Abstract
The composition and spatial organization of the mucosal flora in biopsy specimens from patients with inflammatory bowel disease (IBD; either Crohn's disease or ulcerative colitis), self-limiting colitis, irritable-bowel syndrome (IBS), and healthy controls were investigated by using a broad range of fluorescent bacterial group-specific rRNA-targeted oligonucleotide probes. Each group included 20 subjects. Ten patients who had IBD and who were being treated with antibiotics were also studied. Use of nonaqueous Carnoy fixative to preserve the mucus layer was crucial for detection of bacteria adherent to the mucosal surface (mucosal bacteria). No biofilm was detectable in formalin-fixed biopsy specimens. Mucosal bacteria were found at concentrations greater than 109/ml in 90 to 95% of IBD patients, 95% of patients with self-limiting colitis, 65% of IBS patients, and 35% of healthy controls. The mean density of the mucosal biofilm was 2 powers higher in IBD patients than in patients with IBS or controls, and bacteria were mostly adherent. Bacteroides fragilis was responsible for >60% of the biofilm mass in patients with IBD but for only 30% of the biofilm mass in patients with self-limiting colitis and <15% of the biofilm mass in patients with IBS. In contrast, bacteria which positively hybridized with the probe specific for Eubacterium rectale-Clostridium coccoides accounted for >40% of the biofilm in IBS patients but for <15% of the biofilm in IBD patients. In patients treated with (5-ASA) or antibiotics, the biofilm could be detected with 4,6-diamidino-2-phenylindole but did not hybridize with fluorescence in situ hybridization probes. A Bacteroides fragilis biofilm is the main feature of IBD. This was not previously recognized due to a lack of appropriate tissue fixation. Both 5-ASA and antibiotics suppress but do not eliminate the adherent biofilm.
It is estimated that more than 1 million Americans have ulcerative colitis or Crohn's disease (CD), the two most common forms of inflammatory bowel disease (IBD). CD can occur anywhere in the digestive tract, often with inflammatory lesions spreading deep into the layers of affected tissues. Ulcerative colitis (UC) usually affects only the mucosa of the large intestine and rectum. UC, CD, and self-limiting colitis (slc) all cause diarrhea, with or without accompanying bleeding. However, CD and UC are chronic inflammatory diseases, which makes them different from slc (which is mostly caused by infectious agents) and from irritable bowel syndrome (IBS; which is mainly symptomatic and which is not accompanied by inflammation).
It is hypothesized that IBD results from an aberrant immune response against intestinal bacteria that results in inflammatory damage to intestinal tissues. Many clinical and experimental observations strongly implicate intestinal bacteria in the pathogenesis of IBD (9). However, despite intensive research, neither the mechanisms by which bacteria affect the development of IBD nor the disease-specific changes in the intestinal flora have been determined (9, 25). Using microbial culture validated by quantitative PCR with subsequent cloning and sequencing, we have previously shown that colon biopsy specimens from healthy humans without IBD are nearly free of bacteria once they are washed of fecal contents (31). In contrast, the concentrations of bacteria in biopsy specimens from IBD patients are high even after they are washed. The numbers of bacteria adherent to the mucosal surface (mucosal bacteria) increase progressively from slc to UC to CD, being 2 powers higher than those in healthy controls. However, all bacteria identified were of fecal origin, and no differences in the compositions between different patient and control groups could be identified (31).
The term biofilm has been used to describe a structured community of bacterial cells enclosed in a self-produced polymeric matrix and adherent to an inert or living surface (6). The mucosal bacteria found in patients with IBD can thus be regarded as intestinal biofilms. Biofilms, with their inherent resistance to antibiotics and host immune attack, have increasingly been identified as sources of many recalcitrant bacterial infections. These include periodontal disease, endocarditis, chronic obstructive lung disease, and foreign body-related infections (6, 7). The impact of biofilms on the pathogenesis of IBD is unknown.
Previous investigations of bacterial involvement in IBD have mainly been based on comparative studies of cultured bacterial isolates. It is likely that the intestinal flora is structurally organized (25). Sampling of the intestinal contents prior to culture or gene-based identification disrupts the structural organization of the bacteria within the gut and may disguise the complex interactions between the intestinal flora and the host. Fluorescence in situ hybridization (FISH) combines the molecular identification of bacteria with the direct visualization of the relationships between the bacteria and the mucosa, providing a significant advantage over culture, PCR, and histological methods alone. In this study, the composition and spatial organization of the mucosal bacteria in patients with IBD and controls were investigated by FISH with a broad range of bacterial group-specific rRNA-targeted probes.
MATERIALS AND METHODS
Patients.
All patients were investigated for medical indications and gave informed consent for additional biopsies according to the protocol approved by the Ethics Commission of Charite Hospital. Groups of 20 consecutive patients for whom a complete medical history was available and who underwent colonoscopy at the Charite Hospital were enrolled. Groups consisted of patients with CD, UC, slc, or IBS and healthy controls. The accepted criteria for UC, CD, IBS, and controls were used (5, 19, 23, 31). Slc was defined as a first-time colitis that macroscopically and histologically resembled infectious-like colitis and that had an appearance and histology atypical for IBD. However, the overall clinical history at the endpoint of this study was too short for final conclusions, and no follow-up colonoscopy was available.
None of these patients had been treated with antibiotics in the previous 2 months. Patients currently treated with 5-aminosalicylic acid (5-ASA; mesalamine), prednisolone, or azathioprine were accepted. Patients receiving other therapies were excluded. We studied an additional 10 IBD patients (5 with CD and 5 with UC) who had received antibiotics (metronidazole and ciprofloxacin) at the time of the colonoscopy. Six of these patients received antibiotics 1 day prior to the colonoscopy. The remaining four patients were on antibiotics for 4 to 12 days prior to the colonoscopy. The clinical data for the patients studied are summarized in Table 1.
