Abstract
Real-time PCR and fluorescence in situ hybridization (FISH) were evaluated as rapid methods for the diagnosis of bacterial meningitis and compared to standard diagnostic procedures. For PCR, a LightCycler approach was chosen, implementing eubacterial and specific PCR assays for the most relevant bacteria. For FISH, a similar probe set containing eubacterial and specific probes was composed of published and newly designed probes. Both methods were evaluated by use of cerebrospinal fluid (CSF) samples from patients with suspected bacterial meningitis. For all microscopy- and culture-positive samples (n = 28), the eubacterial PCR was positive. In addition, all identifiable pathogens were detected with specific PCR assays, according to an algorithm based on the Gram stain. The FISH method detected the pathogen in 13 of 18 positive samples. While the FISH method remained negative for all microscopy- and culture-negative samples (n = 113), the eubacterial PCR was positive for five of these samples. Sequencing of the amplicon revealed the presence of Neisseria meningitidis, Streptococcus agalactiae, and Haemophilus influenzae in three of these five samples. In addition, samples with discordant results by culture and microscopy were successfully investigated by PCR (10 samples) and FISH (5 samples). In conclusion, PCR is a highly sensitive tool for rapid diagnosis of bacterial meningitis. FISH is less sensitive but is useful for the identification of CSF samples showing bacteria in the Gram stain. Based on our results, an approach for laboratory diagnosis of meningitis including PCR and FISH is discussed.
Acute purulent infection of the meninges is the most common infection of the central nervous system, with a yearly incidence of about 3 in 100,000 inhabitants in industrialized countries (8, 22, 24). More than 95% of cases of bacterial meningitis are caused by one of the following bacteria: Neisseria meningitidis, Streptococcus pneumoniae, Streptococcus agalactiae, Staphylococcus spp., Escherichia coli, Haemophilus influenzae, and Listeria monocytogenes (22, 24). The identification of the pathogen from cerebrospinal fluid (CSF) usually takes 1 to 2 days by culture. Moreover, culture frequently remains negative, especially if the CSF is taken after initiation of antimicrobial therapy (8). Since the outcome of infection depends highly on an early initiation of adequate therapy (8, 26), new rapid diagnostic methods are urgently needed (25).
In the recent past, PCR assays have been developed for the specific detection of bacteria causing meningitis such as N. meningitidis (16, 21), S. pneumoniae (11, 29), and S. agalactiae (10). Several studies have demonstrated the usefulness of eubacterial broad-range PCRs for the diagnosis of bacterial meningitis (3, 6, 13, 14, 23). However, most published PCR protocols were either time-consuming or did not facilitate species diagnosis of the bacterial pathogens. Therefore, in this study, a broad-range real-time eubacterial LightCycler PCR assay for the detection of all relevant bacteria causing meningitis was evaluated. In addition, a panel of previously published species- and genus-specific real-time LightCycler PCR assays (30) was evaluated by use of CSF samples for the first time.
Fluorescence in situ hybridization (FISH) using fluorescently labeled oligonucleotide probes complementary to unique target sites on the rRNA has already been successfully implemented in the field of clinical microbiology. Examples are the detection and identification of pathogens in blood cultures, tissues, and cell cultures (2, 7, 12, 17, 19, 27). FISH is a cheap and quick method that does not require costly technical equipment. It has a low risk of contamination, since it lacks a nucleic acid amplification step. Its utility for diagnosis of bacterial meningitis has, however, not been determined yet. Therefore, we defined a FISH probe set for the detection of bacterial pathogens in cerebrospinal fluid that includes previously published probes (2, 7, 12, 17, 27) as well as newly designed probes for the detection of N. meningitidis, E. coli, and Staphylococcus spp.
The two novel molecular approaches, the real-time PCR and the FISH technique, were evaluated on 141 clearly defined CSF samples, including 28 microscopy- and culture-positive and 113 microscopy- and culture-negative samples. In addition, 10 samples with discordant microscopy and culture results were included.
