Abstract
In 1996, a disease outbreak occurred at a captive breeding facility in Idaho, causing anorexia, dehydration, and diarrhea or sudden death in 72 of 110 Northern aplomado falcons (Falco femoralis septentrionalis) from 9 to 35 days of age and in 6 of 102 peregrine falcons (Falco peregrinus) from 14 to 25 days of age. Sixty-two Northern aplomado and six peregrine falcons died. Epidemiologic analyses indicated a point source epizootic, horizontal transmission, and increased relative risk associated with cross-species brooding of eggs. Primary lesions in affected birds were inclusion body hepatitis, splenomegaly, and enteritis. The etiology in all mortalities was determined by molecular analyses to be a new species of adenovirus distantly related to the group I avian viruses, serotypes 1 and 4, Aviadenovirus. In situ hybridization and PCR demonstrated that the virus was epitheliotropic and lymphotropic and that infection was systemic in the majority of animals. Adeno-associated virus was also detected by PCR in most affected falcons, but no other infectious agents or predisposing factors were found in any birds. Subsequent to the 1996 epizootic, a similar disease caused by the same adenovirus was found over a 5-year period in orange-breasted falcons (Falco deiroleucus), teita falcons (Falco fasciinucha), a merlin (Falco columbarius), a Vanuatu peregrine falcon (Falco peregrinus nesiotes), and gyrfalcon × peregrine falcon hybrids (Falco rusticolus/peregrinus) that died in Wyoming, Oklahoma, Minnesota, and California. These findings indicate that this newly recognized adenovirus is widespread in western and midwestern North America and can be a primary pathogen in different falcon species.
Adenoviruses are diverse pathogens that generally exhibit low levels of virulence and have host ranges limited to one or several closely related species. Adenovirus infection has been identified by ultrastructural or molecular methods in fish, birds, reptiles, mammals, and amphibia, and virus has been isolated from at least 40 vertebrate species (2, 11, 19, 38, 53). Most natural infections are subclinical or manifest mild, transient signs limited to the intestinal, renal, ocular, or respiratory systems, and significant illness develops only in conjunction with other viral or bacterial pathogens, with toxin exposure, or in immunocompromised individuals (32, 40, 47). Occasionally, however, malefic disease outbreaks caused by emergent viral strains, cross-species transmission, or high-dose infection of young, naive animals occur. In these cases, systemic inflammation and tissue damage develop, and fatality rates can reach 70 to 90% (18, 21).
Prior to 2003, the family Adenoviridae was divided by host range and antibody reactivity into two genera: the genus Mastadenovirus for mammalian adenoviruses and the genus Aviadenovirus for avian viruses (8). Now, four genera (Mastadenovirus, Aviadenovirus, Atadenovirus, and Siadenovirus) have been proposed, and viruses have been provisionally reclassified based on nucleotide and predicted amino acid similarities of homologous genes (2, 11, 12). Within Mastadenovirus and Aviadenovirus, viruses have been further subdivided into subgenera and groups based on shared neutralizing epitopes (2, 19). In most cases, these serotype groupings closely follow patterns of tissue tropism and pathogenicity (2, 30, 40). For example, in humans, 51 serotypes of adenovirus have been identified and classified into six subgenera. Subgenus B is most virulent and is associated with cardiopulmonary disorders in infants and urinary tract infections in adults; subgenera A and F cause mild gastroenteritis; and subgenus D has affinity for the eye and is frequently isolated in association with AIDS (10).
For birds, detailed molecular and cellular studies comparable to analyses with humans have been done on the type species of group I (fowl adenovirus 1 [FAV-1] of chickens), group II (hemorrhagic enteritis virus [HEV] of turkeys), and group III (egg drop syndrome [EDS] virus of chickens) viruses of poultry (8, 37, 40). FAV-1, which includes the chicken embryo lethal orphan (CELO) strain, is probably the best characterized of any avian adenovirus and has been shown experimentally to have features similar to many human adenoviruses, including ligand activity for the human coxsackievirus-adenovirus receptor (CAR) and production of a peptide that inhibits apoptosis by binding the human retinoblastoma protein (28, 49). In chickens, FAV-1 is tropic for hepatocytes, pancreatic acinar cells, and gizzard epithelium and is typically an opportunistic pathogen, causing disease in young animals that are coinfected with more aggressive agents or otherwise immunosuppressed (8, 19, 40). However, it can by itself produce severe systemic illness and death in some galliform species, such as quail and turkeys (18, 21, 22). EDS virus and HEV similarly display host-dependent levels of virulence. EDS virus induces little or no disease in waterfowl but causes significant reproductive abnormalities in chickens, while HEV is nonpathogenic in pheasants and produces life-threatening enteritis in turkeys and guinea fowl (9, 37, 40).
Falcons are birds of prey comprising 38 species and several subspecies in the genus Falco. Hybrid birds have been developed, usually by falconers, from captive breeding of different species and exist throughout North America and Europe. Most natural populations of falcons were decimated in the early 20th century by loss of habitat, pesticide exposure, poachers, and illness (5, 54). Because of the scarcity of these animals, little is known about etiologies of naturally occurring diseases. In this study, we characterize the epidemiology, pathology, and gene sequences of a novel adenovirus that caused high morbidity and mortality in a large group of Northern aplomado falcons (Falco femoralis septentrionalis) and sporadic deaths in three other falcon species, a peregrine falcon subspecies, and peregrine × gyrfalcon hybrid falcons in the United States.
MATERIALS AND METHODS
Epidemiology of the Northern aplomado falcon 1996 epizootic.
In 1996, the Peregrine Fund World Center for Birds of Prey in Boise, Idaho, was a closed captive rearing facility for raptorial species, including Northern aplomado falcons (Falco femoralis septentrionalis) and peregrine falcons (Falco peregrinus). The goal of the center was to raise threatened or endangered raptors for reintroduction into native habitats. All adult falcons at the facility were separated from new hatches, which were maintained in a brooder room and hand reared. Northern aplomado and peregrine falcons were always kept separately, except for the use of peregrine falcons to brood some aplomado falcon eggs for a portion of the hatching season. All falcons were fed eviscerated domestic chicken (Gallus gallus) and quail (Coturnix coturnix japonica). Domestic pigeons (Columba livia) were used in rural areas adjacent to the World Center by several falconers who may have had access to the falcon brooder room around the time of the outbreak. In the spring of 1996, 110 Northern aplomado falcons were hatched at the Peregrine Fund World Center for Birds of Prey in Boise, Idaho. These animals comprised the study population used for epidemiologic analyses of a disease outbreak that affected 72 Northern aplomado falcons in June 1996. Aplomado falcons were considered affected by the epizootic if they exhibited anorexia, diarrhea, or sudden death or had characteristic viral lesions upon postmortem examination. An epidemic histogram was constructed that compared the number of dead birds to the date of death. The median incubation period (MIP) was determined from the time of the index case until the median time of occurrence of the primary cluster of cases in the epizootic histogram. A second estimate of the MIP was obtained by calculating the median date of occurrence of the primary cases and the median date of occurrence of the secondary cases. Case fatality was determined as a measure of disease occurrence among aplomado falcons and was the proportion of clinically affected aplomado falcons that died before 90 days of age. An attack rate table was constructed to determine the relative risk of aplomado falcons affected by the epizootic when the incubating species was a Northern aplomado falcon or a peregrine falcon.
Animals.
Ninety-seven animals had tissues analyzed by PCR. Sixty-two were Northern aplomado falcons, and six were peregrine falcons that died in the June 1996 outbreak at the World Center in Boise, Idaho. Nine were domestic chickens and seven were quail used as food for Northern aplomado and peregrine falcons at the World Center during the 1996 outbreak. Four were domestic pigeons used by falconers in regions adjacent to the World Center during the 1996 outbreak. Two animals were adult orange-breasted falcons (Falco deiroleucus) from a private facility in Wyoming which also housed other raptors. These two falcons died in August 1997. One animal was an adult peregrine falcon subspecies, known as a Vanuatu peregrine falcon (Falco peregrinus nesiotes), from a private home in San Diego, Calif., that also housed peregrine falcons. This animal died in December 2001. Two animals were juvenile teita falcons (Falco fasciinucha), and two juvenile animals were gyrfalcon × peregrine falcon hybrids (Falco rusticolus/peregrinus) that were cohoused in a facility in Oklahoma and died in February 2002. One animal was a juvenile merlin (Falco columbarius) from Minnesota that died in April 2003. Two animals were adult Northern aplomado falcons that died in 2002 and were from the San Diego Zoo's Wild Animal Park in Escondido, Calif.
Postmortem examination.
Complete necropsies were performed on all 97 animals. Histology was done on all animals except the teita falcons, gyrfalcon × peregrine falcons, and merlin. Samples of all organs from the other 93 animals were immersion fixed in 10% neutral buffered formalin, routinely processed, embedded in paraffin, sectioned, stained with hematoxylin and eosin (HE) for histology, and examined by board-certified veterinary pathologists. The severity of microscopic lesions was scored from 0 to 4, with 0 representing no lesions, 1 representing minimal changes (≤2 lesions per 20 ×400 fields), 2 representing mild changes (3 to 6 lesions per 20 ×400 fields), 3 representing moderate changes (7 to 10 lesions per 20 ×400 fields), and 4 representing severe changes (≥11 lesions per 20 ×400 fields). A lesion was defined as the presence of necrosis, inflammation, lymphoid atrophy, or hyperplasia. The presence and number of intranuclear inclusions were not part of the lesion scoring system. In addition to samples for histology, unfixed portions of liver, spleen, lung, brain, intestine, kidney, heart, and bone marrow from 24 Northern aplomado falcons, 4 peregrine falcons, the Vanuatu peregrine falcon, 9 chickens, 7 quail, and 4 pigeons were frozen at −80C for DNA and RNA extractions.
