 | B (14) | 1.55 | A (15) |
| 10 | 84-91 | Mammal | B | fetus | 1.55 | A (15) | 1.55 | A (15) |
| 11 | 84-94 | Mammal | B | fetus | 1.55 | A (15) | 1.60 | E (16) |
| 12 | 84-104 | Mammal | B | fetus | 1.54 | G (15) | 1.59 | F (15) |
| 13 | 84-107 | Mammal | B | fetus | 1.55 | A (15) | 1.55 | A (15) |
| 14 | 84-112 | Mammal | A | venerealis | 1.58 | H (17) | 1.85 | G (17) |
| 15 | 99-257 | Mammal | A | venerealis | 1.64 | I (18) | 1.50 | H (14) |
| 16 | 85-388 | Reptile | A | fetus | 1.80 | J (10) | NAb | NA |
| 17 | 85-389 | Reptile | A | fetus | 1.44 | K (8) | NA | NA |
| 18 | 03-427 | Reptile | A | fetus | 1.72 | L (9) | NA | NA |
| 19 | 85-388 | Reptile | A/B | fetus | 1.83 | M (10) | NA | NA |
Strain 99-256 is from the American Type Culture Collection (ATCC 33561) and has been considered a C. fetus subsp. venerealis strain. However, the present study provides evidence that it is C. fetus subsp. fetus rather than C. fetus subsp. venerealis, based on RAPD and PFGE.
NA, not applicable, due to poor digestion.
RAPD-PCR.
RAPD-PCRs were performed using 10 different primers (Table 2). The PCR conditions for primers 1254, 1281, 1283, 1290, 1247, D8635, and D14307 (1), primers ERIC-1 and ERIC-2 (8), and primer OPA-11 (16) were all as originally described.
TABLE 2.
RAPD identification of C. fetus strains
| Primer | Sequence (5′ → 3′) | Reference | Distinguishingd
|
||
|---|---|---|---|---|---|
| Cf vs-Cva | M vs Rb | A vs Bc | |||
| 1281 | AACGCGCAAC | 1 | N | D | N |
| 1283 | GCGATCCCCA | 1 | N | D | N |
| 1290 | GTGGATGCGA | 1 | N | D | N |
| D8635 | GAGCGGCCAAAGGGAGCAGAC | 1 | D | D | N |
| 1247 | AAGAGCCCGT | 1 | D | D | D |
| D14307 | GGTTGGGTGAGAATTGCACG | 1 | D | D | D |
| 1254 | CCGCAGCCAA | 1 | N | D | N |
| OPA-11 | CAATCGCCGT | 16 | D | D | N |
| ERIC-1 | ATGTAAGCTCCTGGGGATTCA | 8 | N | D | D |
| ERIC-2 | AAGTAAGTGACTGGGGTGAGCG | 8 | D | D | D |
Cf, C. fetus subsp. fetus (n = 13 strains); Cv, C. fetus subsp. venerealis (n = 2 strains).
M, mammalian C. fetus strain (n = 15); R, reptile C. fetus strain (n = 4).
A, type A C. fetus strain (n = 12); B, type B C. fetus strain (n = 6).
N, not distinguishable; D, distinguishable.
PFGE profiling.
The preparation of bacterial DNA-agarose samples and subsequent enzyme digestion for PFGE was as described previously (5). DNA was digested with SmaI or SalI (New England Biolabs [NEB], Beverly, MA), and fragments were electrophoretically separated in 1% agarose gels at 14°C with the following variations according to the particular range of markers used: for low-range PFG Marker (NEB), 6 V/cm and switch times from 1 to 12 s for 15 h; for MidRange I PFG Marker (NEB), switch times ramped from 1 to 25 s for 24 h; and for Lambda Ladder PFG Marker (NEB), 4.5 V/cm and switch times ramped from 5 to 120 s for 48 h.
DNA-DNA hybridization. Dot blot DNA-DNA hybridizations were performed with 5 ng of whole bacterial genomic DNA. The probes consisted of fluoresceinated genomic DNA prepared from C. fetus subsp. fetus mammalian strain 23D or C. fetus subsp. fetus strain 85-388 of reptile origin. Each sample of chromosomal DNA was heat denatured at 100°C for 5 min and applied to a positively charged nylon membrane, and the DNA was immobilized by UV irradiation. Labeling of the probes and the hybridization reactions were performed based on the protocol supplied with the Random Primer Fluorescein Labeling Kit with antifluorescein-AP (Perkin-Elmer Life Sciences, Inc., Boston, MA).