MATERIALS AND METHODS
Bacterial strains used as control strains for determination of the analytical sensitivity and specificity included Staphylococcus aureus (ATCC 25923), Staphylococcus epidermidis (ATCC 12228), S. pneumoniae (ATCC 49619), S. agalactiae (ATCC 13813), Staphylococcus pyogenes (ATCC 12344) Enterococcus faecalis (ATCC 29212), Haemophilus influenzae (ATCC 49247), Haemophilus parainfluenzae (ATCC 33392), E. coli (ATCC 25922), and N. meningitidis (ATCC 13077). The E. coli probe (Esco 473) was evaluated on reference strains of E. coli (ATCC 25921, ATCC 25922, and ATCC 35218) and Klebsiella oxytoca (ATCC 43893) and on clinical isolates of E. coli (11), Morganella sp. (4), Shigella spp. (2), Salmonella enterica serovar Enteritidis (1), Klebsiella spp. (2), Serratia sp. (1), Citrobacter sp. (1), Yersinia sp. (1), Pseudomonas aeruginosa (4), Hafnia sp. (1), and Proteus spp. (3). The newly designed Staphylococcus spp.-specific probe (Staph 698) was tested on the reference strains of S. aureus (ATCC 25923 and ATCC 4330), S. epidermidis (ATCC 12228), S. agalactiae (ATCC 13813), S. pyogenes (ATCC) 12344 S. pneumoniae (ATCC 49619) and on clinical isolates of S. aureus (3), S. epidermidis (13), coagulase-negative staphylococci other than S. epidermidis (8), Enterococcus spp (8)., S. agalactiae (1), S. pneumoniae (1), Streptococcus salivarius (1), S. pyogenes (1), and Bacillus sp. (1). The probe for N. meningitidis was evaluated on N. meningitidis reference strains of serogroups A, B, C, E, L, X, Z, and W135 (ATCC 13090, ATCC 13102, ATCC 13113, ATCC 35558, ATCC 43828, ATCC 35560, ATCC 35561, ATCC 35561, and ATCC 35559); clinical isolates of N. meningitidis serogroup B, serogroup Y (17), serogroup C (10), serogroup Z (1), and serogroup W135 (1); a reference strain of Neisseria gonorrhoeae (ATCC 19424); clinical isolates of N. gonorrhoeae (30); reference strains of other neisseriae (ATCC 51483, ATCC 23970, ATCC 33926, ATCC 19696, ATCC 43768, ATCC 29256, ATCC 49275, ATCC 51223, ATCC 13120, ATCC 49377, ATCC 29315, ATCC 25295, ATCC 14686, ATCC 14685, ATCC 49930, and ATCC 14687); and 31 reference strains of nonneisseriae, including Bordetella spp. (ATCC 511541, ATCC 51783, and ATCC 35086), Rhodococcus spp. (ATCC 184, ATCC 33609, and ATCC 13808), and Vibrio spp. (ATCC 14126, ATCC 17802, ATCC 27562, ATCC 33564, and ATCC 33809).
Determination of analytical sensitivity.
Serial 10-fold dilutions of control strains were prepared in pooled culture-negative CSF samples from patients without suspicion of bacterial meningitis. Quantities of viable bacteria were counted by plating the dilutions on sheep or chocolate agar. Slides for FISH and DNA samples for PCR were prepared as described below.
Clinical specimens.
For the clinical evaluation of PCR and FISH, CSF samples from patients with suspected bacterial meningitis were used which had been sent to our laboratory for routine diagnostics. Approximately 250 μl of each CSF sample was inoculated on sheep blood agar and chocolate agar (Oxoid, Germany) and incubated at 36°C in room air supplemented with 5% CO2 for 72 h. Identification of organisms isolated from CSF was achieved by Api (Api NH, Api 20 strep, Api Coryne, Api NH, Api CH50 Bacillus; BioMerieux, Germany). For staphylococci, diagnosis was based on typical morphology (color and hemolysis, etc.), positive catalase reaction, clumping factor, aurease detection, and growth on mannitol-salt-agar.
Preparation of microscopic slides.
Microscopic samples for gram staining and FISH were prepared by cytocentrifugation of 75 μl of CSF to a glass slide (1,200 rpm, 10 min) and subsequent fixation in 70% methanol for 3 min.
Gram staining was performed according to standard procedures, with carbolfuchsin as a counterstain. The amounts of bacteria, granulocytes, and lymphocytes were documented in a semiquantitative way in five grades (many, moderate, few, very few, or not present).
DNA isolation for PCR analysis.
A minimum of 50 μl of CSF of microscopy-negative samples and 10 μl of microscopy-positive samples or 200 μl of spiked CSF samples for determination of analytical sensitivity was used for isolation of DNA. The DNA was prepared using the QIAamp mini kit (QIAGEN, Hilden, Germany) according to the instructions of the manufacturer. Total DNA was eluted in 50 μl of AE buffer (QIAGEN) and stored at −20°C until measurement in the PCR.
Eubacterial LightCycler PCR.
For detection of a broad spectrum of bacteria, a eubacterial 16S rRNA gene LightCycler PCR assay was established, using primers RW01 (5′-AACTGGAGGAAGGTGGGGAT-3′) and DG74 (5′-AGGAGGTGATCCAACCGCA-3′), which amplify a 360- to 380-bp fragment at the 5′ end of the 16S rRNA gene (5). The amplification mixture of the PCR consisted of 2 μl of 10× reaction mix (LightCycler Fast-Start master DNA, SYBR green; Roche Diagnostics, Germany), 4 mM MgCl2, 0.5 μM concentrations of each primer, and 2 μl of template DNA in a final volume of 20 μl. Samples were amplified as follows: an initial denaturation step at 95°C for 10 min and 40 cycles of denaturation at 95°C for 0 s, annealing at 50°C for 10 s, and elongation at 72°C for 20 s. The temperature transition rate was 20°C/s. The generation of target amplicons for each sample was monitored during the elongation steps at 530 nm (LightCycler channel F1). Samples positive for the target gene were identified at the cycle number where the fluorescence exceeded the background. This cycle number, expressed as the crossing point (Cp), was determined automatically by the LightCycler software (version 3.5) according to the second derivative maximum method. Following the amplification phase, a melting curve analysis (starting at 50°C) was performed with a temperature transition rate of 0.1°C/s. A positive (1 ng of E. coli ATCC 25922 DNA) and a negative control, consisting of all PCR reagents described above and 2 μl of PCR grade water (included in the Fast-Start master mix kit; Roche Diagnostics) instead of template DNA, were included in each PCR run. The master mix was prepared in a PCR cabinet with HEPA-filtered circulating air and intermittent UV irradiation, and the DNA of clinical samples as well as that of the controls was added in a separate room.
Species- and genus-specific LightCycler PCR assay.