Eggs.
Sixteen unhatched Northern aplomado falcon eggs and six unhatched peregrine falcon eggs were harvested from brooder rooms at the World Center during the June 1996 epizootic and were frozen at −80C for molecular studies.
Poultry adenovirus serotypes.
Frozen or lyophilized tissue culture lysates for each of the 12 chicken serotypes (serotypes 1 to 12) of group I adenovirus were obtained from S. Dhillon, Avian Health and Food Safety Laboratory, Puyallup, Wash., and the SPAFAS Charles River Diagnostics Laboratories (Storrs, Conn.) for comparative molecular analyses.
DNA and RNA extractions and cDNA synthesis.
DNA was extracted from frozen tissues, eggs, and poultry adenovirus lysates for all animals by using the QIAGEN tissue kit according to the manufacturer's tissue sample protocol, except that the recommended amounts of sample were first placed with the lysis buffer in 1.5-ml screw-cap FastPrep vials containing ceramic beads and lysed by agitation in a FastPrep shaker (Q-BIOgene, Carlsbad, Calif.) at a speed of 4 to 5.5 for 40 to 60 s, after which the lysate was transferred to a clean Eppendorf tube for continuation of the QIAGEN protocol. DNA was extracted from formalin-fixed, paraffin-embedded samples of liver, lung, spleen, and intestine from those animals for which frozen tissues were not available by using the same protocol as above, except that samples were first deparaffinized with successive washes in octane, xylene, and 100% ethyl alcohol and then air dried. Total RNA was extracted from frozen tissues from Northern aplomado and peregrine falcons and the Vanuatu peregrine falcon by using TRIzol (Invitrogen Life Technologies, Carlsbad, Calif.). cDNA was synthesized from DNase-treated total RNA using the Superscript System (Invitrogen Life Technologies) with random hexamers except for the Pneumovirinae PCR listed below.
PCR.
Seventeen PCR protocols were performed on DNA from frozen and paraffin-embedded tissues or cDNA. The PCR protocols and expected sizes of the amplicons are listed below. For all reactions, 50 to 500 nanograms of DNA was added to a 25-μl reaction mixture containing 10 mM Tris (pH 8.0), 50 mM KCl, 5 mM MgCl2, 200 μM each of dATP, dCTP, dGTP, and dTTP, 50 picomoles of each primer, and AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA) at a final concentration of 0.05 U/μl. PCRs were used to amplify the following regions. (i) For the avian adenovirus 3′ hexon region, oligonucleotide primers to conserved pedestal areas at the 3′ region of the hexon gene were used as previously described (36) with several modifications. The sense primer (AACGTCAAYCCCTTCAACCACC) and antisense primer (TTGCCTGTGGCGAAAGGCG) were used at 95°C for 6 min, followed by 40 cycles of 95°C for 45 seconds, 48°C for 1 min, and 72°C for 1 min, and then 72°C for 8 min to generate a 1.3-kb product. (ii) For the avian adenovirus 5′ hexon region, a sense primer (GIGGRCCITCITTYAARCC) to a conserved pedestal area at the 5′ region of the hexon gene and an antisense primer (GTAAGTAACCAGATCGAAGGTG) specific for the falcon adenovirus 3′ hexon region were used at 95°C for 6 min, followed by 40 cycles of 95°C for 45 seconds, 52°C for 1 min, and 72°C for 2 min, and then 72°C for 8 min to generate a 2-kb product. (iii) For the avian adenovirus penton base, the sense primer (TRGGHGTYYARKTKGGGKKB) and antisense primer (GGNADADNADBTBCRYRCHAYVAC) were used at 95°C for 6 min, followed by 40 cycles of 95°C for 45 seconds, 51°C for 1 min, and 72°C for 1 min, and then 72°C for 8 min to generate a 608-bp product. (iv) For avian adenovirus DNA polymerase, the sense primer (VHHBRTBTCRCAWTCVMMBRSCC) and the antisense primer (GWCVMGRGCMTTYDYVWSHGAGTGG) were used at 95°C for 6 min, followed by 40 cycles of 95°C for 45 seconds, 52°C for 30 seconds, and 72°C for 1 min, and then 72°C for 8 min to generate a 237-bp product. (v) For the falcon adenovirus-specific penton base/hexon intervening region, the sense primer (CAATGTAGACGATGAAGACGGAGAG) and antisense primer (CACTGTAAGGCTTGAAAGACGGT) were used at 95°C for 6 min, followed by 45 cycles of 95°C for 45 seconds, 53°C for 1 min, and 72°C for 2 min, and then 72°C for 8 min to generate a 2.7-kb product. (vi) For the falcon adenovirus-specific hexon region, the sense primer (AAGAAACACCACAACAGGG) and the antisense primer (GTAAGTAACCAGATCGAAGGTG) were used at 95°C for 6 min, followed by 35 cycles of 95°C for 45 seconds, 54°C for 1 min, and 72°C for 1 min, and then 72°C for 8 min to generate a 385-bp product. (vii) For Herpesviridae, nested degenerate primers that target consensus regions of the DNA polymerase gene of all herpesviruses were used as previously described (51) to generate a 225-bp product. (viii) For Circoviridae, nested degenerate primers that target consensus regions of the rep protein for all circoviruses were used as previously described (29) to generate an approximately 240-bp product. (ix) For Coronaviridae, primers that target a conserved area of the polymerase gene of all coronaviruses were used as previously described (27) to generate a 405-bp product. (x) For Pneumovirinae, one set of primers that target consensus regions of the fusion protein of all Newcastle disease viruses and a second set of primers that target the matrix protein of avian pneumovirus were used as previously described (1) to generate 309- and 631-bp products, respectively. (xi) For Parvovirinae, primers that target consensus areas of the capsid protein for parvoviruses, including adeno-associated virus (AAV) or dependovirus, were used as previously described (16) to generate a 255-bp product. (xii) For Avipolyomavirus, primers that target the t/T antigen region of avian polyomavirus were used as previously described (24) to generate a 310-bp product. (xiii) For Influenzavirus A, primers that target consensus regions of the matrix protein of all influenza A viruses were used as previously described (14) to generate a 244-bp product. (xiv) For Salmonella, primers that target conserved regions of a sipB-sipC gene fragment of all Salmonella spp. were used as previously described (44) to generate a 250-bp product. (xv) For Campylobacter, primers that target conserved areas of the 16S rRNA gene were used as previously described (34) to generate a 297-bp product. (xvi) For Mycoplasma, primers that target conserved regions of the 16S rRNA gene were used as previously described (52) to generate a 210-bp product. (xvii) For beta-actin, the sense primer (CCCCCGTGCTGTGTTCCCATCTATCG) and antisense primer (GGGTGCTCCTCAGGGGCTACTCTCAG) were used at 95°C for 6 min, followed by 35 cycles of 95°C for 45 seconds, 58°C for 1 min, and 72°C for 1 min, and then 72°C for 8 min to generate a 450-bp product.
DNA manipulation and sequencing.
PCR products of the expected sizes from representative tissues and cases were purified and either directly sequenced or cloned using the TOPO TA cloning kit (Invitrogen). Sequencing reactions were performed using the CEQ DTCS (dye terminator cycle sequencing) Quick-Start kit (Beckman Coulter, Fullerton, CA). Sequences were acquired using a CEQ 2000XL capillary sequencer (Beckman Coulter). Sequence analysis and alignments were conducted using the MacVector v. 7.0 and AssemblyLIGN v. 1.0.9 software packages (Accelrys, San Diego, CA). Sequence data were compared to the GenBank database using the basic local alignment search tool.
Southern hybridization and detection.
Southern blotting of all PCR products to positively charged nylon membranes (Amersham Pharmacia Biotech, Piscataway, NJ, or Millipore, Bedford, MA) was performed with a PosiBlot pressure apparatus (Stratagene, La Jolla, CA). DNA was fixed to the membrane using a Stratalinker (Stratagene, La Jolla, CA). DNA probes were generated by labeling with digoxigenin using the PCR DIG Probe Synthesis kit (Roche Molecular Biochemicals, Indianapolis, IN) and specific oligomers or PCR-amplified plasmids containing cloned and sequenced gene segments. Hybridization and detection were performed by using reagents of the DIG High Prime DNA Labeling and Detection kit (Roche Molecular Biochemicals, Indianapolis, IN) and exposing the treated blots to Kodak X-Omat LS X-ray film (Eastman Kodak Co., Rochester, NY).
In situ hybridization.