TABLE 1.
Clinical characteristics of patients studied
| Characteristic | CD patients | UC patients | Slc patients | IBS patients | Controls | IBD patients on antibiotics |
|---|---|---|---|---|---|---|
| No. of females/no. of males | 9/11 | 11/9 | 8/12 | 14/6 | 13/7 | 6/4 |
| Mean age (yr) | 32.5 | 44.4 | 37.3 | 47.8 | 46.2 | 39.1 |
| No. of patients receiving: | ||||||
| 5-ASA therapy | 5 | 16 | 8 | |||
| Prednisolone | 12 | 8 | 8 |
Biopsy specimens.
The biopsy specimens for FISH were taken from the ileum, ascending and sigmoid colon, and if possible, macroscopically noninflamed tissues. The biopsy specimens were fixed in nonaqueous Carnoy solution (22) for 2 h and then processed and embedded into paraffin blocks by standard techniques. Four-micrometer sections were placed on SuperFrost slides (R. Langenbrinck, Emmendingen, Germany) for the FISH studies.
Additional biopsy specimens from these same locations were also fixed in formalin and submitted for histopathologic evaluation. The methods for the processing and embedding of Carnoy solution- and formalin-fixed biopsy specimens in paraffin were identical. After completion of the pathological evaluation, the formalin-fixed sections were further investigated by FISH. Unless specified otherwise, all FISH studies were performed on tissues fixed in Carnoy solution.
FISH.
Oligonucleotide probes were synthesized; and a carbocyanite dye (Cy3), fluorescein isothiocyanate (FITC), or Cy5 fluorescent dye (MWG Biotech, Ebersberg, Germany) was added at the 5′ end. Forty domain-, group-, and species-specific FISH probes were applied (Table 2).
TABLE 2.
FISH probesa
| Probe name | Target | Reference |
|---|---|---|
| Eub338 | Virtually all bacteria, kingdom Bacteria (Eubacteria) | 1 |
| Alf1b | Alpha subclass of class Proteobacteria | 20 |
| Beta42a | Beta subclass of class Proteobacteria | 20 |
| Gam42a | Gamma subclass of class Proteobacteria (including Enterobacteriaceae) | 20 |
| Ebac | Enterobacteriaceae | 4 |
| Ec1531 | Escherichia coli | 26 |
| Y16s-69 | Yersinia species | 32 |
| Srb385 | Sulfate-reducing bacteria | 2 |
| Hpy-1 | Helicobacter pylori epsilon subclass of Proteobacteria | 8 |
| Arc1430 | Arcobacter sp. epsilon subclass of Proteobacteria | 29 |
| HGC | Class Actinobacteria (gram-positive bacteria with high G + C contents) | 27 |
| LGC | Firmicutes (gram-positive bacteria with low G + C contents) | 24 |
| Sfb | Segmented filamentous bacteria | 33 |
| Erec482 | Eubacterium rectale-Clostridium coccoides group | 11 |
| Lach | Subgroup (including Lachnospira multipara) | 14 |
| Ehal | Subgroup (including Eubacterium hallii) | 14 |
| Chis150 | Clostridium histolyticum group | 11 |
| Clit135 | Clostridium lituseburense group (including Clostridium difficile) | 11 |
| Lab158 | Lactobacillus and Enterococcus group | 12 |
| Strc493 | Streptococcus group | 11 |
| Enc131 | Enterococcus spp. and other | 10 |
| Efaec | Enterococcus faecalis, Enterococcus sulfuricus | 15 |
| Ato291 | Atopobium, Coriobacterium, Eggerthella, and Collinsella spp. | 13 |
| Cor653 | Coriobacterium group | 13 |
| Ecyl | Eubacterium cylindroides and other | 14 |
| Phasco | Phascolarctobacterium faecium group | 14 |
| Veil | Veillonella group | 14 |
| Rbro, Rfla | Ruminococcus bromii, Ruminococcus flavefaciens, and other | 14 |
| UroA, UroB | Ruminococcus obeum-like bacteria (subgroup of Eubacterium rectale) | 35 |
| Ser1410 | Genus Brachyspira | 16 |
| Bif164 | Bifidobacterium | 18 |
| CF319a | Cytophaga-Flavobacteria group | 21 |
| Bac303 | Bacteroides-Prevotella group | 21 |
| Bfra602 | Bacteroides fragilis group | 11 |
| Bdis656 | Bacteroides distasonis group | 11 |
| Fprau | Fusobacterium prausnitzii group | 30 |
| Arch915 | Archaea | 20 |
| Non338 | Nonsense probe used to test for nonspecific binding | 20 |
The formamide concentration and hybridization temperature were chosen to achieve the optimal stringency, as described in the references. Additional hybridizations by use of a permeation step with lysozyme for 15, 30, and 60 min were performed in parallel for detection of gram-positive bacteria. For convenience, the numbers within the probe name that identify the location of the probe sequence within the ribosomal RNA genes are not repeated in the text.
In situ quantification of mucosal bacteria.
The bacteria were visualized by FISH and 4,6-diamidino-2-phenylindole (DAPI) staining with an e600 fluorescence microscope (Nikon, Tokyo, Japan) and photodocumented with a DXM1200 camera and software (Nikon). The enumeration was performed when the hybridization signals were clear and morphologically distinguishable as bacterial cells by at least triple-color identification with universal and group-specific FISH probes and DAPI stain and by the absence of cross-hybridization or hybridization with the Non338 nonsense probe.
Quantification of the bacteria and assessment of the population structure were performed within mucus adjacent to the biopsy specimen surface at locations that showed no signs of mechanical disruption of the epithelial layer. The autofluorescence background of the human tissue allowed tissue structures and the mucus layer to be well visualized. Nevertheless, for each biopsy specimen staining with at least one or two additional stains (Alcian blue or hematoxylin and eosin) was performed.