For identification of meningeal pathogens in microscopy-positive samples, specific hybridization probe LightCycler PCR assays were run (Table 1) using primers and probes published previously (20, 30). Identification of enterococci and streptococci was achieved in the same PCR by melting curve analysis, since streptococci show a melting temperature of <60°C, E. faecalis shows a melting temperature of 70°C, and E. faecium shows a melting temperature of 63°C in the PCR assay (30). PCR assays to be run on each sample were chosen according to the result of microscopy (morphology and Gram stain characteristics) by following an algorithm based on the Gram stain (see Table 3). This algorithm also considers the presence of gram-negative rods, although there were no samples with gram-negative rods included in this study. The PCR assays for the detection of E. coli, Enterobacteriaceae, Haemophilus influenzae, and Pseudomonas aeruginosa were published previously (30). Amplification mixtures consisted of 2 μl of 10× reaction mix (LightCycler Fast-Start master DNA hybridization probes; Roche Diagnostics), 4 mM MgCl2, 0.5 μM concentrations of each primer, 0.2 μM concentrations of each oligonucleotide probe, and 2 μl of template DNA in a final volume of 20 μl. Samples were amplified as described above but with an annealing temperature of 54°C for the N. meningitidis-specific assay. The generation of target amplicons was monitored between the annealing and the elongation steps at 640 nm. Following the amplification phase, a melting curve analysis (starting at 40°C) was performed with a temperature transition rate of 0.1°C/s. Positive (the respective American Type Culture Collection strain) and negative controls, as described above, were included in each PCR run. The master mix was prepared in a PCR cabinet with HEPA-filtered circulating air and intermittent UV irradiation, and the DNA of clinical samples as well as that of the controls was added in a separate room.
TABLE 1.
Primers and probes used for detection of meningeal pathogens by PCR
| Target organism | Primer pair | Probes | Reference(s) |
|---|---|---|---|
| Staphylococcus spp. | RW01/DG74 | Staph FL/LC | 5, 30 |
| Staphylococcus aureus | Sa442-F/Sa442-R | Sa442-HP-1/-2 | 20 |
| Staphylococcus epidermidis | MasFs/epiR1 | Epi FL/LC | 30 |
| Streptococcus agalactiae | Sag59/Sag190 | STB-F-HP-1/-2 | 10, 30 |
| Streptococcus spp. | RW01/DG74 | EntFL/LCa | 30 |
| Enterococcus spp. | RW01/DG74 | EntFL/LCa | 30 |
| Neisseria meningitidis | MenIS-1/-8 | MenIS_HP-1/-4 | 30 |
Differentiation of Streptococcus spp. and Enterococcus spp. was done by melting curve analysis, as described in Materials and Methods.
TABLE 3.
Algorithm used for the FISH and PCR approaches
| Result of Gram stain | Organism(s) tested for by:
|
|
|---|---|---|
| PCR assays | FISH (probe type) | |
| Gram-positive cocci in clusters | Staphylococcus aureus | Staphylococcus aureus (PNA) |
| Staphylococcus spp. | Staphylococcus spp. (PNA) | |
| Staphylococcus epidermidis | ||
| Gram-positive cocci in chains and diplococci | Enterococcus/Streptococcus spp. | S. pneumoniae (DNA) |
| S. agalactiae | S. agalactiae (DNA) | |
| Gram-negative cocci | Neisseria meningitidis | Neisseria meningitidis (DNA) |
| Gram-negative rods | E. coli | E. coli (DNA) |
| Haemophilus influenzae | Haemophilus influenzae (DNA) | |
| No bacteria visible | Eubacteria | Eubacteria (PNA) |
DNA sequencing.
PCR products of the eubacterial 16S rRNA gene PCR were sequenced with an Abi Prism 310 Genetic Analyzer (Abi Prism; Biosystems, Warrington, England) using a dye terminator cycle sequencing ready reaction kit (Abi Prism). The primers DG74 and RW01 (10 pmol each, see above) were used for sequencing of the forward and reverse sequences.
Selection and design of FISH probes.
The previously published FISH assays were implemented as described previously (7, 12, 17, 18, 27) (Table 2). New DNA probes were designed for N. meningitidis and E. coli using the ARB program package, freely available at http://www.arb-home.de/. Oligonucleotide probes were synthesized and directly 5′ labeled with 5(6)-carboxyfluorescein-N-hydroxysuccinimideester or the hydrophilic sulfoindocyanine fluorescent dye Cy3 (Thermo Hybaid, Germany). All probes were controlled with regard to probe specificity by checking publicly available 16S rRNA sequences included in the ARB database and by a National Center for Biotechnology Information BLAST search (1). The probes were further evaluated on reference strains and clinical isolates as listed above.
TABLE 2.