Sections (5 μm) of selected formalin-fixed, paraffin-embedded tissues on Platinum slides (Mercedes Medical, Sarasota, FL) were deparaffinized, hydrated, treated with trypsin Digest-All (Zymed Laboratories, South San Francisco, Calif.) at 37°C for 10 min, washed in Tris-buffered saline (TBS), heated at 98°C for 12 min in TBS, placed immediately in 4°C TBS, and prehybridized with Dig Easy Hyb Granule solution (Roche Diagnostics Corp., Indianapolis, IN) at 42°C for 1 h. A 385-bp cloned and sequenced segment of the falcon adenovirus hexon gene obtained from the falcon adenovirus-specific hexon region PCR was labeled with digoxigenin using an incorporation method (Roche Diagnostics Corp.) and used in Dig Easy Hyb Granule solution for hybridization at 42°C for 16 h at 50 picomoles of labeled probe per ml of hybridization solution. Slides were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 0.1% sodium dodecyl sulfate (SDS) twice for 5 min at room temperature (RT), followed by two washes in 0.5× SSC with 0.1% SDS at 68°C for 15 min. Slides were blocked in blocking solution (Roche Diagnostics Corp.) for 30 min, washed in TBS, treated with anti-digoxigenin antibody (Roche Diagnostics Corp.) diluted 1:5,000 (150 mU/ml) in TBS for 1 h at RT, washed in TBS, treated with nitroblue tetrazolium chloride-5-bromo-4-chloro-3-indolylphosphate, toluidine salt (NBT/BCIP) (Roche Diagnostics Corp.) for 10 min, washed in TBS and distilled water, counterstained with Gill's hematoxylin (Surgipath Instrumentation Inc., Richmond, IL), and mounted with crystal mount (Biomeda Corp., Foster City, CA). Duplicate control slides received an identical treatment except that no labeled probe was added to the hybridization solution. For selected sections, this protocol was used after immunohistochemical staining for T lymphocytes as described below.
Immunohistochemistry.
Sections (5 μm) of selected formalin-fixed, paraffin-embedded tissues on Platinum slides (Mercedes Medical) were deparaffinized and hydrated. For apoptotic DNA, slides were treated with mouse anti-human single-stranded DNA (Chemicon International Corp., Temecula, CA) after formamide denaturation as described elsewhere (15). For CD3 staining, slides were heated to 98°C in 0.1 M citrate buffer (pH 6.0) for 20 min, allowed to cool to RT, blocked for endogenous peroxidase with Peroxo-block (Zymed Laboratories), washed in phosphate-buffered saline (PBS), and blocked with Cas Block (Zymed Laboratories) for 30 min at RT. Slides were treated with a culture supernatant of a rat anti-CD3ɛ monoclonal antibody that is routinely used to detect T lymphocytes in animal tissues (a kind gift from P. F. Moore, University of California, Davis) diluted 1:10 in PBS at RT for 1 h, washed, treated with a biotinylated goat anti-rat antibody (Zymed Laboratories) at a 1:400 dilution in PBS for 30 min at RT, washed, treated with streptavidin-peroxidase (Zymed Laboratories) at a 1:400 dilution in PBS for 30 min at RT, and washed. Assessment of cross-reactivity of the CD3 and CD79 antibodies was done by observing specific staining patterns in normal intestine, spleen, thymus, lymph node, brain, liver, and skin. Control slides did not receive primary antibody. All slides were treated with diaminobenzidine tetrahydrochloride (DAB) (Zymed Laboratories) for 1 to 5 min at RT, washed, dehydrated, and mounted.
Phylogenetic analyses.
Sequences were aligned with the MacVector ClustalW program (Accelrys). Phylogenetic relationships were estimated by using the program Paup 4.0 (48) with the neighbor-joining and maximum-likelihood methods with the Kimura-2 parameter. Bootstrap values were counted as percentages over 1,000 replicates.
Nucleotide sequence accession numbers.
Twenty-one sequences were submitted to GenBank. The 6.2-kb fragment of the falcon adenovirus obtained from Northern aplomado falcons has GenBank accession bankit 642046; the 240-bp fragment of the falcon adenovirus polymerase gene has GenBank accession bankit 646116; the 1.2-kb fragments of the chicken adenoviruses have GenBank accession bankit 631751 for FAV-1, bankit 631761 for FAV-2, bankit 642906 for FAV-3, bankit 642908 for FAV-4, bankit 631767 for FAV-5, bankit 642912 for FAV-6, bankit 631773 for FAV-7, bankit 631775 for FAV-8, bankit 631777 for FAV-9, bankit 642924 for FAV-10, bankit 631783 for FAV-11, and bankit 642934 for FAV-12; the 350-bp fragment of the adenovirus hexon region from teita falcons, a merlin, and gyrfalcon × peregrine falcon hybrids has GenBank accession bankit 631785; the 350-bp fragment of the adenovirus hexon region from orange-breasted falcons, Northern aplomado falcons, peregrine falcons, and a Vanuatu peregrine falcon has GenBank accession bankit 642936; the 201-bp fragment of the adeno-associated viruses has GenBank accession bankit 642940 for peregrine falcons and orange-breasted falcons, bankit 631787 for teita falcons and gyrfalcon × peregrine falcons, bankit 642944 for the merlin, and bankit 631789 and bankit 642946 for the two Northern aplomado falcon strains.
RESULTS
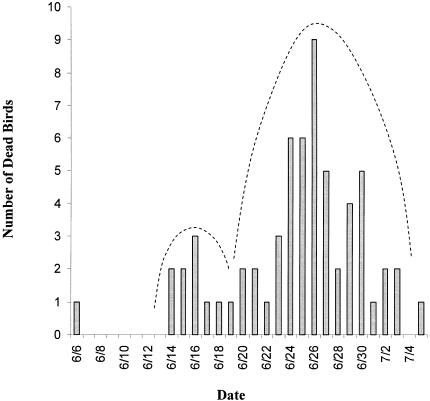
In June 1996, a disease outbreak lasting approximately 33 days occurred at the Peregrine Fund World Center for Birds of Prey in Boise, Idaho. Seventy-two of 110 Northern aplomado falcon hatchlings were affected. Clinical signs consisted of anorexia (18%), diarrhea (15%), anorexia and diarrhea (28%), or sudden death (31%). The index case was a 22-day-old Northern aplomado falcon hatchling that became ill and died on 6 June 1996. A primary cluster of 10 cases followed by a secondary cluster developed over the next 5 weeks (Fig. 1). The MIPs for the first and second cluster of cases were 11 and 10 days, respectively. Sixty-two Northern aplomado falcons died in the outbreak. The median age at death was 20 days (range, 6 to 40 days), and the case fatality rate was 86.1%. The attack rate for aplomado falcons incubated by peregrine falcons was 85.7% compared to 59.3% for aplomado falcons incubated by aplomado falcons. The relative risk was 1.5, indicating that birds incubated by peregrine falcons were 1.5 times more likely to become infected than those incubated by aplomado falcons. During the June 1996 outbreak, six hatchling peregrine falcons aged 14 to 25 days also became ill and died at the World Center. Subsequent to and unrelated to the World Center outbreak, two adult orange-breasted falcons in Wyoming, two juvenile teita falcons, two juvenile gyrfalcon × peregrine falcons, and a juvenile merlin in a bird-of-prey facility in Minnesota, and an adult Vanuatu peregrine falcon from a private home in San Diego, Calif., became ill with diarrhea and died.
FIG. 1.
Epizootic histogram of the 1996 adenovirus outbreak in Northern aplomado falcons showing point source followed by two clusters of mortalities.
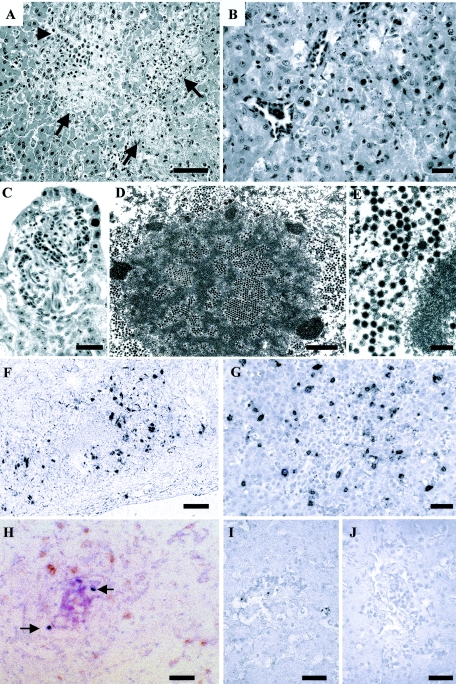
The primary lesion in affected falcons was multifocal random coagulation necrosis and inflammation in the liver. Necrotic foci ranged from one or several hepatocytes to 100- to 300-μm-diameter areas and had variable numbers of peripheral inflammatory cell infiltrates comprised chiefly of mononuclear leukocytes. Aggregates of inflammatory cells were also adjacent to portal areas, some of which had necrosis of the limiting plate. Intact hepatocytes along the borders of necrotic areas, the biliary epithelium, as well as, in some cases, hepatocytes within normal parenchyma had karyomegaly with peripherally marginated chromatin and intranuclear basophilic or eosinophilic inclusion bodies in many birds. Transmission electron microscopy demonstrated 70- to 90-nm nonenveloped icosahedral viral particles occasionally arranged into paracrystalline arrays (Fig. 2). Multifocal necrosis and inflammation with mucosal epithelial intranuclear inclusion bodies were also present in the small intestines in several cases and were occasionally associated with hemorrhages. Inflammation and necrosis were less commonly present in the lung, kidney, pancreas, and trachea and were usually associated with intranuclear inclusion bodies. The brains of several animals had mild inflammation but no necrosis or inclusion bodies. The mucosal epithelium of the ventriculus often had inclusion bodies but no other changes.