16S rRNA sequence analyses.
Two C. fetus subsp. venerealis strains, 99-257 and 84-112, were analyzed using the universal 16S rRNA gene primers 8F and 1510R (22) and were compared with previously published sequences of other C. fetus strains involved in this study (29, 33). Sequence analyses were performed with the Genetics Computer Group programs (Madison, WI).
Nucleotide sequence accession number.
The nucleotide sequences of 16S rRNA for strains 84-112 and 99-257 have been deposited in GenBank under accession number AY864915.
RESULTS
RAPD.
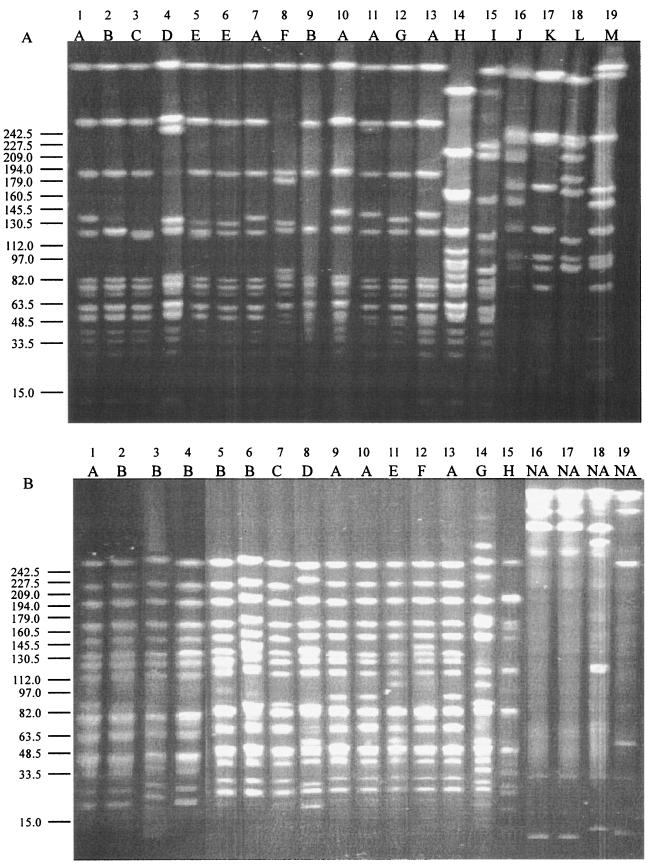
To better detect DNA diversity at the level of the whole genome among different C. fetus strains, we screened the 19 isolates with 10 different primers in RAPD analyses (Table 2 and Fig. 1 and data not shown). Use of each of the 10 primers resulted in distinguishing the mammalian and reptilian C. fetus strains (Fig. 1 and data not shown), use of five of the primers permitted distinguishing between C. fetus subsp. fetus and C. fetus subsp. venerealis, and four of the primers differentiated type A and type B strains. The use of primer ERIC-2, 1247, or D14307 each permitted distinguishing C. fetus subsp. fetus and C. fetus subsp. venerealis, type A and type B, and mammalian C. fetus and reptilian C. fetus strains (Fig. 1 and Table 2).
FIG. 1.
RAPD profiles of 19 C. fetus strains generated using primers 1254 (A) and ERIC-1 (B). The lane numbers representing the strains correspond to the strain numbers in Table 1.
PFGE.
