Abstract
Loop-mediated isothermal amplification (LAMP) is a novel nucleic acid amplification method in which reagents react rapidly and efficiently, with a high specificity, under isothermal conditions. We used a LAMP assay for the detection of herpes simplex virus type 1 (HSV-1), herpes simplex virus type 2 (HSV-2), and varicella-zoster virus (VZV). The virus specificities of primers were confirmed by using 50 HSV-1, 50 HSV-2, and 8 VZV strains. The assay was performed for 45 min at 65°C. The LAMP assay had a 10-fold higher sensitivity than a PCR assay. An analysis of nucleotide sequence variations in the target and primer regions used for the LAMP assay indicated that 3 of 50 HSV-1 strains had single nucleotide polymorphisms. No HSV-2 or VZV strains had nucleotide polymorphisms. Regardless of the sequence variation, there were no differences in sensitivity with the HSV-1-specific LAMP assay. To evaluate the application of the LAMP assay for clinical diagnosis, we tested clinical samples from 40 genital herpes patients and 20 ocular herpes patients. With the LAMP assay, 41 samples with DNA extraction and 26 direct samples without DNA extraction were identified as positive for HSV-1 or HSV-2, although 37 samples with DNA extraction and just one without DNA extraction were positive by a PCR assay. Thus, the LAMP assay was less influenced than the PCR assay by the presence of inhibitory substances in clinical samples. These observations indicate that the LAMP assay is very useful for the diagnosis of HSV-1, HSV-2, and VZV infections.
Herpes simplex virus types 1 (HSV-1) and 2 (HSV-2) and varicella-zoster virus (VZV) are alphaherpesviruses that infect, establish lifelong latency in, and subsequently reactivate from human sensory neuronal ganglia (1, 20). The reactivation of a latent HSV or VZV infection can occur spontaneously or in association with physical or emotional stress and immune suppression. Following reactivation from latent ganglion reservoirs, each of these herpesviruses may cause significant clinical symptoms in the individual and may spread to uninfected persons. Symptomatic VZV reactivation is an infrequent, usually once-in-a-lifetime event in 10% of the population that results in zoster (shingles), while HSV-1 and HSV-2 reactivation occurs frequently and results in numerous symptomatic and asymptomatic recurrences.
HSV-1 and HSV-2 infections and even some VZV infections cause similar clinical symptoms, for example, cutaneous vesicles, keratitis, and acute retinal necrosis (ARN), and the causative agent cannot be distinguished based on the clinical features. However, different clinical courses and prognoses are caused by each virus. HSV-2 genital herpes tends to recur, but HSV-1 genital herpes does not (7). VZV ARN is severer and has a worse prognosis than HSV ARN (4). The identification of the virus is important for the determination of treatment and for an understanding of the clinical progress and prognosis; therefore, an effective laboratory method is urgently needed for the diagnosis of HSV-1, HSV-2, and VZV infections.
At present, molecular methods such as dot blotting, in situ hybridization, and PCR have been developed for pathogen detection. In particular, PCR has been used for numerous recent applications for pathogen detection and pathology research. However, these methods require precision instruments for amplification and DNA extraction, which are the major obstacles to the widespread use of these methods in relatively small-scale clinical laboratories such as those in private clinics. Recently, a new, rapid, and sensitive technique called loop-mediated isothermal amplification (LAMP) was developed (13). LAMP is a nucleic acid amplification method which relies on autocycling strand-displacement DNA synthesis performed by the Bst DNA polymerase large fragment. The amplification products are stem-loop DNA structures with several inverted repeats of the target and structures with multiple loops. The LAMP reaction can be conducted under isothermal conditions ranging from 60 to 65°C by using only one type of enzyme and four primers recognizing six distinct regions. The most important merit of this method is that no denaturation of the DNA template is required (11). Moreover, when two primers, termed loop primers, are added, the LAMP reaction time can be shortened (12). With this modification, the LAMP method is able to amplify the target DNA in a shorter time than and with an extremely high specificity compared to the PCR method. Moreover, the LAMP method produces a large amount of amplified product, resulting in easier detection, such as detection by visual judgment based on the turbidity or fluorescence of the reaction mixture (10). Recently, several investigators have reported the rapid identification of Mycobacterium (6), human herpesvirus 6 (5), human herpesvirus 7 (22), West Nile virus (14), and the severe acute respiratory syndrome coronavirus (3) and have commended the usefulness of the LAMP assay.
For the present study, we used a LAMP assay as the detection system for the diagnosis of HSV-1, HSV-2, and VZV infections. The sensitivity, specificity, and applicability of this method for the direct detection of HSV-1, HSV-2, and VZV from clinical specimens were also evaluated.
MATERIALS AND METHODS
Cells and viruses.