FIG. 2.
Photomicrographs. (A) Northern aplomado falcon with multifocal hepatic coagulation necrosis (arrows) and mononuclear leukocyte infiltration. Arrowhead indicates bile ductule. The sample was stained with HE. Bar, 198 μm. (B) Peregrine falcon liver with karyomegaly, intranuclear inclusions, and hepatocellular necrosis characterized by cell separation and karyorrhexis. HE staining was used. Bar, 96 μm. (C) Northern aplomado falcon small intestine with intranuclear inclusions and cellular degeneration. HE staining was used. Bar, 96 μm. (D) Liver cell from Northern aplomado falcon with intranuclear adenovirus particles arranged into arrays, clusters of viral particles that have distended the nuclear membrane, and peripheral margination of chromatin, viewed under a transmission electron microscope. Bar, 1.62 μm. (E) Northern aplomado falcon liver nucleus with icosahedral adenovirus particles (diameter, 70 to 90 nm) with electron-dense nucleoid, viewed under a transmission electron microscope. Bar, 185 nm. (F) In situ hybridization of Northern aplomado falcon liver for adenovirus by use of NBT/BCIP with Mayer's hematoxylin counterstain. Bar, 164 μm. (G) Immunostaining for CD3 in an area of inflammation and necrosis of a Northern aplomado falcon liver by using DAB with Mayer's hematoxylin counterstain. Bar, 96 μm. (H) In situ hybridization for falcon adeonovirus with NBT/BCIP (pink) and immunostaining for CD3 with DAB (brown) in a Northern aplomado falcon liver. Arrows show double-stained cells (Mayer's hematoxylin counterstain). Bar, 78 μm. (I and J) Peregrine falcon liver immunostaining (with DAB and Mayer's hematoxylin counterstain) for apoptotic DNA showing small amounts of positive reactivity (I) and no reactivity (J). Bar, 164 μm.
Splenomegaly was a consistent finding and was characterized by expansion of perivascular and parenchymal reticular or histiocytic cells. Variable levels of lymphoid atrophy and lympholysis accompanied the histiocytic hyperplasia. Intranuclear inclusion bodies were present in mononuclear cells resembling lymphocytes but were infrequent. The bursa often had moderate to severe lymphoid atrophy and lympholysis. Intranuclear inclusion bodies were present in overlying mucosal epithelium but were not seen in lymphocytes. Several Northern aplomado falcon chicks with splenic and bursal lymphoid atrophy also had multifocal thrombosis, foci of heterophils in parenchymal organs, and numerous intranuclear inclusions and karyomegaly in the liver but no other hepatic changes.
In addition to the hepatic, enteric, and splenic lesions described above, the Vanuatu falcon had multifocal arteritis, thrombosis, and chronic systemic arteriosclerosis.
Mean scores of the severity of microscopic lesions (necrosis, inflammation, lymphoid atrophy, or hyperplasia) and the presence of viral inclusion bodies in all organs for Northern aplomado, peregrine, orange-breasted, and Vanuatu peregrine falcons are listed in Table 1.
TABLE 1.
Summary of scored microscopic lesions and PCR results for falconsa,b
| Organ | Microscopic-lesion score
|
No. of birds with intranuclear inclusion bodies/total no.
|
No. of PCR-positive birds/total no.
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Falcon adenovirus-specific hexon region
|
Parvovirinae
|
All other pathogens
|
||||||||||||||||||
| Apl | Per | OB | V | Apl | Per | OB | V | Apl | Per | OB | V | Apl | Per | OB | V | Apl | Per | OB | V | |
| Liver | 2.82 | 3.37 | 1.5 | 3 | 48/62 | 5/8 | 2/2 | 0/1 | 51/62 | 5/8 | 1/2 | 1/1 | 39/62 | 3/8 | 2/2 | 0/1 | 0/62 | 0/8 | 0/2 | 0/1 |
| Spleen | 3.11 | 3.33 | 2 | 2 | 34/62 | 2/8 | 0/2 | 0/1 | 13/14 | 2/3 | 0/2 | 1/1 | 5/14 | 2/3 | 2/2 | 1/1 | 0/14 | 0/3 | 0/2 | 0/1 |
| Bursa of Fabricius | 2.48 | 1.67 | NA | NA | 15/62 | 0/8 | NA | NA | NE | NE | NA | NA | NE | NE | NA | NA | NE | NE | NA | NA |
| Intestine | 0.72 | 0.83 | 4 | 0 | 22/62 | 1/8 | 0/2 | 0/1 | 8/15 | 3/6 | 1/2 | 0/1 | 6/15 | 2/8 | NE | 1/1 | 0/15 | 0/8 | NE | 0/1 |
| Lung | 0.62 | 0.37 | 0 | 1 | 5/62 | 0/8 | 0/2 | 0/1 | 9/10 | 0/1 | NE | NE | 5/10 | 1/1 | NE | 0/1 | 0/10 | 0/1 | NE | 0/1 |
| Kidney | 0.45 | 0.17 | 0 | 1 | 10/62 | 1/8 | 1/2 | 1/1 | 10/13 | 0/2 | NE | NE | 5/13 | 1/2 | NE | NE | 0/13 | 0/2 | NE | NE |
| Pancreas | 0.38 | 0.25 | 0 | 0 | 3/62 | 0/8 | 0/2 | 0/1 | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE |
| Trachea | 0.14 | 0 | 0 | 0 | 3/62 | 0/8 | 0/2 | 0/1 | 1/1 | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE |
| Brain | 0.04 | 0 | 0 | 0 | 0/62 | 0/8 | 0/2 | 0/1 | 7/9 | 0/8 | NE | NE | 3/9 | 0/8 | NE | NE | 0/9 | 0/8 | NE | NE |
| Bone marrow | 0 | 0 | 0 | 0 | 0/62 | 0/8 | 0/2 | 0/1 | 9/12 | NE | NE | NE | 3/12 | NE | NE | NE | 0/12 | NE | NE | NE |
| Heart | 0 | 0 | 0 | 0 | 0/62 | 0/8 | 0/2 | 0/1 | 0/3 | 0/2 | NE | NE | 0/3 | 0/2 | NE | NE | 0/3 | 0/2 | NE | NE |
| Skin | 0 | 0 | 0 | 0 | 0/62 | 0/8 | 0/2 | 0/1 | 3/3 | NE | NE | NE | 0/3 | NE | NE | NE | 0/3 | NE | NE | NE |
| Eye | 0 | 0 | 0 | 0 | 0/62 | 0/8 | 0/2 | 0/1 | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE |
| Esophagus | 0 | 0 | 0 | 0 | 0/62 | 0/8 | 0/2 | 0/1 | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE |
| Crop | 0 | 0 | 0 | 0 | 0/62 | 0/8 | 0/2 | 0/1 | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE |
| Ventriculus | 0 | 0 | 0 | 0 | 17/62 | 4/8 | 1/2 | 0/1 | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE |
| Proventriculus | 0 | 0 | 0 | 0 | 0/62 | 0/8 | 0/2 | 0/1 | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE |
| Thymus | 0 | 0 | 0 | 0 | 0/62 | 0/8 | 0/2 | 0/1 | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE |
| Adrenal gland | 0 | 0 | 0 | 0 | 0/62 | 0/8 | 0/2 | 0/1 | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE |
| Thyroid gland | 0 | 0 | 0 | 0 | 0/62 | 0/8 | 0/2 | 0/1 | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE |
| Reproductive tract | 0 | 0 | 0 | 0 | 0/62 | 0/8 | 0/2 | 0/1 | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE | NE |
| Egg | NE | NE | NA | NA | NE | NE | NA | NA | 0/16 | 0/6 | NA | NA | 0/16 | 0/6 | NA | NA | 0/16 | 0/6 | NA | NA |
Except for teita falcons, gyrfalcons, and merlin.
Abbreviations: Apl, Northern aplomado falcon; Per, peregrine falcon; OB, orange-breasted falcon; V, Vanuatu falcon; NA, not applicable; NE, not examined.
In situ hybridization using a probe specific for the falcon adenovirus hexon gene demonstrated virus in hepatocytes and biliary epithelium and scattered positive cells in the mucosal epithelium of the ventriculus and small intestine, renal tubular and parabronchiolar epithelium, splenic mononuclear leukocytes, and pancreatic acinar cells. Intravascular leukocytes and adipocytes were infrequently positive. Positive staining was consistently intranuclear, except in some hepatocytes which also had detectable levels of virus in the cytoplasm (Fig. 2).