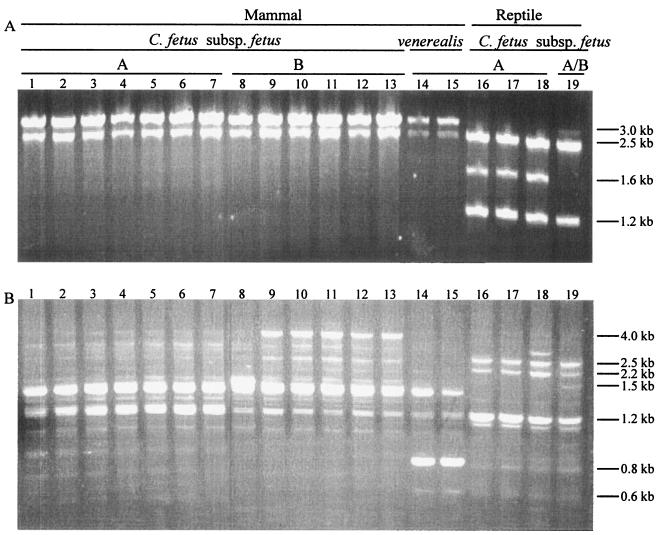
The SmaI and SalI PFGE patterns of the 19 C. fetus strains are shown in Table 1 and Fig. 2. SmaI digestion produced 8 to 18 fragments with sizes varying from 3 kb to 436 kb (Fig. 2A and data not shown). Among the 13 mammalian C. fetus subsp. fetus strains, there exist seven SmaI digestion patterns designated A to G. The two C. fetus subsp. venerealis (H and I) and four reptile origin C. fetus (J to M) strains each showed a different SmaI pattern. Based on the restriction patterns, we calculated the genome size for each strain. Using SmaI digestion, the 13 mammalian C. fetus subsp. fetus genomes varied from 1.42 to 1.60 Mb (mean, 1.52 ± 0.06), the 2 C. fetus subsp. venerealis genomes were 1.58 and 1.64 Mb (mean, 1.61 ± 0.05), and the 4 reptile strains were 1.80, 1.44, 1.72, and 1.83 Mb (mean, 1.70 ± 0.18). For the 13 mammalian C. fetus subsp. fetus strains, results of the SalI digestion confirmed the genomic size (1.52 ± 0.05). SalI digestion of the whole genome demonstrated six patterns for the 13 mammalian C. fetus subsp. fetus strains and two patterns for the 2 C. fetus subsp. venerealis strains. However, each of the four reptile origin C. fetus subsp. fetus strains showed a SalI digestion profile totally different from that for the mammalian C. fetus subsp. fetus strains; these appear to be due to incomplete digestion. The genome sizes calculated from both SmaI and SalI digestion showed intrastrain variation, in part because some of the digested fragments showed very similar migration characteristics and were not readily distinguishable.
FIG. 2.
PFGE profiles of 19 C. fetus strains digested with SmaI (A) and SalI (B). MidRange I PFG Marker was used. The lane numbers correspond to the strain numbers shown in Table 1.
DNA-DNA hybridization.
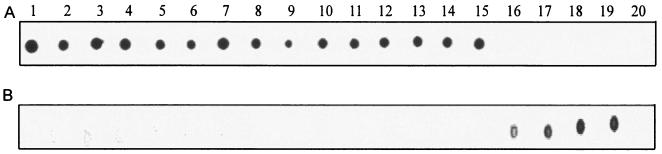
The C. fetus subsp. fetus 23D genome probe showed a high degree of hybridization with all 13 mammal C. fetus subsp. fetus strains and with the 2 C. fetus subsp. venerealis strains but much less with the 4 reptile C. fetus strains. In contrast, the probe based on reptile strain 85-388 showed strong hybridization to all four reptile strains but none to any of the mammalian C. fetus subsp. fetus and C. fetus subsp. venerealis strains. As expected, neither of the probes showed any significant hybridization signal with the control Campylobacter jejuni strain 11168 (Fig. 3). The hybridization experiments were each performed twice, yielding similar results.
FIG. 3.
Dot blot DNA-DNA hybridization with fluoresceinated genomic DNA probes prepared from DNAs of bovine (mammalian) strain 23D (A) and turtle (reptile) strain 85-388 (B). The lane numbers correspond to the strain numbers in Table 1. Lane 20 represents C. jejuni strain 111168 as a negative control. The homologous hybridizations are in lanes 4 and 19 for 23D (panel A) and 85-388 (panel B), respectively.
16S rRNA analyses.