Photographs of three different microscopic fields were taken under high power (magnification, ×1,000) for each hybridization. The three fields represented bacteria attached to the mucosal surface (adherent bacteria), bacteria scattered within the mucus layer (mucus-scattered bacteria), and bacteria in the outer portions of the mucus (mucus ceiling bacteria).
For the purposes of quantification, adherent bacteria were defined as bacteria that lined 50 μm of the epithelial border (±1 μm of the epithelial border contained within a field 2 by 50 μm) below the intact mucus layer. Mucus-scattered bacteria were calculated within a square field of 10 by 10 μm which was placed within mucus at the maximal concentration of bacteria next to the epithelial surface. Mucus ceiling bacteria were enumerated within a field 5 by 20 μm which was placed within the maximal concentration of the mucus ceiling layer but at least 10 μm away from the epithelial surface.
The mean concentration of mucosal bacteria was defined as the mean concentration of adherent, mucus-scattered, and mucus ceiling bacteria in a region of maximal developed biofilm that covered at least 10% of the intact epithelial circumference of the biopsy specimen section. Two investigators counted the bacteria independently.
The quantification of bacteria was based on the following model. A 10-μl suspension of bacteria with a concentration of 107 cells per ml applied to a glass surface in a circle of 1 cm results in 40 cells per average microscopic field of 200 μm in diameter at a magnification of ×1,000 (3). One bacterium would thus be found in an area of 785 μm2. The sections used for FISH evaluation are 4 μm thick. One bacterium averages 0.8 μm in size. Under these assumptions, a surface of 100 μm2 containing 1 (or 100) bacterial cells will correspond to a concentration of 0.4 ×109 (or 0.4 × 1011) cells per ml. Two hundred fifty bacterial cells per 100 μm2 does not allow the visual distinction of a single bacterium, but spaces between bacteria can still be seen; cases such as this were therefore assigned a concentration of 1011 cells per ml. A homogeneous carpet with no empty spaces between them occurs as bacterial counts increase to 2,500 cells/100 μm2; these cases were assigned a concentration of 1012 cells per ml.
Evaluation of the spatial interrelationship of bacteria, assessment of cross-hybridization, and potential cosuppression of FISH probes.
The spatial organization and the composition of the biofilm were evaluated by using a cumulative multistep extension analysis which included detection of bacteria with single FISH probes (orange fluorescence; Cy3) and DAPI DNA counterstain (blue fluorescence). For each biopsy specimen, probes that positively hybridized with more than 1% of the bacteria that were visualized by DAPI were further combined with each other in pairs and triplets and applied simultaneously in a single hybridization. This permitted a three- or four-color analysis of the population structure within the same microscopic field (Cy3, FITC, Cy5, and DAPI stain orange, green, dark red, and blue, respectively). The hybridizations with different probe combinations were cumulatively extended until the positions of all relevant bacterial groups in relation to each other and their relative concentrations were clarified.
When probes specific for unrelated bacterial groups hybridized with the same bacteria, the hybridization stringency was adjusted until a clear differentiation of the bacterial groups was possible. Probes that cross-hybridized even under high-stringency conditions were excluded from the evaluation, and the enumeration results for single bacterial groups were corrected.
Statistics.
Mean values and standard deviations were calculated from the bacterial counts. By using analysis of variance, a P value of <0.05 was considered significant.
RESULTS
Mucosal bacteria were found at concentrations greater than 109 bacteria/ml in 93% of IBD patients, 95% of patients with slc, 65% of IBS patients, and 35% of healthy controls (Table 3). The concentrations of mucosal bacteria in CD patients were 2 powers higher than those in IBS patients and the controls (P < 0.001). The adherent components were most prominent in IBD (UC or CD) patients. With an exception of two patients with slc, adherent bacteria wrapping the entire circumference of the intact colonic epithelium were observed exclusively in CD and UC patients (Fig. 1). Mucus-scattered and mucus ceiling bacteria predominated in the IBS and control groups.
TABLE 3.
Occurrence, concentrations, distribution of the mucosal bacteria, and the percentage of bacteria accessible by FISH
| Characteristic | CD patients | UC patients | Slc patients | IBS patients | Controls | IBD patients on antibiotics |
|---|---|---|---|---|---|---|
| No. of patients with concn of mucosal bacteria greater than 109/ml | 19 | 18 | 19 | 13 | 7 | 0a (3) |
| No. of patients with concn of adherent bacteria greater than 1010/ml | 18 | 14 | 6 | 2 | 0 | 0 |
| Mean concn of mucosal bacteria (1010/ml) ± SEMb,c | 9.1 ± 20.2 | 0.26 ± 0.31 | 0.31 ± 0.16 | 0.09 ± 0.18 | 0.02 ± 0.06 | 0.02 |
| Ileum | 12.4 | 0.28 | 0.25 | 0.15 | 0.004 | |
| Ascending colon | 6.8 | 0.21 | 0.39 | 0.04 | 0.02 | |
| Sigmoid colon | 8.2 | 0.3 | 0.29 | 0.07 | 0.04 | |
| Mean concn of mucosa adherent bacteria (1010/ml)c | 11.2 | 0.31 | 0.35 | 0.07 | 0.02 | 0.02 |
| Mean concn of mucus spread bacteria (1010/ml)c | 2.4 | 0.08 | 0.34 | 0.11 | 0.03 | 0 |
| Mean concn of mucus ceiling bacteria (1010/ml)c | 13.9 | 0.41 | 0.23 | 0.10 | 0.02 | >0a |
| Mean % of intact epithelial surface covered with mucosal bacteriac | 85 | 52 | 52 | 21 | 7 | Unclear |
| Mean % of epithelial surface showing bacterial adherencec | 52 | 35 | 14 | <5 | <5 | Unclear |
| % of patients with mucosal bacteria poorly or not accessible to probe Eub338 | 25 | 80 | 0 | 0 | 0 | 100 |
Bacteria were enumerated based exclusively on DAPI staining (six patients). In only three patients about 5% of DAPI-stained bacteria hybridized with FISH probes. A comparison with the other groups was therefore not possible.