Immunohistochemistry for CD3 showed that inflammatory cell infiltrates in hepatic, brain, and pulmonary lesions were comprised of approximately 25 to 50% T lymphocytes. No increases in T lymphocyte numbers were seen in intestinal lesions. T-cell-dependent areas of the spleen were decreased in some animals. Immunohistochemistry for CD3 followed by in situ hybridization for the falcon adenovirus demonstrated that many T lymphocytes within hepatic lesions and some within vessels in the lamina propria of the small intestine had intranuclear virus. Immunostaining for apoptotic DNA indicated that only a small proportion (less than 10%) of the cell death and destruction in hepatic and intestinal lesions was caused by apoptosis, whereas approximately 30 to 50% of lytic cells in the spleen and bursa of Fabricius stained for apoptotic DNA. Scattered positive nuclei or nuclear remnants were present in the kidney, and no apoptosis was seen in the brain, lung, or pancreas (Fig. 2).
PCRs using degenerate primers for conserved regions of the avian adenovirus hexon, penton base, and polymerase genes and primers specific for the falcon hexon/penton base were done on tissues from eight Northern aplomado falcons. Assembly of sequence data from cloned amplicons produced a 6.2-kb contiguous fragment containing the penton base, core 1and 2, pVI, and hexon genes and a 240-bp fragment of the polymerase gene. Sequences were identical in all eight animals and were used to develop the falcon adenovirus-specific hexon region PCR, which generated 385-bp amplicons that were cloned and sequenced from an additional six Northern aplomado falcons, four peregrine falcons, two orange-breasted falcons, the Vanuatu peregrine falcon, two teita falcons, two gyrfalcon × peregrine hybrid falcons, and a merlin. Sequence analyses of the hexon regions from these birds showed the presence of two genotypes with 98.7% identity. The teita falcons, gyrfalcon × peregrine falcons, and merlin had one hexon genotype, while the Northern aplomado falcons, peregrine falcons, orange-breasted falcons, and Vanuatu peregrine falcon had a different genotype. All animals in the study were tested by Southern blot hybridization of PCR products, which showed 56 of 62 Northern aplomado falcons and 6 of 6 peregrine falcons as positive for the adenovirus (data not shown). DNA samples from all chickens, quail, and pigeons, the 12 serotypes of group I avian adenovirus isolated from poultry, and all falcon eggs were negative by the falcon adenovirus-specific hexon region PCR.
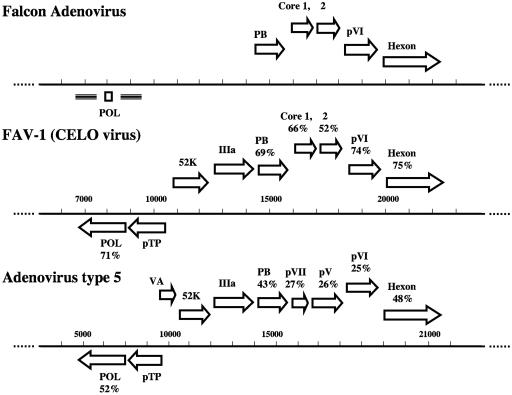
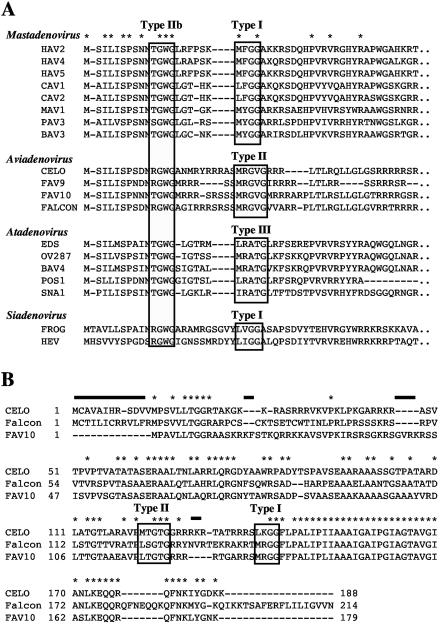
The falcon adenovirus 6.2-kb and 240-bp fragments had an overall G+C content of 48.5% and contained six open reading frames. The organization of genes in the 6.2-kb segment was similar to that of other mastadenoviruses and aviadenoviruses (Fig. 3). Comparisons of DNA and predicted amino acid sequence data for the falcon adenovirus genes and other adenoviruses showed highest overall similarity to FAV-1, at 64% nucleotide and 68% amino acid identity. The falcon adenovirus, like FAV-1, had a continuous gap of 64 residues in the middle of the penton base when aligned with human adenovirus 5 (GenBank accession no. M22141) and lacked both RGD and LDV motifs. The predicted protein for the falcon adenovirus core 1 gene was arginine rich (31% arginine residues), like the FAV-1 core 1 protein (33% arginine residues), and contained type II [(M/L/I)XgX′G] and type IIb (NTGW′G) protease signal sequences (Fig. 4) (12). However, the falcon homologue of the core 2 gene had markedly lower similarity to the FAV-1 core 2 gene, at 57% nucleotide and 52% amino acid identity. The low similarity was most pronounced near the amino terminus, whereas most of the sequence near the carboxy terminus, including type I and type II protease cleavage sites, was conserved. The falcon adenovirus homologue of core 2 protein had even lower similarity with FAV-10 at the amino terminus, including substantial insertions and deletions (Fig. 4).
FIG. 3.
Genomic organization of falcon adenovirus, FAV-1 (CELO), and human adenovirus 5 early and central regions. Percentages of similarity of FAV-1 and human adenovirus 5 predicted amino acid sequences to falcon adenovirus sequences are given above the respective genes. Pol, polymerase; pTP, preterminal protein; VA, virus-associated RNA; 52K, 52-kDa protein; PB, penton base.
FIG. 4.
(A) Alignment of predicted amino acid sequences for the core 1 gene from adenoviruses representative of the proposed four genera in the family Adenoviridae. Protease target sites (types I, II, IIb, and III) are within shaded boxes. Asterisks indicate conserved residues. Viruses (with GenBank accession numbers in parentheses) are as follows: HAV2, human adenovirus 2 (J01917); HAV4, human adenovirus 4 (U70921); HAV5, human adenovirus 5 (M73260); CAV1, canine adenovirus 1 (Y07760); CAV2, canine adenovirus 2 (U77082); MAV1, mouse adenovirus 1 (U95843); PAV3, porcine adenovirus 3 (AF083132); BAV3, bovine adenovirus 3 (AF030154); CELO virus (U46933); FAV-9 (AF083975); FAV-10 (L08450); FALCON, falcon adenovirus; EDS virus (Y09598); OV287, ovine adenovirus 287 (U40837); BAV4, bovine adenovirus 4 (AF036092); POS1, possum adenovirus 1 (AF249333); SNA1, snake adenovirus 1 (AY082603); FROG, frog adenovirus (AF224336); HEV (AF074946). (B) Alignment of predicted amino acid sequences for the core 2 gene from CELO virus, falcon adenovirus, and fowl adenovirus 10. Protease target sites (types I and II) are within shaded boxes. Solid black lines indicate insertion and deletion sequences conserved between falcon adenovirus and CELO virus. Asterisks indicate conserved residues.
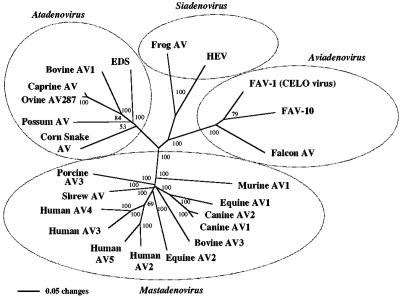
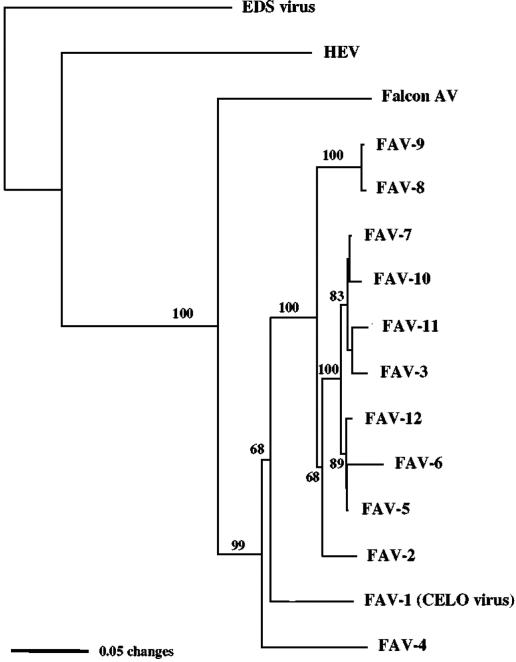
Unrooted phylograms using neighbor-joining or maximum-likelihood methods with the Kimura-2 parameter for DNA and predicted amino acid sequences of the polymerase and hexon genes from various previously characterized adenoviruses placed the falcon adenovirus within the genus Aviadenovirus and grouped HEV in Siadenovirus and EDS virus in Atadenovirus. Additional rooted phylograms using sequences obtained from cloned amplicons generated by the avian adenovirus 3′ hexon region PCR for the 12 chicken serotypes demonstrated that the falcon adenovirus is a distinct species most closely related to FAV-1 and fowl adenovirus serotype 4 (FAV-4). The falcon adenovirus sequence formed the first bifurcation on the Aviadenovirus branch in the unrooted phylogram and was the basal lineage in the rooted phylogram comparing group I avian viruses (Fig. 5 and 6). Phylograms derived from the two falcon hexon genotypes were comparable.
FIG. 5.