Our previous study compared 16S rRNA sequences among strains, including C. fetus subsp. fetus type A (84-32) and type B (84-107), C. fetus subsp. venerealis (99-256), and four reptile strains (85-387, 85-388, 85-389, and 03-427) (29, 33). However, strain 99-256 (ATCC 33561), used as a representative C. fetus subsp. venerealis strain in the previous study (29), was found to be identical in 16S rRNA sequence to C. fetus subsp. fetus. Based on the 16S rRNA sequence and on the present RAPD and PFGE studies, we believe that strain 99-256 should be considered C. fetus subsp. fetus. To better understand the genetic relationship between C. fetus subsp. fetus and C. fetus subsp. venerealis, we sought to determine the 16S rRNA sequences of two other C. fetus subsp. venerealis strains. Within the 1,484-bp 16S rRNA determined, the two C. fetus subsp. venerealis strains (99-257 and 84-112) showed 100% sequence identity to the type A and type B C. fetus subsp. fetus strains from mammals (29). Our results showed that C. fetus subsp. fetus and C. fetus subsp. venerealis had identical 16S rRNA sequences, which is consistent with reports using other determinants (21).
DISCUSSION
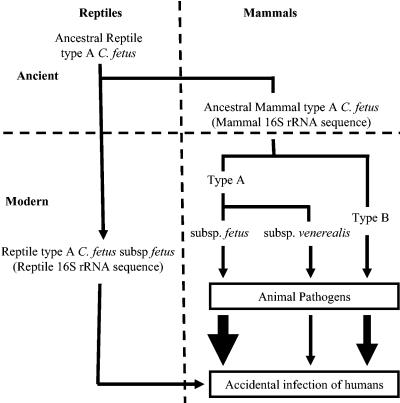
In the present study, we continued to investigate (6, 10, 12, 21, 25, 29, 35) the genetic differences within the species C. fetus, including isolates that were C. fetus subsp. fetus or C. fetus subsp. venerealis, type A or type B, and of mammal or reptile origin, using methods based on RAPD, PFGE, DNA-DNA hybridization, and 16S rRNA analyses. A schematic of the proposed ancestral relationships (Fig. 4) will be discussed below.
FIG. 4.
Schematic of proposed ancestral relationships and pathogenicities for C. fetus, based on the prior (28) and present studies. C. fetus strains colonize a variety of animals and/or are pathogens of these species. Infection of humans usually reflects direct contact with such animals or exposure to C. fetus-contaminated foods. The widths of the arrows indicate the relative frequencies of human infection with these strains.
Discrimination between C. fetus subsp. fetus and C. fetus subsp. venerealis is usually based on the epidemiologic setting and whether an isolate is able to grow in the presence of 1% glycine (27, 28). C. fetus subsp. fetus usually causes sporadic abortion in cattle and sheep and also causes most human infections (4, 14). In contrast, C. fetus subsp. venerealis is mostly restricted to cattle, causing venereal campylobacteriosis, resulting in infertility; human infection is rare (25). The two C. fetus subspecies can be identified by specific PCR (17). The PFGE results for the two C. fetus subsp. venerealis strains studied showed patterns different from one another in both the SmaI and SalI digestions, which differed from those of the C. fetus subsp. fetus strains, and the RAPD results demonstrated that 5 of 10 primers were able to distinguish C. fetus subsp. venerealis strains from C. fetus subsp. fetus strains. Our results showed that C. fetus subsp. fetus and C. fetus subsp. venerealis had identical 16S rRNA sequences, which is consistent with other reports (21). Comparing the present and prior (20, 29) studies, as well as the DNA-DNA hybridization results, indicates that these subspecies are very closely related. The close relatedness between the two subspecies is consistent with previous observations (21, 34). Strain 99-256 (ATCC 33561) had been classified as C. fetus subsp. venerealis, but our typing provides evidence that it is actually C. fetus subsp. fetus, illustrating the difficulty of separating the strains based on phenotype.