P < 0.001 by analysis of variance. The P value gives the difference between groups with inflammation (CD, UC, and slc patients) and groups without colonic inflammation (IBS patients and controls).
Mean value for all colonic biopsy specimens of the same patient.
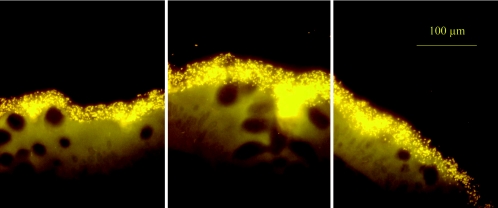
FIG. 1.
Three adjacent microscopic fields of the ascending colon of an untreated CD patient show a biofilm containing adherent Bacteroides fragilis (visualized with the Bfra-Cy3 probe). The biofilm completely covers the mucosal surface and enters the crypts. The epithelial tissue structures are not stained; however, they are well visualized due to autofluorescence.
Occurrence, composition, and spatial arrangement of different bacterial groups within the mucosal biofilm. (i) Occurrence.
Bacteria that hybridized with probes Erec, Bac (Bfra, Bdis), and Fprau were detected in each of the patients not treated with antibiotics. The occurrence of other bacterial groups was variable and had no obvious dependence on disease category (Table 4).
TABLE 4.
Occurrence of different bacterial groups within mucus of patients with elevated concentrations of mucosal bacteriaa
| Bacterial group found
| ||||
|---|---|---|---|---|
| Always | In up to 80% of patients | In 40 to 60% of patients | In 20 to 40% of patients | In less than 20% of patients or singularly |
| Eubacterium rectale | Atoprobium-Coriobacterium | Enterobacteriacae | Clostridium histolyticum | Brachyspira |
| Bacteroides-Prevotella | Ruminococcus bromii | Lachnospira multipara | Clostridium lituseburense | |
| Bacteroides fragilis | Enterococcus faecalis | Bifidobacterium | Ruminococcus flavefaciens | Ruminococcus obeum-like bacteria |
| Bacteroides distasonis | Firmicutes | Streptococcus | ||
| Fusobacterium prausnitzii | Enterococcus | Veillonella | ||
| Actinobacteria | Arcobacter | Lactobacillus and Enterococcus | ||
| Phascolarctobacterium faecium | Cytophage flavobacteria | |||
| Eubacterium hallii | ||||
An elevated concentration was >109 cells/ml.
(ii) Maximal observed percentage of single bacterial groups within the biofilm.
The probes specific for Eubacterium rectale (Erec), Bacteroides fragilis (Bac and Bfra), Brachyspira (Ser), and members of the family Enterobacteriaceae and Escherichia coli (Ebac and Ec) could hybridize with more than 50% of the whole Eubacteria kingdom-positive population within the biofilm.
The probes specific for the Clostridium histolyticum (Chis), Lachnospira multipara (Lach), Veillonella (Veil), and Ruminococcus bromii (Rbro) usually hybridized with ≤1% of the bacterial population. However, in some patients with self-limiting colitis, the abundance of these bacterial groups reached 10 to 20% of the biofilm mass. This was seen in three patients with the Rbro probe, two patients with the Chis probe, one patient with the Veil probe, and one patient with the Lach probe.
The probes specific for Bacteroides distasonis (Bdis), Atopobium (Ato), Coriobacterium (Cor), Eubacterium hallii (EHal), Lactobacillus (Lab), Bifidobacterium (Bif), Ruminococcus flavefaciens (Rfla) and Streptococcus (Strc) most frequently hybridized with less than 1% of the mucosal bacteria; and these bacteria never represented >5% of the whole population.
Probes HGC, LGC, Enc, Ecyl, Phasco, and Clit most frequently hybridized with less than 0.1% of the population. Only in a single patient with slc did they hybridize with about 1% of the bacterial population.
Bacteria that hybridized with probes Arc, UroA, and UroB were seen only singularly within one biopsy specimen. Probes Y16s-69, Sfb, Hpy-1, Dss658, and Aer failed to give signals that were different from the background fluorescence seen with the nonsense probe.
Probes Arch915, Gam42a, Alpf 1b, Beta42a, and Srb385 showed a high grade of cross-hybridization and were excluded from the evaluation.
(iii) Disease-related differences in occurrence and composition of the mucosal flora.
Bacteroides was dominant in biopsy specimens from IBD patients, especially in patients with Crohn's disease. Bacteria that hybridized with the Bac-Bfra probe represented up to 80% of all mucosal bacteria in some IBD patients. In contrast, the mean prevalence of Bacteroides spp. was lower than 15% in patients with IBS. The biofilm in patients with IBS was mainly composed of bacteria that hybridized with the Erec probe (Table 5; Fig. 2). In some IBS patients, bacteria that hybridized with the Erec probe represented up to 90% of all bacteria that positively hybridized with the universal Eub338 probe.
TABLE 5.
Mean percentage of different bacterial groups within the mucosal biofilm
| Parameter | CD patients | UC patients | Slc patients | IBS patients | Controls |
|---|---|---|---|---|---|
| Mean % of the following bacterial groups within biofilm: | |||||
| Bacteroides-Prevotellaa (P < 0.001)b | 71 ± 20 | 62 ± 25 | 40 ± 19 | 20 ± 18 | 20 ± 11 |
| Bacteroides fragilisa (P < 0.001)b | 60 ± 21 | 30 ± 17 | 31 ± 16 | 14 ± 10 | 16 ± 12 |
| Eubacterium rectale-Clostridium coccoidesa (P < 0.001)b | 14 ± 13 | 5 ± 4 | 18 ± 15 | 48 ± 19 | 32 ± 13 |
| Fusobacterium prausnitziia (P > 0.05)b | 6 ± 8 | 7 ± 7 | 9 ± 7 | 13 ± 12 | 10 ± 8 |
| Enterococcus faecalisa | 4 | 1 | 3 | 1 | 0.1 |
| No. (%) of patients positive for E. colic | 10 (53) | 8 (44) | 9 (47) | 5 (41) | 1 (25) |
| Mean (maximum) concn of E. colic | 11 (60) | 5 (10) | 9 (48) | 4 (10) | <1 |
Bacterial groups detected in each patient with mucosal bacteria.