Phylogram of hexon gene sequences from mammals, birds, reptiles, and an amphibian using the neighbor-joining method with the Kimura-2 parameter. Bootstrap values are given at branch nodes. GenBank accession numbers are as follows: caprine adenovirus (AV), AF207660; possum AV, AF338822; shrew AV, AF258784; FAV-1 (CELO virus), U46933; porcine AV3, U34592; ovine AV287, U40839; murine AV1, MB1889; EDS virus, Y09598; equine AV2, L80007; equine AV1, L79955; canine AV2, U77082; canine AV1, U55001; bovine AV3, AF030154; human AV2, AJ293904; human AV3, X76549; human AV4, X84646; human AV5, AD5002; FAV-10, U26221; HEV, AF074946; frog AV, AF224336; corn snake AV, AY082603.
FIG. 6.
Phylogram of hexon gene sequences from the falcon adenovirus and poultry group I adenoviruses, serotypes 1 to 12, using the neighbor-joining method with the Kimura-2 parameter. HEV (GenBank accession no. AF074946) and EDS virus (GenBank accession no. Y09598) sequences were used as outgroups. Bootstrap values are given at branch nodes.
PCRs for Herpesviridae, Circoviridae, Pneumovirinae, Influenzavirus A, Coronavirus, Salmonella, Campylobacter, and Mycoplasma were negative for all falcons and all falcon eggs. PCR for capsid protein of Parvovirinae was positive for 49 of 62 Northern aplomado falcons, 5 of 6 peregrine falcons, both orange-breasted falcons, both teita falcons, the gyrfalcon × peregrine falcon hybrids, the merlin, and the Vanuatu peregrine falcon as determined by cloning and sequencing of amplicons and Southern blot hybridization. Liver, spleen, intestines, lung, and kidney were commonly positive for parvovirus; bone marrow and brain were infrequently positive (Table 1). Sequence data showed two distinct AAVs (aplomado falcon AAV1 and AAV2) in Northern aplomado falcons. Aplomado falcon AAV1 and AAV2 had nucleotide identities of 87% with each other and 83% and 79%, respectively, with a chicken AAV (GenBank accession no. AY186198). Only one AAV sequence was found in peregrine, orange-breasted, and Vanuatu peregrine falcons; it was identical to AAV1 of the Northern aplomado falcon. The merlin had a distinct AAV sequence that had 97% to 87% similarity to other falcon AAVs. The teita falcons and gyrfalcon × peregrine falcons had the same AAV, which had 83% to 86% identity to other falcon AAVs. PCR for Parvovirinae was negative for all falcon eggs. PCR results for pathogens for animals in the study are summarized in Table 1.
DISCUSSION
Aviadenoviruses have a worldwide distribution, and outbreaks of disease have been reported in 31 species of birds, including 8 native and 5 vagrant species of North America (38, 40). In the genus Falco, adenovirus infection has been documented in a merlin, 7 American kestrels (Falco sparverius), and 13 Mauritius kestrels (Falco punctatus) (13, 42, 45). These studies called attention to the danger of adenoviruses in captive falcons but were limited to morphological descriptions of viral particles and lesions in affected birds. The source, genotype, involvement of copathogens, and interspecies communicability of falconid adenoviruses have not been reported. In our study, we found adenovirus in Northern aplomado, peregrine, orange-breasted, Vanuatu, gyrfalcon × peregrinem and teita falcons and a merlin. All birds died in captivity in the United States, and all were cohoused with other species of falcons.
Comparative molecular analyses indicated that the falcon adenovirus is a new species in the genus Aviadenovirus with closest similarity to the group I members FAV-1 and FAV-4. In our phylograms, group II (HEV) and III (EDS virus) avian viruses were assigned to the genera Siadenovirus and Atadenovirus, respectively. These findings are consistent with previous comprehensive phylogenetic analyses of the family Adenoviridae and support the hypothesis that adenoviruses underwent long-term coadaptation with their hosts. Exceptions are some members of the genus Atadenovirus, such as EDS virus, that made major interclass host switches resulting in heightened virulence (2, 12). Estimating the host origin of the falcon adenovirus, even at the ordinal level, was difficult due to the lack of sequence data for adenoviruses in other birds, such as psittacines, pigeons, and ratites. However, the taxonomic affinity of the falcon adenovirus with FAV-1, especially for functional motifs such as putative integrin binding sites and protease cleavage domains, suggests that the virus should be classified in the genus Aviadenovirus and may provide some predictive value in understanding its biology.
The epidemiology of adenovirus infection in Northern aplomado falcons in many ways resembled that of FAV-1 in quail, where young birds (less than 35 days of age) are affected, the incubation period is approximately 6 to 10 days, mortality rates range from 60 to 87%, and virus is highly contagious between chicks but lacks a vertical component (21, 23, 40). FAV-1 in chickens, in contrast, has low morbidity and mortality but high infectivity and widespread distribution derived from both bird-to-bird and bird-to-embryo transmission (40). Analyses of the adenovirus outbreak in Northern aplomado falcons revealed a point source epizootic with an incubation period of 10 to 11 days, a case fatality rate of 86.1%, and a predilection for birds less than 35 days of age. The pattern of morbidity and mortality in the outbreak was consistent with horizontal transmission. PCR analyses for adenovirus on unhatched Northern aplomado eggs collected during the epizootic and in situ hybridization of tissues from diseased chicks indicated that neither vertical transmission or transmission across oviposited eggs occurred and that horizontal infection was the only method of virus spread. These similarities to disease in quail with FAV-1 raise doubt as to the likelihood that the falcon adenovirus is indigenous to Northern aplomado falcons.
The source of the adenovirus in each disease episode was not determined, but most evidence pointed toward one or several members of the genus Falco as possible reservoirs. In a previous report of adenovirus in Mauritius kestrels, turkey poults and domestic chicks fed to the birds were suspected as origins of the causative virus (13). PCR analyses of frozen chicken and quail fed to the Northern aplomado chicks at the time of the World Center outbreak and characterization of the hexon genes from all known chicken group I serotypes eliminated food as a possible source. Regional pigeon populations that may have had contact with some falcons were similarly excluded by PCR and histology. In all of our cases, birds that died were housed in facilities with different species of clinically normal falcons. The increased attack rate and relative risk of adenovirus infection in Northern aplomado falcon chicks hatched from eggs incubated by foster peregrine falcons at the World Center raised suspicion about the peregrine falcon as a reservoir. Gyrfalcons, which are native to North America and winter in some parts of the midwestern United States, where many of the falcon outbreaks occurred, may also be a source of virus (5). Gyrfalcons were a parental species of gyrfalcon × peregrine falcon hybrids housed with the orange-breasted and teita falcons that became infected and died.
Two adenovirus DNA sequences were detected in falcons in our study. Based on the chronology of the disease outbreaks and the high percentage of identity between the two sequences, it is likely the genotypes represented coexistent strains of a single virus species. Strain variants have been well recognized among avian adenoviruses due to the high mutability of the hexon gene which encodes most neutralizing epitopes (10, 19, 40). Strain-related changes of as little as 1% to 2% in the hexon gene of chicken adenovirus isolates can be correlated with substantial differences in virulence (19, 40). The two falcon adenovirus genotypes differed by 1.3%. Unfortunately, morphological comparisons between birds affected by the two strains could not be made because no pathology data were available for the teita falcons, the gyrfalcon × peregrine falcon hybrids, and the merlin. PCR evaluations for other potential disease-causing agents, however, indicated that both genotypes were primary pathogens.
Infection of chickens with group I adenoviruses usually requires the presence of a copathogen, such as birnavirus, circovirus, or mycoplasma, or exposure to mycotoxins for clinical disease to develop (19, 40). In humans, predisposing factors are compromised immunity (AIDS, cancer irradiation or chemotherapy, or organ transplantation), concurrent infections with other agents, and idiopathic individual susceptibility (2). In our study, AAV was the only infectious agent other than adenovirus found in affected falcons and was detected in 78% of birds. AAVs are replication-defective parvoviruses that frequently accompany adenovirus infections and have been reported in mammals, reptiles, and birds (3, 26, 39). They are generally considered nonpathogenic, although some serotypes have been associated with reproductive abnormalities in humans and others have been shown to modify cell cycle progression in vitro, either inducing apoptosis or inhibiting it (25, 35, 43). In birds, latent infections are widespread, and several studies have suggested that AAV can modify humoral immunity and the pathogenicity of adenovirus infections in vivo (39, 57). Our data showed no correlation between the severity or distribution of lesions in falcons and the presence of AAV or a particular genotype of AAV. However, it remains possible that AAV could have influenced the pathogenesis of disease in infected birds. Other recognized copathogens or primary causes of inclusion body hepatitis (herpesvirus, parvovirus, polyomavirus), immunosuppression (circovirus, paramyxovirus), enteritis (salmonella, campylobacter, coronavirus), or pneumonia (mycoplasma, pneumovirus, influenza virus) in birds were excluded by PCR, and the falcon adenovirus was not detected in clinically normal Northern aplomado falcons unassociated with the 1996 outbreak. At this time, it appears that adenovirus was the primary cause of systemic disease in all of the falcon outbreaks.