Probes used for detection of meningeal pathogens by FISH
| Target organism | Probea | Type | % [FA]b | Sequence (5′-3′) | Reference |
|---|---|---|---|---|---|
| Escherichia coli | Esco 473 | DNA | 30 | GCG GGT AAC GTC AAT GAG C | This study |
| Haemophilus influenzae | Haeinf | DNA | 30 | CCG CAC TTT CAT CTT CCG | 7 |
| Neisseria meningitidis | NeMe183c | DNA | 30 | CCT GCT TTC TCT CTC AAG A | This study |
| S. agalactiae | Saga | DNA | 20 | GTA AAC ACC AAA CMT CAG CG | 27 |
| S. pneumoniae | Spn | DNA | 20 | GTG ATG CAA GTG CAC CTT | 12 |
| Staphylococcus spp. | Staph 698 | PNA | 30 (PNA mix) | CTC CAT ATC TCT GCG | This study |
| S. aureus | Stau | PNA | 30 (PNA mix) | GCT TCT CGT CCG TTC | 17 |
| Bacterial kingdom | EUB 338 | DNA | 20-40 | GCT GCC TCC CGT AGG AGT | 2 |
| Bac-Uni1 | PNA | 30 (PNA mix) | CTG CCT CCC GTA GGA | 17 |
All probes are directed to complementary sequences of the 16S subunit of the ribosome.
Concentration of formamide used in hybridization buffer. Additional substances were added for DNA-FISH as described in references 2,15 and for PNA FISH as described in reference 17.
To be used in combination with competitor (5′-CCTGCTTTCCCTCTCAAGA-3′).
Previously published PNA (peptic nucleic acid) probes targeting all bacteria (Bac-Uni1) and S. aureus were obtained from Henrik Stendal from AdvanDx (17) (Table 2). In addition, a genus-specific PNA probe covering all staphylococci was designed and evaluated as described above. It was synthesized and directly 5′ labeled with 5(6)-carboxyfluorescein-N-hydroxysuccinimideester by Metabion (Munich, Germany).
In situ hybridization with DNA probes was performed at 46°C using the hybridization and washing buffers described elsewhere (2, 15). The stained samples were rapidly air dried, embedded, and examined with a Zeiss Axioplan 2 microscope. For the newly developed probes, the optimal hybridization stringency was determined for each probe by increasing the formamide concentrations in the hybridization buffer in increments of 5 to 10% and by lowering the salt concentration accordingly in the wash buffers. The final hybridization conditions are stated in Table 2. In situ hybridization with PNA probes was performed at 56°C as described elsewhere (17).
FISH of clinical samples was conducted according to an algorithm based on the Gram stain (Table 3). FISH with DNA probes was always performed with a specific Cy3-labeled probe in combination with a fluorescein isothiocyanate (FITC)-labeled eubacterial probe (EUB 338) (Table 2) and the 4′,6′-diamidino-2-phenylindole (DAPI) stain (Hoechst, Germany). FITC-labeled PNA probes were implemented in combination with the DAPI stain. Samples were regarded positive if three typical bacterial structures producing positive results in all implemented staining techniques were detected.
Statistical analysis.
Receiver operating characteristic (ROC) analysis was done by Statistical Analysis Software, version 8.2, SAS Institute.
RESULTS
Analytical sensitivity of the methods.
The analytical sensitivity of the methods was determined using serial dilutions of bacterial suspensions in CSF. The eubacterial SYBR green PCR showed a very high sensitivity and even detected less than 25 CFU/ml CSF, using the Neisseria meningitidis and S. epidermidis reference strains. Serial dilutions of the DNA of these strains in sterile saline solution revealed positive PCR signals exceeding the negative control in samples with concentrations of as low as 10 fg DNA/PCR. The specific PCR assays with sequence-specific hybridization probes differed in sensitivity: the N. meningitidis-specific assay detected less than 25 CFU of N. meningitidis/ml spiked CSF, the Enterococcus/Streptococcus-specific assay detected 250 CFU of S. pyogenes/ml spiked CSF, and the S. aureus-specific assay detected 4,000 CFU S. aureus/ml spiked CSF (means of the results from three experiments). The analytical sensitivity of the other PCR assays in CSF was not determined. Data regarding the sensitivity of all PCR assays in sterile saline can be found in reference 30.
The analytical sensitivity of the FISH was comparable to that of the Gram stain (approximately 105 bacteria/ml CSF for all probes).
Evaluation of microscopy- and culture-negative CSF samples.
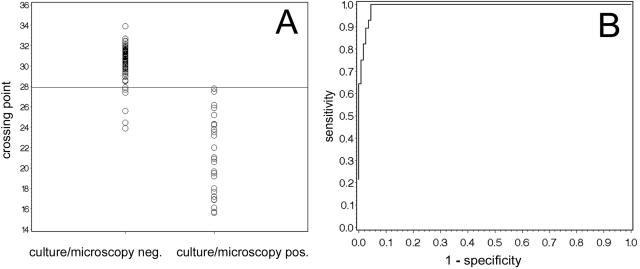
One hundred thirteen CSF samples that remained negative by microscopy and culture were investigated by eubacterial PCR and FISH. When using the eubacterial SYBR green PCR, a background level of bacterial DNA was detectable in most PCR runs, which can be quantified by the Cp in the LightCycler software. To determine a reliable cutoff value for the discrimination of positive and negative samples, we performed a ROC analysis of the Cp values using the microscopy- and culture-negative samples as “true negatives” and the microscopy- and culture-positive samples as “true positives.” By this ROC analysis, samples with a Cp of <27.9 are scored as positive (Fig. 1). In addition, by investigating serial dilutions of the S. epidermidis reference strain, direct sequencing of the amplicon led to identification of the species in all samples with a Cp of <28.0. Applying the cutoff value of 27.9 for the Cp's of the negative samples, 108/113 samples were negative in the eubacterial PCR. In five PCR-positive samples, the PCR product was further characterized by DNA sequencing. In three of the five samples, sequencing of the amplicon revealed the presence of DNA of common causative agents of bacterial meningitis, i.e., Haemophilus influenzae (Cp of 25.6) in a patient with a CSF shunt and suspected bacterial meningitis following surgical removal of an adenocarcinoma of the nose, N. meningitidis (Cp of 23.9) in a patient with clinical meningococcal disease that was later confirmed by blood culture, and S. agalactiae (Cp of 24.5) in a newborn with suspected meningitis. In the remaining two samples (Cp of 27.4 and 27.7), however, the DNA could not be characterized any further, probably due to the presence of a mixture of DNA in these samples and the low concentration of DNA. Four of the five PCR-positive samples contained moderate or higher numbers of granulocytes, while only 14 of the 113 negative samples contained comparable numbers of granulocytes. All five corresponding patients received broad-spectrum antimicrobial therapy for at least 48 h before sampling of the CSF.