Differences between type A and type B C. fetus strains exist not only in lipopolysaccharide composition (19, 23), but also in the sap genes encoding the SLPs (7). Previous studies, based on partial recA and sapD gene sequence analyses, also suggested that type A and type B strains showed genetic differences greater than those between C. fetus subsp. fetus and venerealis (29). Similarly, the current RAPD analysis showed that use of 4 of the 10 primers studied identified differences between type A and type B strains. However, the SLPs of both types are antigenically cross-reactive (36), and there is 100% identity at the 16S rRNA level, consistent DNA-DNA hybridization, and similar SmaI and SalI digestion PFGE patterns. In total, these findings indicate that the mammalian-origin type A and type B C. fetus strains have shared ancestry and have not had major divergence at the genomic level. The data suggest that type A strains are most closely related to the distant C. fetus ancestors and that type B strains arose during their development in the mammalian niche, possibly in the gastrointestinal tract (29).
Of the four reptile strains studied, three were directly isolated from a reptile (15) and one (03-427), which was isolated from a human patient with bacteremia, has 16S rRNA and partial sapD sequences identical to those of the other three reptile strains (33). Although all four strains of reptile origin showed antigenic cross-reactivity with the antiserum raised against SLP of a strain of mammal origin, analyses of the 16S rRNA sequence, the housekeeping gene recA, and the C. fetus-specific gene sapD indicated substantial divergence from the mammalian C. fetus strains. In the present study, all 10 primers used for RAPD analyses showed differences between the reptile and mammalian strains. In the PFGE analyses, each of the four reptile strains showed a different SmaI digestion pattern, which differed from that of the mammalian strains. The results of the DNA-DNA hybridization indicated significant genomic divergence between the mammal and reptile isolates. In total, based on the multiple analyses described above, we conclude that reptile and mammal C. fetus strains have levels of genetic divergence higher than that between C. fetus subsp. fetus and C. fetus subsp. venerealis and between the type A and type B strains; the reptile strains may represent a new C. fetus subsp. fetus. Based on both the previous and current study, the reptilian C. fetus strains appear quite distinct from mammalian C. fetus strains. These might well represent a new subspecies, but to confirm that hypothesis, more studies are needed.
The results of these and prior studies are consistent with the hypothesis that C. fetus of a form related to type A strains was present in ancient reptiles and that this lineage has continued to the present (Fig. 4). Our previous studies indicating that all mammalian C. fetus strains (both C. fetus subsp. fetus and C. fetus subsp. venerealis and both type A and type B strains) had identical 16S rRNA sequences and nearly identical recA and sapD sequences provided evidence that the divergence between type A and type B strains occurred prior to that between C. fetus subsp. fetus and C. fetus subsp. venerealis (29). The 16S rRNA, recA, and sapD sequences all indicate that the reptilian strains are highly divergent from the mammalian strains. The present study confirms that reptilian C. fetus strains also show great divergence, based on the RAPD, PFGE, and DNA-DNA hybridization results. The data suggest that when mammals arose, C. fetus evolved with them into the present type A strains. The development of the C. fetus subsp. venerealis and type B C. fetus subsp. fetus strains likely represents more recent events. Analysis of full C. fetus genomes should permit improved exploration of these hypotheses.
Acknowledgments
We thank Floyd E. Dewhirst for helpful comments and review of the manuscript.
This work was supported in part by grant R01 AI24145 from the National Institutes of Health and by the Medical Research Service of the Department of Veterans Affairs.
REFERENCES
- 1.Akopyanz, N., N. O. Bukanov, T. U. Westblom, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg, R. L., J. W. Jutila, and B. D. Firehammer. 1971. A revised classification of Vibrio fetus. Am. J. Vet. Res. 32:11-22. [PubMed] [Google Scholar]
- 3.Blaser, M. J. 1998. Campylobacter fetus emerging infection and model system for bacterial pathogenesis at mucosal surfaces. Clin. Infect. Dis. 27:256-258. [DOI] [PubMed] [Google Scholar]
- 4.Blaser, M. J., and B. M. Allos. 2005. Campylobacter jejuni and related species, p. 2548-2557. In G. L. Mandell, R. G. Douglas, Jr., and J. E. Bennett (ed.), Principles and practice of infectious diseases, 6th ed. John Wiley & Sons, Inc., New York, N.Y.