The P value gives the difference between groups with inflammation (CD, UC, and slc patients) and groups without colonic inflammation (IBS patients and controls).
Bacterial groups detected in subsets of patients positive for mucosal bacteria.
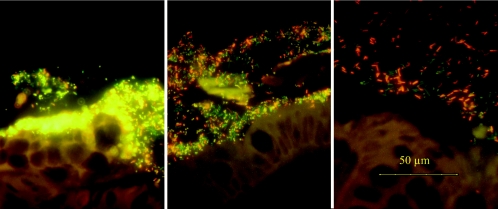
FIG. 2.
Triple-color FISH identifies organisms present in biofilms covering the mucosae in patients with CD (left), self-limiting colitis (middle), and IBS (right). Bacteroides fragilis (Bfra-Cy3 probe) appears yellowish on a green background; the Eubacterium rectale group (Erec-Cy5 probe) appears dark red. All other bacteria that hybridize exclusively with the universal probe (probe Eub-FITC) appear green. There is a striking increase in Bacteroides fragilis concentrations from patients with IBS to patients with IBD, along with changes in bacterial localization.
The biofilm in patients with self-limiting colitis was more complex. The Eubacterium rectale and Bacteroides fragilis groups together averaged approximately two-thirds of the biomass. The remaining third was composed of a highly variable mix of different bacterial groups, which were nearly unique to each patient. Nevertheless, bacteria that hybridized with probes Chis (n = 2), Rbro (n = 3), Lach (n = 1), and Veil (n = 1) constituted more than 10% of the bacterial population only in patients with slc. In all other patients, the concentrations of these bacterial groups were ≤1%. Similarly, the concentrations of bacteria that hybridized with probes Ehal, Lab, Bifi, and Rfla reached the 5% level in at least one patient with slc but in none of the other groups.
No other disease-related differences in concentrations and occurrence were apparent. This could be due to the small number of patients and the broad range of different species covered by single probes. For example, in most cases E. coli was observed at concentrations ranging from 0.1% to 5%. These low concentrations of “E. coli bacteria” were woven as less numerous components in biofilms and showed no signs of adhesion. However, in five patients (three with CD and two with slc), the concentration of Ec probe-positive bacteria reached up to 50%, and the bacteria were tightly attached to the epithelial surface as a nearly homogenous band, probably indicating another subtype of E. coli.
(iv) Spatial organization of the mucosal flora. (a) Adherent bacteria.
Probes specific for the following six bacterial groups hybridized with bacteria adherent to the mucosa: Bacteroides fragilis, Enterobacteriaceae-E. coli, Brachyspira, Eubacterium rectale, Fusobacterium prausnitzii, and Enterococcus faecalis (Table 4). Two types of adherence were observed: coat- or string-like adhesion and a patchy adhesion.
Enterobacteriacea-E. coli (Ebac or Ec probe) and Brachyspira (Ser probe) were sporadically found. A coat of adhesive E. coli was found in three CD patients, one UC patient, and two patients with self-limiting colitis. Brachyspira adherence was observed in one UC patient and one patient with slc.
In contrast, Bacteroides adherence (probe Bac-Bfra) was the main feature of IBD. It was observed in at least one location in 90% of CD patients and 60% of UC patients. A broad, highly concentrated bacterial band was typical for CD. A bead-like or string adhesion was most often seen in UC patients. However, both kinds of adhesion could occur even within the same IBD patient. Despite the significantly more frequent occurrence and the higher concentrations of Bacteroides fragilis in patients with IBD (P < 0. 001), the Bacteroides fragilis adherence was not disease specific, since it was also observed in 10 patients with slc, 2 IBS patients, and 1 healthy control.
The adherence of Eubacterium rectale, Fusobacterium prausnitzii, and Enterococcus faecalis was patchy in most cases (data not shown). Either the bacteria were woven into the band-like biofilm as less numerous components or they were primarily adherent as homogeneous patches interposed with islands of other bacterial groups.
(b) Bacteria invading the mucosa.
The epithelial barrier was intact in all patients and was not eroded by the presence of the adherent bacteria. Bacteria within tissue were often seen at biopsy sites that were mechanically damaged but not at the sites where the epithelial layer was intact (data not shown). For this reason, tissue invasion was impossible to discriminate from contamination by fecal bacteria that invaded tissue cracks caused by the mechanical pressure of the biopsy forceps while they were dragged through the endoscopic canal.
(c) Intracellular bacteria.
Intracellular bacteria were found in single epithelial cells of patients with CD (n = 5), UC (n = 3), and self-limiting colitis (n = 3). Intracellular bacteria were most often located in epithelial cells next to the basal membrane (Fig. 3) and comprised Bacteroides fragilis and Eubacterium rectale populations. A maximum of one to two epithelial cells containing bacteria could be observed.