FIG. 1.
ROC analysis of the eubacterial 16S rRNA gene PCR. (A) Distribution of the crossing points of the individual measurements; (B) ROC analysis. neg., negative; pos., positive.
FISH analysis using the FITC-labeled eubacterial PNA probe (Bac-Uni1) in combination with the DAPI stain was negative in all of these culture- and gram-negative samples.
Evaluation of microscopy- and culture-positive CSF samples.
Samples that were positive by microscopy and culture were investigated by PCR and FISH according to an algorithm based on the Gram stain (Table 3). Regarding PCR, all 28 samples investigated were positive in the eubacterial SYBR green PCR (see above) (Fig. 1). In addition, we could correctly identify 26 of the 28 bacteria to the genus or species level, respectively, by the specific hybridization probe PCR assays (Table 4). The two samples that could not be identified by specific hybridization probes contained gram-positive rods (Corynebacterium sp. and Bacillus sp.). However, by eubacterial 16S rRNA gene PCR and subsequent sequencing of the amplicon, we could identify the strains as Corynebacterium jeikeium and Bacillus cereus (98.8 and 100% homology to GenBank accession no. X84250 and AY138271, respectively).
TABLE 4.
Diagnostic sensitivity of FISH and PCR
| Bacterial species isolated from CSF samples | No. of positive samples/total no. of samples tested by:
|
|
|---|---|---|
| FISH | PCR | |
| Neisseria meningitidis | 1/2 | 4/4 |
| Streptococcus pneumoniae | 4/4 | 7/7b |
| Streptococcus agalactiae | 0/0 | 2/2 |
| Staphylococcus aureus | 1/2 | 2/2 |
| Staphylococcus epidermidis | 4/6a | 7/7 |
| Coagulase-negative staphylococci other than S. epidermidis | 1/2a | 3/3 |
| Enterococcus faecalis | 1/1c | 1/1d |
| Bacillus spp. | 0/0 | 1/1c |
| Corynebacterium spp. | 1/1c | 1/1c |
| Total | 13/18 | 28/28 |
For coagulase-negative staphylococci, a genus-specific Staphylococcus FISH probe was implemented.
S. pneumoniae was identified as Streptococcus spp. by the Enterococcus/Streptococcus-specific PCR.
Positive with the eubacterial FISH probe or the eubacterial PCR, respectively.
Positive in the Enterococcus/Streptococcus-specific PCR and identified as E. faecalis by melting point analysis (30).
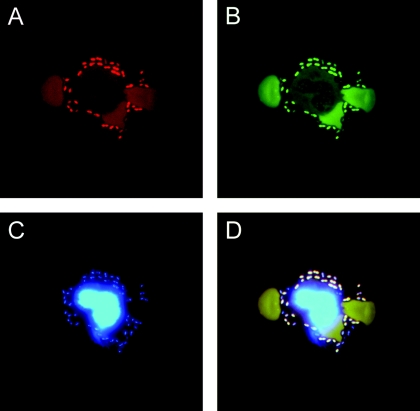
Due to the limited amount of cerebrospinal fluid, only 18 positive samples were available for FISH analysis. FISH led to detection of bacteria in 13 of these samples (Table 4; Fig. 2). In 11 cases, the organism could be identified to the species level. Two samples were positive with the eubacterial probe but contained bacteria that were not covered by specific probes (Enterococcus faecalis and Corynebacterium spp.).
FIG. 2.
FISH detection of pneumococci in a CSF sample from a patient. The sample was stained by an S. pneumoniae-specific Cy3-labeled probe (Spn) (A) in combination with the FITC-labeled eubacterial probe (EUB 338) (B). As a further control, the DNA stain DAPI was implemented, which stained bacteria as well as the nucleus of a granulocyte (C). The overlay (D) demonstrates that the bacteria fluoresce in all channels, resulting in a white color, while the weak autofluorescence of the erythrocytes in the FITC and Cy3 channel results in a brown color.
Evaluation of samples with discordant microscopy and culture results.
Ten samples with discordant results in microscopy and culture were included in the study. Nine of these samples contained moderate or higher numbers of granulocytes, indicating bacterial meningitis. The remaining sample still contained a few granulocytes. Three samples which were positive by microscopy but remained negative by culture were available for PCR analysis. They were investigated according to the algorithm in Table 3. In two samples which showed gram-positive diplococci with the Gram stain, the Enterococcus/Streptococcus-specific PCR was positive for streptococci and the S. agalactiae-specific PCR was negative. In the third sample, which showed gram-negative diplococci with the Gram stain, the N. meningitidis-specific PCR was positive. In the first patient, who had visible gram-positive diplococci in the CSF, pneumococci were cultured in a blood culture taken at the same day as the CSF sample. In the other patient, the probable pneumococcal meningitis could not be confirmed any further. The patient with gram-negative diplococci in the CSF sample did show clinical signs of disseminated meningococcal disease. Only the CSF sample of the first patient was available for FISH. This sample was positive with both the EUB probe and the S. pneumoniae-specific probe.