- 5.de Lencastre, H., E. P. Severina, R. B. Roberts, B. N. Kreiswirth, A. Tomasz, and the BARG Initiative Pilot Study Group. 1996. Testing the efficacy of a molecular surveillance network: methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecium (VREF) genotypes in six hospitals in the metropolitan New York City area. Microb. Drug Resist. 2:343-351. [DOI] [PubMed] [Google Scholar]
- 6.Duim, B., P. A. Vandamme, A. Rigter, S. Laevens, J. R. Dijkstra, and J. A. Wagenaar. 2001. Differentiation of Campylobacter species by AFLP fingerprinting. Microbiology 147:2729-2737. [DOI] [PubMed] [Google Scholar]
- 7.Dworkin, J., M. K. Tummuru, and M. J. Blaser. 1995. Segmental conservation of sapA sequences in type B Campylobacter fetus cells. J. Biol. Chem. 70:15093-15101. [DOI] [PubMed] [Google Scholar]
- 8.Endtz, H. P., C. W. Ang, N. van Den Braak, B. Duim, A. Rigter, L. J. Price, D. L. Woodward, F. G. Rodgers, W. M. Johnson, J. A. Wagenaar, B. C. Jacobs, H. A. Verbrugh, and A. van Belkum. 2000. Molecular characterization of Campylobacter jejuni from patients with Guillain-Barre and Miller Fisher syndromes. J. Clin. Microbiol. 38:2297-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Euzéby, J. P. 1997. List of bacterial names with standing in nomenclature: a folder available on the Internet (URL: http://www.bacterio.cict.fr/). Int. J. Syst. Bacteriol. 47:590-592. [DOI] [PubMed] [Google Scholar]
- 10.Fujita, M., S. Fujimoto, T. Morooka, and K. Amako. 1995. Analysis of strains of Campylobacter fetus by pulsed-field gel electrophoresis. J. Clin. Microbiol. 33:1676-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia, M. M., M. D. Eaglesome, and C. Rigby. 1983. Campylobacters important in veterinary medicine. Vet. Bull. 53:793-818. [Google Scholar]
- 12.Gorkiewicz, G., G. Feierl, C. Schober, F. Dieber, J. Kofer, R. Zechner, and E. L. Zechner. 2003. Species-specific identification of Campylobacters by partial 16S rRNA gene sequencing. J. Clin. Microbiol. 41:2537-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grogono-Thomas, R., M. J. Blaser, M. Ahmadi, and D. G. Newell. 2003. Role of S-layer protein antigenic diversity in the immune responses of sheep experimentally challenged with Campylobacter fetus subsp. fetus. Infect. Immun. 71:147-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerrant, R. L., R. G. Lahita, W. C. Winn, Jr., and R. B. Roberts. 1978. Campylobacteriosis in man: pathogenic mechanisms and review of 91 bloodstream infections. Am. J. Med. 65:584-592. [DOI] [PubMed] [Google Scholar]
- 15.Harvey, S., and M. C. Greenwood. 1985. Isolation of Campylobacter fetus from a pet turtle. J. Clin. Microbiol. 21:260-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez, J., A. Fayos, M. A. Ferrus, and R. J. Owen. 1995. Random amplified polymorphic DNA fingerprinting of Campylobacter jejuni and C. coli isolated from human faeces, seawater and poultry products. Res. Microbiol. 146:685-696. [DOI] [PubMed] [Google Scholar]
- 17.Hum, S., K. Quinn, J. Brunner, and S. L. On. 1997. Evaluation of a PCR assay for identification and differentiation of Campylobacter fetus subspecies. Aust. Vet. J. 75:827-831. [DOI] [PubMed] [Google Scholar]
- 18.Karenlampi, R. I., T. P. Tolvanen, and M. L. Hanninen. 2004. Phylogenetic analysis and PCR-restriction fragment length polymorphism identification of Campylobacter species based on partial groEL gene sequences. J. Clin. Microbiol. 42:5731-5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moran, A. P., D. T. O'Malley, T. U. Kosunen, and I. M. Helander. 1994. Biochemical characterization of Campylobacter fetus lipopolysaccharides. Infect. Immun. 62:3922-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.On, S. L. W. 2001. Taxonomy of Campylobacter, Arcobacter, Helicobacter and related bacteria: current status, future prospects and immediate concerns. J. Appl. Microbiol. 90:1S-15S. [DOI] [PubMed] [Google Scholar]