The character and distribution of lesions in falcons overall resembled those described for FAV-1 more than those for any other avian adenovirus. Inclusion body hepatitis is the most characteristic lesion of FAV-1 infection and consists of random necrosis, mononuclear leukocyte infilration, and hepatocellular Cowdry type A or B inclusion bodies (19, 40, 53). The falcons in our study had these changes and additionally developed inclusion bodies in biliary duct epithelium unassociated with necrosis. Adenovirus infection of biliary epithelium has been reported in several mammals, including humans, but is very uncommon (4, 56). In birds, it has been described only in quail infected with FAV-1 (23). Infection of the ventriculus and the kidneys is common for both FAV-1 and FAV-4 and usually causes ulceration, erosion, and fibrillation of koilin or tubular degeneration and necrosis, respectively (6, 33). In falcons, epithelia of the ventriculus and renal tubules, like biliary epithelium, had viral inclusions but few or no lesions. Unlike chickens with FAV-4, falcons did not have hydropericardium and cardiac necrosis, and in contrast to the report of adenovirus infection in Mauritius kestrels, vasculitis was present in only one of our animals, the Vanuatu falcon (6, 13, 19, 40). The reason for the different morphological expression of disease in the Vanuatu falcon was not clear but may have been related to the presence of multifocal chronic arteriosclerosis in this animal.
The collective morphological and molecular findings from our study provided some basic insight into the pathogenesis of falcon adenovirus infection. A close association between the development of necrosis and inflammation and the presence of adenovirus, as demonstrated by detection of inclusion bodies, in situ hybridization, and PCR was seen. This and immunostaining for apoptotic DNA indicated that most tissue damage resulted from a direct cytolytic effect of virus rather than programmed cell death. The minor role of apoptosis is not surprising, since many adenoviruses, including FAV-1, have genes that encode antiapoptotic peptides (7). By contrast, HEV in turkeys causes intestinal tissue damage chiefly via apoptosis. This effect can be abrogated by cyclosporine or thalidomide treatment and appears to result from immune dysregulation secondary to infection of B lymphocytes (37, 47). Due to the lack of an effective antibody for B lymphocytes in falcons, we could not determine if the falcon adenovirus infected B cells. Falcon T lymphocytes were shown to be infected, but the consequences were not clear. One possibility is that infection may have allowed systemic spread of virus through cell-associated viremia. Additionally, T cells may have been a site of viral latency, such as occurs in humans infected with adenovirus (17). Alternatively, infection of T lymphocytes may have caused immune suppression. Some Northern aplomado and peregrine falcon chicks had lymphoid depletion in the spleen and bursa and systemic lesions indicative of secondary bacterial sepsis. In chickens, experimental studies have shown that high inoculation doses of some group I avian viruses cause immune compromise in young animals (31, 53, 55). Significant load differences in virus exposure among falcon chicks almost certainly occurred, and it is possible that birds receiving large amounts of virus may have developed impaired immunity.
The tissue tropism of falcon adenovirus was widespread. For human adenoviruses and the CELO virus, tissue-specific production of receptors, such as CAR and integrins, appear to influence the affinity of virus for different organs. CAR is a normal cell surface product that has high affinity for the penton fibers of many adenoviruses and is utilized in virus-to-cell adhesion (20, 40, 50). Integrins are transmembrane glycoproteins that often act in conjunction with CAR by binding certain ligands on the penton base, thereby allowing adherent adenoviruses to penetrate the cell (32). The penton base of the falcon adenovirus, like that of FAV-1, lacked RGD and LDV recognition motifs for integrin binding, and thus, cell entry must have depended on use of a different receptor (46). It is possible that falcons express a CAR homologous to that present in humans. The falcon adenovirus may, alternatively, utilize another receptor, such as that targeted by human adenovirus 11 (30). FAV-1 has been shown experimentally to bind the human CAR, but it is not known whether galliforms produce a molecule similar to CAR (49). For several mammalian species, the level of CAR production is typically very high in newborn animals and decreases with age (20, 50). If a homologue of CAR was involved in falcon disease, it may explain the increased distribution and pathogenicity we saw in young animals.
Falcons were among the most successful and widely distributed birds of prey until the middle of the 20th century, when populations declined with the increasing use of pesticides and with the loss of habitat to urbanization and agriculture (5, 41, 54). Ongoing captive breeding and release programs have helped introduce endangered falcons back into the wild. However, the assemblage of different avian taxa in confined facilities for prolonged periods considerably raises the risk of cross-species spread of infectious agents. Our data indicate that the falcon adenovirus is a primary pathogen that can infect several species of falcons and is probably widespread throughout western and midwestern North America. Epidemiologic, phylogenetic, and pathogenetic features of the virus in many ways resembled those of FAV-1 and FAV-4 and suggested that falcon husbandry should probably be modified to avoid all direct and indirect contact between different species and subspecies in the genus Falco, even after birds have cleared quarantine. Continued molecular and serologic surveillance of falcons and other birds of prey should help better clarify the biology of this virus and allow implementation of more-effective preventative measures by institutes and groups involved in conservation.
Acknowledgments
This work was supported by the Zoological Society of San Diego. We thank the Beckman Coulter Corporation for donation of the automated capillary DNA sequencer and high-speed centrifuge used in these studies. We also thank Charles and Shirley Sykes of San Diego for financial support of our laboratory.
Technical assistance was provided by Laura Richman, April Gorow, and Yvonne Cates.
REFERENCES
- 1.Ali, A., and D. L. Reynolds. 2000. A multiplex reverse transcription-polymerase chain reaction assay for Newcastle disease virus and avian pneumovirus (Colorado strain). Avian Dis. 44:938-943. [PubMed] [Google Scholar]
- 2.Benko, M., and B. Harrach. 2003. Molecular evolution of adenoviruses. Curr. Top. Microbiol. Immunol. 272:3-35. [DOI] [PubMed] [Google Scholar]
- 3.Bossis, I., and J. A. Chiorini. 2003. Cloning of an avian adeno-associated virus (AAAV) and generation of recombinant AAAV particles. J. Virol. 77:6799-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brundler, M. A., N. Rodriguez-Baez, R. Jaffe, A. G. Weinberg, and B. B. Rogers. 2003. Adenovirus ascending cholangiohepatitis. Pediatr. Dev. Pathol. 6:156-159. [DOI] [PubMed] [Google Scholar]
- 5.Cade, T. J. 1982. The falcons of the world. Cornell University Press, Ithaca, N.Y.
- 6.Chandra, R., S. K. Shukla, and M. Kumar. 2000. The hydropericardium syndrome and inclusion body hepatitis in domestic fowl. Trop. Anim. Health Prod. 32:99-111. [DOI] [PubMed] [Google Scholar]
- 7.Chiocca, S., A. Baker, and M. Cotten. 1997. Identification of a novel antiapoptotic protein, GAM-1, encoded by the CELO adenovirus. J. Virol. 71:3168-3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiocca, S., R. Kurzbauer, G. Schaffner, A. Baker, V. Mautner, and M. Cotten. 1996. The complete DNA sequence and genomic organization of the avian adenovirus CELO. J. Virol. 70:2939-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowen, B. S., H. Rothenbacher, L. D. Schwartz, M. O. Braune, and R. L. Owen. 1988. A case of acute pulmonary edema, splenomegaly, and ascites in Guinea fowl. Avian Dis. 32:151-156. [PubMed] [Google Scholar]
- 10.Crawford-Miksza, L., and D. P. Schnurr. 1996. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 70:1836-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davison, A. J., K. M. Wright, and B. Harrach. 2000. DNA sequence of frog adenovirus. J. Gen. Virol. 81:2431-2439. [DOI] [PubMed] [Google Scholar]
- 12.Farkas, S. L., M. Benko, P. Elo, K. Ursu, A. Dan, W. Ahne, and B. Harrach. 2002. Genomic and phylogenetic analyses of an adenovirus isolated from a corn snake (Elaphe guttata) imply a common origin with members of the proposed new genus Atadenovirus. J. Gen. Virol. 83:2403-2410. [DOI] [PubMed] [Google Scholar]
- 13.Forbes, N. A., G. N. Simpson, R. J. Higgins, and R. E. Gough. 1997. Adenovirus infection in Mauritius kestrels (Falco punctatus). J. Avian Med. Surg. 11:31-33. [Google Scholar]