Seven samples which were negative by microscopy but positive by culture were available for PCR (Table 5). The eubacterial PCR was positive for all samples, and the N. meningitidis PCR correctly detected two cases of meningococcal meningitis (Table 5). Due to the minimal amount of sample, only four of these samples were available for FISH. The eubacterial PNA FISH probe (Bac-Uni1) in combination with the DAPI stain was positive for three samples (Table 5). After knowing the culture result, we also performed specific PCR and FISH probe assays for the identified species, the results of which are shown in Table 5.
TABLE 5.
Results of samples with negative microscopy but positive culture resultsa
| Culture result | Result by PCR assay
|
Result by FISH probe
|
|||
|---|---|---|---|---|---|
| Eubacterial | Neisseria meningitidis | Specific | Eubacterial PNA | Specific | |
| Neisseria meningitidis | + | + | + (N. meningitidis) | − | − (N. meningitidis) |
| Neisseria meningitidis | + | + | + (N. meningitidis) | NA | NA |
| Staphylococcus aureus | + | − | + (Staphylococcus spp.) − (S. aureus) | + | + (S. aureus) |
| Staphylococcus aureus | + | − | + (Staphylococcus spp.) − (S. aureus) | + | + (S. aureus) |
| Staphylococcus aureus | + | − | + (Staphylococcus spp.) − (S. aureus) | + | + (S. aureus) |
| Staphylococcus epidermidis | + | − | + (Staphylococcus spp.) + (S. epidermidis) | NA | NA |
| Coagulase-negative staphylococcus | + | − | + (Staphylococcus spp.) | NA | NA |
+, positive; −, negative; NA, not available.
DISCUSSION
Bacterial meningitis is a life-threatening disease which requires rapid diagnosis and antimicrobial therapy. Novel molecular methods offer the opportunity to establish a quick and reliable diagnosis. We evaluated real-time PCR and FISH for their applicability to the diagnosis of bacterial meningitis.
For PCR analysis, a real-time LightCycler approach was chosen to maximize the speed of diagnostics. A eubacterial broad-range PCR assay, based on a previously published primer pair (5), was adapted to a real-time LightCycler PCR protocol using SYBR green as fluorescent dye. In comparison to specific hybridization probes, SYBR green offered the advantage that its fluorescence interferes to a lesser extent with sequencing reactions and thus allowed direct sequencing of the LightCycler PCR products. The eubacterial SYBR green PCR proved highly sensitive for the diagnosis of bacterial DNA in CSF samples. Due to this high analytical sensitivity of the eubacterial PCR and the ubiquity of bacterial DNA in reagents and plastic materials, etc., a background level of DNA was detected in most clinical samples. Therefore, we performed a ROC analysis of the crossing points of the measurements and determined a cutoff level for interpretation of samples. By applying this cutoff value, we even detected meningeal pathogens in three microscopy- and culture-negative clinical samples. Considering the clinical symptoms of the patients and the finding of granulocytes in the CSF, these results can be regarded as true positives. False-positive results were obtained in only 2/113 (1.8%) samples. Previously published hybridization probe LightCycler PCR assays (30) were, in addition, evaluated in terms of their utility to detect bacterial pathogens in CSF samples for the first time (Tables 1 and 3). The analytical sensitivity of the specific PCR assays in CSF was comparable to that previously published for DNA preparations in physiological saline (30). Among all assays, the Staphylococcus spp.-specific PCR and the S aureus-specific PCR had the lowest analytical sensitivities. This may be a disadvantage of the assays, but, on the other hand, it lowers the risk of false-positive results due to traces of contaminating staphylococcal DNA. However, due to the low sensitivity of the S. aureus-specific PCR, three CSF samples with negative microscopy but cultural detection of S. aureus had false-negative results with the S. aureus-specific PCR assay (Table 5). Nevertheless, in microscopy-positive samples, all specific PCR assays showed a diagnostic sensitivity of 100%, detecting all respective pathogens.
FISH has already been successfully implemented for the rapid detection and differentiation of various pathogens (7, 9, 12). It had, however, not been evaluated on CSF samples before, probably because no probes for N. meningitidis had been available. In this study, a probe for N. meningitidis was newly introduced. Because the previously published probes for E. coli (9) showed rather weak fluorescence in preliminary experiments (data not shown), a new probe with strong binding activity was designed by consulting a publication on the accessibility of the ribosome (4). The newly designed probe showed bright fluorescence and only cross-reacted with Shigella species, which is inevitable, since Shigella and E. coli have identical 16S rRNA sequences. Due to the extreme rareness of Shigella as a cause of meningitis and the similar therapeutic options, this cross-reactivity is not regarded as relevant for the investigation of CSF. Also, with regard to sensitivity, we decided to implement PNA probes for the detection of staphylococci. FISH of staphylococci with DNA probes requires extensive permeabilization treatments and only stained about 50% of the bacterial cells (data not shown). PNA probes penetrate the cell walls more easily, since they are electrically neutral and shorter. In addition to a previously published probe for S. aureus, we designed a new genus-specific probe for staphylococci. This probe has only one mismatch to some Bacillus spp. and therefore cross-reacts with these bacteria. This cross-reactivity was regarded as not relevant, since staphylococci and Bacillus clearly differ in their Gram stain appearance.