- 21.On, S. L., and C. S. Harrington. 2001. Evaluation of numerical analysis of PFGE-DNA profiles for differentiating Campylobacter fetus subspecies by comparison with phenotypic, PCR, and 16S rDNA sequencing methods. J. Appl. Microbiol. 90:285-293. [DOI] [PubMed] [Google Scholar]
- 22.Pei, Z., E. J. Bini, L. Yang, M. Zhou, F. Francois, and M. J. Blaser. 2004. Bacterial biota in the human distal esophagus. Proc. Natl. Acad. Sci. USA 101:4250-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pérez-Pérez, G. I., M. J. Blaser, and J. H. Bryner. 1986. Lipopolysaccharide structures of Campylobacter fetus are related to heat-stable serogroups. Infect. Immun. 51:209-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ray, K. C., Z. C. Tu, R. Grogono-Thomas, D. G. Newell, S. A. Thompson, and M. J. Blaser. 2000. Campylobacter fetus sap inversion occurs in the absence of RecA function. Infect. Immun. 68:5663-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salama, S. M., M. M. Garcia, and D. E. Taylor. 1992. Differentiation of the subspecies of Campylobacter fetus by genomic sizing. Int. J. Syst. Bacteriol. 42:446-450. [DOI] [PubMed] [Google Scholar]
- 26.Sebald, M., and M. Véron. 1963. Base DNA content and classification of vibrios. Ann. Inst. Pasteur (Paris) 105:897-910. [PubMed] [Google Scholar]
- 27.Skirrow, M. B. 1990. Campylobacter and Helicobacter infections of man and animals, p. 531-545. In M. T. Parker and L. H. Collier (ed.), Principles of bacteriology, virology and immunity, 8th ed., vol. 2. Edward Arnold, London, England.
- 28.Smibert, R. M. 1984. Genus Campylobacter, p. 111-118. In N. R. Krieg, and H. G. Holt (ed.), Bergey's manual of systemic bacteriology, vol. 1. The Williams & Wilkins Co., Baltimore, Md. [Google Scholar]
- 29.Tu, Z. C., F. E. Dewhirst, and M. J. Blaser. 2001. Evidence that the Campylobacter fetus sap locus is an ancient genomic constituent with origins before mammals and reptiles diverged. Infect. Immun. 69:2237-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tu, Z. C., C. Gaudreau, and M. J. Blaser. 2005. Mechanisms underlying Campylobacter fetus pathogenesis in humans: surface-layer protein variation in relapsing infections. J. Infect. Dis. 191:2082-2089. [DOI] [PubMed] [Google Scholar]
- 31.Tu, Z. C., J. Hui, and M. J. Blaser. 2004. Conservation and diversity of sap homologues and their organization among Campylobacter fetus isolates. Infect. Immun. 72:1715-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tu, Z. C., T. M. Wassenaar, S. A. Thompson, and M. J. Blaser. 2003. Structure and genotypic plasticity of the Campylobacter fetus sap locus. Mol. Microbiol. 48:685-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tu, Z. C., G. Zeitlin, J. P. Gagner, T. Keo, B. A. Hanna, and M. J. Blaser. 2004. Campylobacter fetus of reptile origin as a human pathogen. J. Clin. Microbiol. 42:4405-4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandamme, P. 2000. Taxonomy of the family Campylobacteraceae, p. 3-26. In I. Nachamkin, and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 35.Wagenaar, J. A., M. A. van Bergen, D. G. Newell, R. Grogono-Thomas, and B. Duim. 2001. Comparative study using amplified fragment length polymorphism fingerprinting, PCR genotyping, and phenotyping to differentiate Campylobacter fetus strains isolated from animals. J. Clin. Microbiol. 39:2283-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, E., M. M. Garcia, M. S. Blake, Z. H. Pei, and M. J. Blaser. 1993. Shift in S-layer protein expression responsible for antigenic variation in Campylobacter fetus. J. Bacteriol. 175:4979-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young, V. B., C. A. Dangler, J. G. Fox, and D. B. Schauer. 2000. Chronic atrophic gastritis in SCID mice experimentally infected with Campylobacter fetus. Infect. Immun. 68:2110-2118. [DOI] [PMC free article] [PubMed] [Google Scholar]