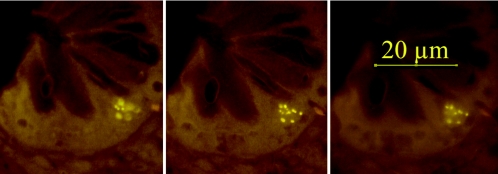
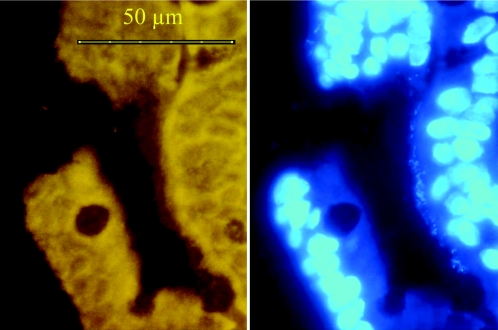
FIG. 3.
Three photos of the same microscopic fields made at different focus levels. Intracellular bacteria (Eub-Cy3 probe) are located in a single epithelial cell at the bottom of the crypt.
(d) Crypt bacteria.
Bacteria within crypts were found in all patients who had bacteria broadly adherent to the mucosal surface (Fig. 1 and 4).
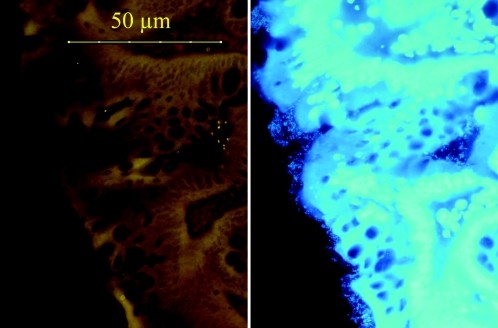
FIG. 4.
Sigmoid colon biopsy specimen from an UC patient treated with 3 g mesalamine orally plus 4-g mesalamine enemas. Only a small number of bacteria located within crypts definitively hybridized with the Eub338 probe universal for bacteria (Cy3 orange fluorescence is on the left). The same microscopic view with DAPI fluorescence revealed a thin but tightly adherent biofilm that was not amenable to FISH probes, that covered the biopsy surface, and that extended deep into the crypt.
The influence of therapy on the mucosal flora.
There were no striking differences in the compositions and concentrations of mucosal bacteria in patients with or without prednisolone or azathioprine treatment (P > 0.19; data not shown).
In patients receiving neither 5-ASA nor antibiotics, 80 to 90% of DAPI-stained bacteria hybridized with the FISH probes, resulting in brilliant pictures (Fig. 1 to 3). In contrast, the fluorescence of the bacteria in 5-ASA-treated patients was weak and was often extinguished while the microscope was focused. Only 1 to 30% of the bacteria that were positive by DAPI staining hybridized with the universal Eub probe in IBD patients treated with 5-ASA (Fig. 4), and the concentrations of the mucosal flora were markedly reduced. The low percentage of bacteria accessible by FISH was thus specific. Patients treated with 5-ASA could be recognized exclusively on the basis of this feature.
No mucosal bacteria could be detected by either FISH or DAPI staining in four IBD patients who had received antibiotics (two of them had received antibiotics 1 day prior to the colonoscopy). Islands of biofilms could be demonstrated in six other antibiotic-treated patients by DAPI staining (Fig. 5). Either none or less than 5% of these bacteria hybridized with universal or other bacterial FISH probes. No difference in the presence of mucosal bacteria was observed between patients receiving antibiotics for 1 day or longer (P > 0.29).
FIG. 5.
Sigmoid colon biopsy specimen from a CD patient who had received antibiotics (metronidazole and ciprofloxacin) the day prior to the colonoscopy. The bacterial biofilm is still seen with the DAPI stain (right) but is not accessible to FISH probes (Eub338 probe, left).
FISH of formalin-fixed biopsy specimens.
Although biofilms were readily observed in Carnoy solution-fixed biopsy specimens, as described above, no biofilm could be observed in formalin-fixed biopsy specimens taken from the same location by either FISH, DAPI, or Gram staining. Only single bacteria were detected scattered in the residual mucus at low concentration (data not shown).
DISCUSSION
The intestinal flora is fascinating for its immense impact on human well-being and is also challenging to analyze because of the lack of relevant data. In the last 100 years, attempts to identify characteristic changes in the intestinal flora in IBD have been inconsistent (9, 25). Most previous investigations were based on studies of bacterial isolates. However, the bacteria within the intestine are spatially organized (34). Sampling of the intestinal contents disrupts the structure of the microbiota. Remarkable progress has been achieved in recent years by in situ identification of bacteria based on rRNA gene analysis. The scientific group of Amann (1, 2, 10, 20, 21, 24, 27, 29) promoted the development of probes and methods for in situ hybridization of environmental microbiota. The scientific group of Welling and Harmsen (11, 12, 13, 14, 15, 18, 32, 35) contributed decisively to the development of FISH probes specific for constituents of the fecal community. Many other groups worldwide have made important contributions to the characterization of diverse fecal bacterial groups. Despite this remarkable progress, the spatial organization of the mucosal flora in patients with IBD has not been systematically investigated. This is important, since IBD is thought to represent an aberrant immune response against enteric bacteria. We developed a cumulative multistep extension approach to address this issue. This approach allowed us to directly compare the locations of different bacterial groups in relation to each other and to the mucosa and to assess potential cross-hybridization.
We found adherent bacterial biofilms in practically all IBD patients who had no recent history of antibiotic or 5-ASA treatment. Although the mucosal bacterial concentrations were higher than 109/ml in 35% of healthy controls and 65% of IBS patients, the mean concentrations of bacteria in both groups were at least 2 powers lower than those in CD patients, and their appearances and compositions were different. The biofilm in untreated IBD patients was thick, dense, and adherent. Bacteroides made up more than two-thirds of the biofilm in patients with IBD. The proportion of the Eubacterium rectale group (detected with the Erec probe) was less than 15%. In contrast, the Eubacterium rectale group was predominant in IBS patients. The biofilm in patients with self-limiting colitis was more diverse, less concentrated, and often loosely attached to the mucosal surface. The Eubacterium rectale and Bacteroides groups together contributed approximately two-thirds of the biomass. Biofilms were found only sporadically in healthy controls and had no recognizable patterns.