- 14.Fouchier, R. A., T. M. Bestebroer, S. Herfst, L. Van Der Kemp, G. F. Rimmelzwaan, and A. D. Osterhaus. 2000. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J. Clin. Microbiol. 38:4096-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frankfurt, O. S., and A. Krishan. 2001. Identification of apoptotic cells by formamide-induced DNA denaturation in condensed chromatin. J. Histochem. Cytochem. 49:369-378. [DOI] [PubMed] [Google Scholar]
- 16.Gao, G. P., M. R. Alvira, L. Wang, R. Calcedo, J. Johnston, and J. M. Wilson. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA 99:11854-11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garnett, C. T., D. Erdman, W. Xu, and L. R. Gooding. 2002. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J. Virol. 76:10608-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guy, J. S., J. L. Schaeffer, and H. J. Barnes. 1988. Inclusion-body hepatitis in day-old turkeys. Avian Dis. 32:587-590. [PubMed] [Google Scholar]
- 19.Hess, M. 2000. Detection and differentiation of avian adenoviruses: a review. Avian Pathol. 29:195-206. [DOI] [PubMed] [Google Scholar]
- 20.Ito, M., M. Kodama, M. Masuko, M. Yamaura, K. Fuse, Y. Uesugi, S. Hirono, T. Okura, K. Kato, Y. Hotta, T. Honda, R. Kuwano, and Y. Aizawa. 2000. Expression of coxsackie and adenovirus receptor in hearts of rats with experimental autoimmune myocarditis. Circ. Res. 86:275-280. [DOI] [PubMed] [Google Scholar]
- 21.Jack, S. W., W. M. Reed, and T. A. Bryan. 1987. Inclusion body hepatitis in bobwhite quail (Colinus virginianus). Avian Dis. 31:662-665. [PubMed] [Google Scholar]
- 22.Jack, S. W., and W. M. Reed. 1990. Further characterization of an avian adenovirus associated with inclusion body hepatitis in bobwhite quail. Avian Dis. 34:526-530. [PubMed] [Google Scholar]
- 23.Jack, S. W., and W. M. Reed. 1990. Pathology of experimentally induced quail bronchitis. Avian Dis. 34:44-51. [PubMed] [Google Scholar]
- 24.Johne, R., and H. Muller. 1998. Avian polyomavirus in wild birds: genome analysis of isolates from Falconiformes and Psittaciformes. Arch. Virol. 143:1501-1512. [DOI] [PubMed] [Google Scholar]
- 25.Kiehl, K., J. R. Schlehofer, R. Schultz, M. Zugaib, and E. Armbruster-Moraes. 2002. Adeno-associated virus DNA in human gestational trophoblastic disease. Placenta 23:410-415. [DOI] [PubMed] [Google Scholar]
- 26.Kim, D. Y., M. A. Mitchell, M. Rudy, R. W. Bauer, R. Poston, and D. Y. Cho. 2002. An outbreak of adenoviral infection in inland bearded dragons (Pogona vitticeps) coinfected with dependovirus and coccidial protozoa (Isospora sp.). J. Vet. Diagn. Investig. 14:332-334. [DOI] [PubMed] [Google Scholar]
- 27.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, et al. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1947-1958. [DOI] [PubMed] [Google Scholar]
- 28.Lehrmann, H., and M. Cotten. 1999. Characterization of CELO virus proteins that modulate the pRb/E2F pathway. J. Virol. 73:6517-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mankertz, A., K. Hattermann, B. Ehlers, and D. Soike. 2000. Cloning and sequencing of columbid circovirus (coCV), a new circovirus from pigeons. Arch. Virol. 145:2469-2479. [DOI] [PubMed] [Google Scholar]
- 30.Mei, Y. F., J. Skog, K. Lindman, and G. Wadell. 2003. Comparative analyses of the genome organization of human adenovirus 11, a member of the human adenovirus species B, and the commonly used human adenovirus 5 vector, a member of species C. J. Gen. Virol. 84:2061-2071. [DOI] [PubMed] [Google Scholar]
- 31.Naeem, K., T. Niazi, S. A. Malik, and A. H. Cheema. 1995. Immunosuppressive potential and pathogenicity of an avian adenovirus isolate involved in hydropericardium syndrome in broilers. Avian Dis. 39:723-728. [PubMed] [Google Scholar]
- 32.Nemerow, G. R., and P. L. Stewart. 1999. Role of alpha integrins in adenovirus cell entry and gene delivery. Microbiol. Mol. Biol. Rev. 63:725-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono, M., Y. Okuda, S. Yazawa, Y. Imai, I. Shibata, S. Sato, and K. Okada. 2003. Adenoviral gizzard erosion in commercial broiler chickens. Vet. Pathol. 40:294-303. [DOI] [PubMed] [Google Scholar]
- 34.Patterson, M. M., M. D. Schrenzel, Y. Feng, S. Xu, F. E. Dewhirst, B. J. Paster, S. A. Thibodeau, J. Versalovic, and J. G. Fox. 2000. Helicobacter aurati sp. nov., a urease-positive Helicobacter species cultured from gastrointestinal tissues of Syrian hamsters. J. Clin. Microbiol. 38:3722-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raj, K., P. Ogston, and P. Beard. 2001. Virus-mediated killing of cells that lack p53 activity. Nature 412:914-917. [DOI] [PubMed] [Google Scholar]
- 36.Raue, R., and M. Hess. 1998. Hexon based PCRs combined with restriction enzyme analysis for rapid detection and differentiation of fowl adenoviruses and egg drop syndrome virus. J. Virol. Methods 73:211-217. [DOI] [PubMed] [Google Scholar]
- 37.Rautenschlein, S., M. Suresh, and J. M. Sharma. 2000. Pathogenic avian adenovirus type II induces apoptosis in turkey spleen cells. Arch. Virol. 145:1671-1683. [DOI] [PubMed] [Google Scholar]
- 38.Ritchie, B. W. (ed.). 1995. Avian viruses: function and control. Wingers Publishing Inc., Lake Worth, Fla.
- 39.Sadasiv, E. C., T. H. Piela, and P. W. Chang. 1989. Detection of latent avian adenovirus-associated virus proteins in chicken cells. Avian Dis. 33:125-133. [PubMed] [Google Scholar]
- 40.Saif, Y. M. 2003. Adenovirus infections, p. 214-251. In Y. M. Saif, H. J. Barnes, J. R. Glisson, A. M. Fadly, L. R. McDougald, and D. E. Swayne (ed.), Diseases of poultry, 11th ed. Iowa State Press, Ames.
- 41.Sandfort, C. (ed.). 1994. Northern aplomado falcon restoration, p. 1-105. The Peregrine Fund, Boise, Idaho.
- 42.Schelling, S. H., D. S. Garlick, and J. Alroy. 1989. Adenoviral hepatitis in a Merlin (Falco columbarius). Vet. Pathol. 26:529-530. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt, M., S. Afione, and R. M. Kotin. 2000. Adeno-associated virus type 2 Rep78 induces apoptosis through caspase activation independently of p53. J. Virol. 74:9441-9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma, V. K., and S. A. Carlson. 2000. Simultaneous detection of Salmonella strains and Escherichia coli 157:H7 with fluorogenic PCR and single-enrichment-broth culture. Appl. Environ. Microbiol. 66:5472-5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sileo, L., J. C. Franson, D. L. Graham, C. H. Domermuth, B. A. Rattner, and O. H. Pattee. 1983. Hemorrhagic enteritis in captive American kestrels (Falco sparverius). J. Wildl. Dis. 19:244-247. [DOI] [PubMed] [Google Scholar]
- 46.Suresh, M., S. St Cyr, and J. M. Sharma. 1995. Molecular cloning and sequence analysis of the penton base genes of type II avian adenoviruses. Virus Res. 39:289-297. [DOI] [PubMed] [Google Scholar]
- 47.Suresh, M., and J. M. Sharma. 1996. Pathogenesis of type II avian adenovirus infection in turkeys: in vivo immune cell tropism and tissue distribution of the virus. J. Virol. 70:30-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swofford, D. L. 1998. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, Mass.
- 49.Tan, P. K., A. I. Michou, J. M. Bergelson, and M. Cotten. 2001. Defining CAR as a cellular receptor for the avian adenovirus CELO using a genetic analysis of the two viral fibre proteins. J. Gen. Virol. 82:1465-1472. [DOI] [PubMed] [Google Scholar]
- 50.Tomko, R. P., R. Xu, and L. Philipson. 1997. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. USA 94:3352-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.VanDevanter, D. R., P. Warrener, L. Bennett, E. R. Schultz, S. Coulter, R. L. Garber, and T. M. Rose. 1996. Detection and analysis of diverse herpesviral species by consensus primer PCR. J. Clin. Microbiol. 34:1666-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Kuppeveld, F. J., J. T. van der Logt, A. F. Angulo, M. J. van Zoest, W. G. Quint, H. G. Niesters, J. M. Galama, and W. J. Melchers. 1992. Genus- and species-specific identification of mycoplasmas by 16S rRNA amplification. Appl. Environ Microbiol. 58:2606-2615. (Erratum, 59: 655, 1993.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vereecken, M., P. de Herdt, and R. Ducatelle. 1998. Adenovirus infections in pigeons: a review. Avian Pathol. 27:333-338. [DOI] [PubMed] [Google Scholar]
- 54.Vitousek, P. M., H. A. Mooney, J. Lubchenco, and J. M. Milillo. 1997. Human domination of earth's ecosystems. Science 277:494-499. [Google Scholar]
- 55.Wang, C. H., and C. M. Chang. 2000. Pathogenicity and gene analysis of adenovirus from pigeons with inclusion body hepatitis. J. Vet. Med. Sci. 62:989-993. [DOI] [PubMed] [Google Scholar]
- 56.Woods, L. W., N. G. Walters, and B. Johnson. 1991. Cholangiohepatitis associated with adenovirus-like particles in a pygmy goat. J. Vet. Diagn. Investig. 3:89-92. [DOI] [PubMed] [Google Scholar]
- 57.Yates, V. J., Y. O. Rhee, and D. E. Fry. 1977. Serological response of chickens exposed to a type 1 avian adenovirus alone or in combination with adeno-associated virus. Avian Dis. 21:408-414. [PubMed] [Google Scholar]