The analytical sensitivity of the FISH method was comparable to that of the Gram stain, which had been expected, since both are slide-based techniques.
A eubacterial probe on a PNA base (Bac-Uni1) was chosen for the investigation of microscopy- and culture-negative samples to cover all bacteria, including those with a thick cell wall, like staphylococci. As expected from the experiments on the analytical sensitivity, the method did not allow the recognition of otherwise undetected pathogens. However, it did not produce any false-positive results, stressing the specificity of the FISH method.
When evaluating microscopy- and culture-positive CSF samples by FISH, 13 of 18 samples were judged positive (Table 4; Fig. 2). In the remaining samples, no bacteria were detected, neither with DAPI nor FISH staining. It is possible that the missed bacteria had a low metabolic activity and therefore a low ribosome content, leading to only faint FISH signals. The negative DNA stain, however, argues for the hypothesis that too-low numbers of bacteria were present on these slides. Thus, the main reason for the relatively high number of FISH-negative samples is probably the uneven distribution of bacteria on the different slides. This is further supported by the fact that all positive samples missed by FISH showed only a low concentration of bacteria on the Gram stain. In addition, FISH was positive in three of four samples that were negative by Gram stain but positive by culture (Table 5).
As a conclusion from this work, we propose an approach for the rapid diagnosis of meningitis that combines FISH and PCR based on the Gram stain. FISH is the method of choice for samples in which multiple bacteria are detected by Gram staining. In these cases, FISH allows the identification of the pathogen within 2 h. Advantages of the FISH technique are the low price and the little demands concerning technical equipment compared to PCR, which requires costly reagents and instruments, such as the LightCycler. The low price also makes FISH interesting for small laboratories, even in less industrialized countries. Due to its relatively low diagnostic sensitivity, the FISH method cannot be recommended for the work-up of samples with a low bacterial load that show no or very few bacteria in the Gram stain. Such cases can rapidly be solved by real-time PCR with very high sensitivity and specificity. For cases with low numbers of bacteria on the Gram stain, we propose implementing specific PCR assays according to the result of the Gram stain according to the algorithm used in this study (Table 3). In cases of negative microscopy, the eubacterial PCR may be combined with the specific PCR for N. meningitis to allow a quick decision on therapy and on prophylaxis of contact persons. Thus, two cases of meningococcal meningitis but negative Gram stain results (Table 5) could be detected in this study. The eubacterial PCR allows further identification of bacterial DNA due to sequencing of the PCR product, as shown previously (28) and in this study. Additionally, the eubacterial PCR can be used for the identification of bacteria that are not covered by the specific primers and probes. Taken together, PCR and FISH have the potential to considerably accelerate the diagnosis of bacterial meningitis. To benefit from the different advantages of PCR and FISH, both methods should be combined in a reasonable way.
Acknowledgments
This work was supported by a grant of the University of Ulm.
We gratefully acknowledge the support provided by the director of our institute, R. Marre. We thank Henrik Stendal from AdvanDx for the support with material and practical advice concerning PNA FISH. We greatly acknowledge the helpful expert advice on DNA FISH given by Matthias Horn and Stephan Schmitz-Esser, Department of Microbial Ecology, University of Vienna. We thank Hayrettin Tumani for supplying CSF samples for the evaluation of the analytical sensitivity and specificity of the methods. We thank Syron Belak for performing the ROC analysis.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balganesh, M., M. K. Lalitha, and R. Nathaniel. 2000. Rapid diagnosis of acute pyogenic meningitis by a combined PCR dot-blot assay. Mol. Cell. Probes 14:61-69. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs, B. M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greisen, K., M. Loeffelholz, A. Purohit, and D. Leong. 1994. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J. Clin. Microbiol. 32:335-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall, L. M., B. Duke, and G. Urwin. 1995. An approach to the identification of the pathogens of bacterial meningitis by the polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 14:1090-1094. [DOI] [PubMed] [Google Scholar]
- 7.Hogardt, M., K. Trebesius, A. M. Geiger, M. Hornef, J. Rosenecker, and J. Heesemann. 2000. Specific and rapid detection by fluorescent in situ hybridization of bacteria in clinical samples obtained from cystic fibrosis patients. J. Clin. Microbiol. 38:818-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussein, A. S., and S. D. Shafran. 2000. Acute bacterial meningitis in adults. A 12-year review. Medicine (Baltimore) 79:360-368. [DOI] [PubMed] [Google Scholar]