The differences between patients with IBD and the other groups of patients would probably be more impressive if the patients had not been treated at the time of the colonoscopy. The biofilm in patients receiving no therapy was highly accessible to FISH. More than 80% of DAPI-stained bacteria hybridized with universal and group-specific probes, providing brilliant pictures, irrespective of the diagnosis. In contrast, the accessibility of the biofilm in some CD patients and many UC patients was strikingly low. Only 1% to 30% of the adherent bacteria visualized by DAPI staining were accessible by FISH probes. This low degree of accessibility of bacteria by FISH was seen in particular in patients receiving 5-ASA. IBD patients not treated with 5-ASA had highly fluorescent biofilms. A similar but more profound reduction of the proportion of bacteria accessible by FISH was observed in patients receiving antibiotics, even with just 1 day of therapy.
FISH with rRNA-targeted probes is dependent on the rRNA content in the individual cells, which is related to metabolic activity. The low ratio of bacteria accessible by FISH in IBD patients treated with 5-ASA and antibiotics indicates that 5-ASA must at least have bacteriostatic activity, in addition to its known anti-inflammatory properties. On the other hand, the persistence of adherent bacteria that are not accessible by FISH in patients treated with either 5-ASA or antibiotics indicates that the biofilm can survive antibacterial substances while staying metabolically silent.
Bacterial involvement in IBD is usually discussed in the context of mucosal barrier changes caused by destructive inflammation. Astonishingly, we found no clear bacterial infiltration of the mucosa in any of the patients. Bacteria were always located above but never below the epithelial layer, even in biopsy specimens with severe inflammation. Submucosal bacteria were seen exclusively in regions next to snatches, rips, and drags of the epithelial surface that were most likely caused by damage of the tissue with the biopsy forceps but not by the inflammation. Similarly, bacteria found in crypts or intracellular locations were secondary to adherence following prolific biofilms and were even found in regions without any signs of inflammation within the same patient. Thus, the crossing of the mucus and bacterial adherence take place before the mucosal destruction.
In contrast to the well-characterized high degree of diversity of the fecal bacteria, the diversity of the mucosal bacteria was astonishing low. Of all bacterial groups investigated, only six were found to be adherent: Bacteroides (probes Bac-Bfra and Bdis), Enterobacteriaceae-E. coli (probe Ebac-Ec) Brachyspira (probe Ser) Fusobacterium prausnitzii (probe Fprau), Eubacterium rectale (probe Erec), and Enterococcus faecalis (probe Efaec). Only four of them were found within the mucus of each patient: Bacteroides fragilis, Bacteroides distasonis, Eubacterium rectale, and Fusobacterium prausnitzii. Although other bacterial groups are commonly found in feces (11, 12, 13, 35), these either were occasionally found within mucosal biofilm in 10% to 50% of the patients or were not detected at all. Obviously, only a small portion of the so-called indigenous intestinal flora can selectively penetrate the mucus or even adhere to the epithelial surface, opening the door for opportunists.
We found only two publications dedicated to the in situ hybridization of mucosal bacteria in patients with IBD based on rRNA-targeted probes (17, 28). In both publications, paraffin sections of formalin-fixed tissues were investigated retrospectively. Only low concentrations of scattered bacteria were identified in those studies, and neither described an adherent biofilm. In our study, the comparison of matched biopsy specimens fixed either in Carnoy solution or in 10% normal buffered formalin showed that the adherent bacterial biofilm is completely lost upon formalin fixation. Thus, archived formalin-fixed paraffin-embedded blocks cannot be used for in situ hybridization studies. Fixation in nonaqueous Carnoy solution, which preserves the mucus layer, appears to be necessary for detection of biofilm in paraffin-embedded tissues.
In summary, the FISH evaluation of the mucosal flora has reached a level of diagnostic significance. We found that an adherent mucosal biofilm mainly composed of Bacteroides fragilis is a prominent feature in patients with IBD. We can now reliably characterize and monitor the mucosal biofilm in the course of IBD and other intestinal diseases. Further studies are needed to determine the mechanisms that regulate biofilm development, propagation, and resistance and the specific role of biofilms in the pathogenesis of intestinal inflammation.
Acknowledgments
We thank the Broad Medical Research Program of The Eli and Edythe L. Broad Foundation for supporting this project.
REFERENCES
- 1.Amann, R., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R., J. Stromley, R. Devereux, R. Key, and D. A. Stahl. 1992. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl. Environ. Microbiol. 58:614-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohnert, J., B. Hübner, and K. Botzenhart. 2002. Rapid identification of Enterobacteriaceae using a novel 23S rRNA-targeted oligonucleotide probe. Int. J. Hyg. Environ. Health 203:77-82. [DOI] [PubMed] [Google Scholar]
- 5.Chapman, R. W., W. S. Selby, and D. P. Jewell. 1986. Controlled trial of intravenous metronidazole as an adjunct to corticosteroids in severe ulcerative colitis. Gut 27:1210-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 7.Costerton, J. W., R. Veeh, M. Shirtliff, M. Pasmore, C. Post, and G. Ehrlich. 2003. The application of biofilm science to the study and control of chronic bacterial infections. J. Clin. Investig. 112:1466-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feydt-Schmidt, A., H. Rüssmann, N. Lehn, A. Fischer, L. Antoni, D. Störk, and S. Koletzko. 2002. Fluorescence in situ hybridization vs. epsilometer test for detection of clarithromycin-susceptible and clarithromycin-resistant Helicobacter pylori strains in gastric biopsies from children. Aliment. Pharmacol. Ther. 16:2073-2079. [DOI] [PubMed] [Google Scholar]
- 9.Fiocchi, C. 1998. Inflammatory bowel disease; etiology and pathogenesis. Gastroenterology 115:182-205. [DOI] [PubMed] [Google Scholar]
- 10.Frahm, E., I. Heiber, S. Hoffman, C. Koob, H. Meier, W. Ludwig, R. Amann, K. H. Schleifer, and U. Obst. 1998. Application of 23S rDNA-targeted oligonucleotide probes specific for enterococci to water hygiene control. Syst. Appl. Microbiol. 21:450-453. [DOI] [PubMed] [Google Scholar]
- 11.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harmsen, H. J., P. Elfferich, F. Schut, and G. W. Welling. 1999. A 16S rRNA-targeted probe for detection of lactobacilli and enterococci in fecal samples by fluorescent in situ hybridization. Microbiol. Ecol. Health Dis. 11:3-12. [Google Scholar]
- 13.Harmsen, H. J., A. C. Wildeboer-Veloo, J. Grijpstra, J. Knol, J. E. Degener, and G. W. Welling. 2000. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl. Environ. Microbiol. 66:4523-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harmsen, H. J., G. C. Raangs, T. He, J. E. Degener, and G. W. Welling. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jansen, G. J., M. Mooibroek, J. Idema, H. J. Harmsen, G. W. Welling, and, J. E. Degener. 2000. Rapid identification of bacteria in blood cultures by using fluorescently labeled oligonucleotide probes. J. Clin. Microbiol. 38:814-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen, T. K., M. Boye, P. Ahrens, B. Korsager, P. S. Teglbjaerg, C. F. Lindboe, and K. Moller. 2001. Diagnostic examination of human intestinal spirochetosis by fluorescent in situ hybridization for Brachyspira aalborgi, Brachyspira pilosicoli, and other species of the genus Brachyspira (Serpulina). J. Clin. Microbiol. 39:4111-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleessen, B., A. J. Kroesen, H. J. Buhr, and M. Blaut. 2002. Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand. J. Gastroenterol. 37:1034-1036. [DOI] [PubMed] [Google Scholar]
- 18.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16SrRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malchow, H., K. Ewe, J. W. Brandes, H. M. Goebell, H. Ehms, H. Sommer, and H. Jesdinsky. 1984. European cooperative Crohn's disease study (ECCDS): results of drug treatment. Gastroenterology 86:249-266. [PubMed] [Google Scholar]
- 20.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K. H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 21.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 22.Matsuo, K., H. Ota, T. Akamatsu, A. Sugiyama, and T. Katsuyama. 1997. Histochemistry of the surface mucous gel layer of the human colon. Gut 40:782-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mearin, F., M. Roset, X. Badia, A. Balboa, E. Baro, J. Ponce, M. Diaz-Rubio, E. Caldwell, M. Cucala, A. Fueyo, and N. J. Talley. 2004. Splitting irritable bowel syndrome: from original Rome to Rome II criteria. Am. J. Gastroenterol. 99:122-130. [DOI] [PubMed] [Google Scholar]
- 24.Meier, H., R. Amann, W. Ludwig, and K. H. Schleifer. 1999. Specific oligonucleotide probes for in situ detection of a major group of gram-positive bacteria with low DNA G+C content. Syst. Appl. Microbiol. 22:186-196. [DOI] [PubMed] [Google Scholar]
- 25.Podolsky, D. K. 2002. Inflammatory bowel disease. N. Engl. J. Med. 347:417-429. [DOI] [PubMed] [Google Scholar]
- 26.Poulsen, L. K, T. R. Licht, C. Rang, K. A. Krogfelt, and S. Molin. 1995. Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin-treated mice. J. Bacteriol. 177:5840-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K. H. Schleifer. 1994. In situ probing of gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology 140:2849-2858. [DOI] [PubMed] [Google Scholar]
- 28.Schultsz, C., F. M. Van Den Berg, F. W. Ten Kate, G. N. J. Tytgat, and J. Dankert. 1999. The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls. Gastroenterology 117:1089-1097. [DOI] [PubMed] [Google Scholar]
- 29.Snaidr, J., R. Amann, I. Huber, W. Ludwig, and K. H. Schleifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suau, A., V. Rochet, A. Sghir, G. Gramet, S. Brewaeys, M. Sutren, L. Rigottier-Gois, and J. Dore. 2001. Fusobacterium prausnitzii and related species represent a dominant group within the human fecal flora. Syst. Appl. Microbiol. 24:139-145. [DOI] [PubMed] [Google Scholar]
- 31.Swidsinski, A., A. Ladhoff, A. Pernthaler, S. Swidsinski, V. Loening-Baucke, M. Ortner, J. Weber, U. Hoffmann, S. Schreiber, M. Dietel, and H. Lochs. 2002. Mucosal flora in inflammatory bowel disease. Gastroenterology 122:44-54. [DOI] [PubMed] [Google Scholar]
- 32.Trebesius, K., D. Harmsen, A. Rakin, J. Schmelz, and J. Heesemann. 1998. Development of rRNA-targeted PCR and in situ hybridization with fluorescently labelled oligonucleotides for detection of Yersinia species. J. Clin. Microbiol. 36:2557-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urdaci, M. C., B. Regnault, and P. A. D. Grimont. 2001. Identification by in situ hybridization of segmented filamentous bacteria in the intestine of diarrheic trout (Oncorhynchus mykiss). Res. Microbiol. 152:67-73. [DOI] [PubMed] [Google Scholar]
- 34.Wilson, M. 2001. Bacterial biofilms and human disease. Sci. Prog. 84:235-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zoetendal, E. G., K. Ben-Amor, H. J. Harmsen, F. Schut, A. D. Akkermans, and W. M. de Vos. 2002. Quantification of uncultured Ruminococcus obeum-like bacteria in human fecal samples by fluorescent in situ hybridization and flow cytometry using 16S rRNA-targeted probes. Appl. Environ. Microbiol. 68:4225-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]