- 9.Jansen, G. J., M. Mooibroek, J. Idema, H. J. Harmsen, G. W. Welling, and J. E. Degener. 2000. Rapid identification of bacteria in blood cultures by using fluorescently labeled oligonucleotide probes. J. Clin. Microbiol. 38:814-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ke, D., C. Menard, F. J. Picard, M. Boissinot, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2000. Development of conventional and real-time PCR assays for the rapid detection of group B streptococci. Clin. Chem. 46:324-331. [PubMed] [Google Scholar]
- 11.Kearns, A. M., R. Freeman, O. M. Murphy, P. R. Seiders, M. Steward, and J. Wheeler. 1999. Rapid PCR-based detection of Streptococcus pneumoniae DNA in cerebrospinal fluid. J. Clin. Microbiol. 37:3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kempf, V. A., K. Trebesius, and I. B. Autenrieth. 2000. Fluorescent in situ hybridization allows rapid identification of microorganisms in blood cultures. J. Clin. Microbiol. 38:830-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotilainen, P., J. Jalava, O. Meurman, O. P. Lehtonen, E. Rintala, O. P. Seppala, E. Eerola, and S. Nikkari. 1998. Diagnosis of meningococcal meningitis by broad-range bacterial PCR with cerebrospinal fluid. J. Clin. Microbiol. 36:2205-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu, J. J., C. L. Perng, S. Y. Lee, and C. C. Wan. 2000. Use of PCR with universal primers and restriction endonuclease digestions for detection and identification of common bacterial pathogens in cerebrospinal fluid. J. Clin. Microbiol. 38:2076-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manz W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Micrbiol. 1:593-600. [Google Scholar]
- 16.Newcombe, J., K. Cartwright, W. H. Palmer, and J. McFadden. 1996. PCR of peripheral blood for diagnosis of meningococcal disease. J. Clin. Microbiol. 34:1637-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliveira, K., G. W. Procop, D. Wilson, J. Coull, and H. Stender. 2002. Rapid identification of Staphylococcus aureus directly from blood cultures by fluorescence in situ hybridization with peptide nucleic acid probes. J. Clin. Microbiol. 40:247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perry-O'Keefe, H., H. Stender, A. Broomer, K. Oliveira, J. Coull, and J. J. Hyldig-Nielsen. 2001. Filter-based PNA in situ hybridization for rapid detection, identification and enumeration of specific micro-organisms. J. Appl. Microbiol. 90:180-189. [DOI] [PubMed] [Google Scholar]
- 19.Poppert, S., A. Essig, R. Marre, M. Wagner, and M. Horn. 2002. Detection and differentiation of chlamydiae by fluorescence in situ hybridization. Appl. Environ. Microbiol. 68:4081-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reischl, U., H. J. Linde, B. Leppmeier, N. Lehn. 2002. Duplex LightCycler PCR assay for the rapid detection of methicillin-resistant Staphylococcus aureus and simultaneous species confirmation, p. 93-105. In U. Reischl, C. Wittwer, and F. Cockerill (ed.), Rapid cycle real-time PCR. Springer Verlag, Berlin, Germany.
- 21.Richardson, D. C., L. Louie, M. Louie, and A. E. Simor. 2003. Evaluation of a rapid PCR assay for diagnosis of meningococcal meningitis. J. Clin. Microbiol. 41:3851-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saez-Llorens, X., and G. H. McCracken, Jr. 2003. Bacterial meningitis in children. Lancet 361:2139-2148. [DOI] [PubMed] [Google Scholar]
- 23.Saravolatz, L. D., O. Manzor, N. VanderVelde, J. Pawlak, and B. Belian. 2003. Broad-range bacterial polymerase chain reaction for early detection of bacterial meningitis. Clin. Infect. Dis. 36:40-45. [DOI] [PubMed] [Google Scholar]
- 24.Schuchat, A., K. Robinson, J. D. Wenger, L. H. Harrison, M. Farley, A. L. Reingold, L. Lefkowitz, and B. A. Perkins. 1997. Bacterial meningitis in the United States in 1995. Active Surveillance Team. N. Engl. J. Med. 337:970-976. [DOI] [PubMed] [Google Scholar]
- 25.Segreti, J., and A. A. Harris. 1996. Acute bacterial meningitis. Infect. Dis. Clin. N. Am. 10:797-809. [DOI] [PubMed] [Google Scholar]
- 26.Stevens, J. P., M. Eames, A. Kent, S. Halket, D. Holt, and D. Harvey. 2003. Long term outcome of neonatal meningitis. Arch. Dis. Child. Fetal Neonatal Ed. 88:F179-F184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trebesius, K., L. Leitritz, K. Adler, S. Schubert, I. B. Autenrieth, and J. Heesemann. 2000. Culture independent and rapid identification of bacterial pathogens in necrotising fasciitis and streptococcal toxic shock syndrome by fluorescence in situ hybridisation. Med. Microbiol. Immunol. (Berlin) 188:169-175. [DOI] [PubMed] [Google Scholar]
- 28.Trotha, R., T. Hanck, W. Konig, and B. Konig. 2001. Rapid ribosequencing—an effective diagnostic tool for detecting microbial infection. Infection 29:12-16. [DOI] [PubMed] [Google Scholar]
- 29.van Haeften, R., S. Palladino, I. Kay, T. Keil, C. Heath, and G. W. Waterer. 2003. A quantitative LightCycler PCR to detect Streptococcus pneumoniae in blood and CSF. Diagn. Microbiol. Infect. Dis. 47:407-414. [DOI] [PubMed] [Google Scholar]
- 30.Wellinghausen, N., B. Wirths, A. R. Franz, L. Karolyi, R. Marre, and U. Reischl. 2004. Algorithm for the identification of bacterial pathogens in positive blood cultures by real-time LightCycler polymerase chain reaction (PCR) with sequence-specific probes. Diagn. Microbiol. Infect. Dis. 48:229-241. [DOI] [PubMed] [Google Scholar]