An African green monkey kidney cell line (Vero) and human embryonic lung (HEL) fibroblast cells were cultivated in Dulbecco modified Eagle's minimum essential medium (DMEM) supplemented with 10% calf serum. We used 50, 50, and 8 clinically isolated strains of HSV-1, HSV-2, and VZV, respectively. HSV-1 strains were isolated from patients with either genital herpes (n = 20), herpetic keratitis (n = 15), or herpetic conjunctivitis (n = 15). All HSV-2 strains were isolated from patients with genital herpes. The source of the VZV strains is unknown. HSV and VZV strains were isolated on Vero cells and HEL cells, respectively, and stocked at −80°C after a few passages. Each virus was identified and confirmed by a PCR method.
DNA extraction.
Virus DNA was extracted from infected cells or clinical samples by proteinase K treatment and phenol-chloroform extraction. After ethanol precipitation, the DNA was suspended in H2O and stored at −20°C until use.
LAMP method.
We used six primers for the LAMP assay, including two outer primers (F3 and B3), a forward inner primer (FIP), a backward inner primer (BIP), and two loop primers (LPF and LPB). The location and sequence of each primer are shown in Fig. 1 and Table 1, respectively. FIP consisted of F1 and F2, and BIP consisted of B1 and B2. F3 and B3 were located outside of F2 and B2. Since additional loop primers increase the amplification efficiency, we also synthesized loop primers (LPF and BPF). HSV-1-, HSV-2-, and VZV-specific primers were designed to target the UL1 to UL2, US4, and ORF62 gene regions, respectively. In order to design virus-specific primers, we selected regions of low homology with the nucleotide sequences of the other two viruses.
FIG. 1.
Location and sequence of LAMP target and primer regions for HSV-1 (A), HSV-2 (B), and VZV (C). The single nucleotide polymorphisms of three HSV-1 strains (the G1-4, conj10, and KH15 strains) are shaded. Arrows indicate the nucleotide bases of strains with polymorphism.
TABLE 1.
Primers used for LAMP
| Virus | Primer | Sequence (5′-3′) |
|---|---|---|
| HSV-1 | F3 | CAGCCACACACCTGTGAA |
| B3 | TCCGTCGAGGCATCGTTAG | |
| FIP(F1-F2) | CCAGACGTTCCGTTGGTAGGTC-ACTTTGACTATTCGCGCACC | |
| BIP(B1-B2) | CCATCATCGCCACGTCGGAC-TCGGCGTCTGCTTTTTGTG | |
| LPF | AAATCCTGTCGCCCTACACAGCGG | |
| LPB | CACCCCGCGACGGGACGCCG | |
| HSV-2 | F3 | TCAGCCCATCCTCCTTCG |
| B3 | GCCCACCTCTACCCACAA | |
| FIP(F1-F2) | CCCTGGTACGTGACGTGTACG-AGTATGGAGGGTGTCGCG | |
| BIP(B1-B2) | AAATGCTTCCCTGCTGGTGCC-CGCCGAGTTCGATCTGGT | |
| LPF | CACGTCTTTGGGGACGGCGGCT | |
| LPB | ATCTGGGACCGCGCCGCGGAGACAT | |
| VZV | F3 | CCAATACGACCACCGGATC |
| B3 | TGGCTTCGTCTCGAGGATTT | |
| FIP(F1-F2) | CTCCCACTGGTACGTCAAGTGG-AGGGTCAAAAACCCTGGC | |
| BIP(B1-B2) | TTCCTCAACATCCCCGACATCG-TACCCGATGGGGGATACC | |
| LPF | CGAAATGTAGGATATAAAGG | |
| BPF | CATCCTTATATTCAAAAGTAGCT |
The LAMP reaction was performed with a Loopamp DNA amplification kit (Eiken Chemical Co., Ltd., Tochigi, Japan). A reaction mixture (12.5 μl) containing a 1.6 μM concentration of each inner primer (FIP and BIP), a 0.2 μM concentration of each outer primer (F3 and B3), a 0.8 μM concentration of each loop primer (LPF and BPF), 2× reaction mix (6.25 μl), Bst DNA polymerase (0.5 μl), and 1.0 μl of DNA sample was incubated at 65°C for 45 min in a Loopamp real-time turbidmeter (LA-200; Teramecs) and then heated above 80°C for 2 min to terminate the reaction. A portion of each amplified product was analyzed in a 2% agarose gel containing 0.5 μg of ethidium bromide per ml. In addition, the specificity of the LAMP-amplified products was further validated by digestion with a restriction enzyme (PstI for HSV-1 amplicons, HaeIII for HSV-2 amplicons, and AluI for VZV amplicons).
PCR.
For an assessment of the sensitivity of the LAMP assay for HSV-1 and HSV-2, PCRs were performed with three primer sets. The first primer set (primer set 1) was B3 and F3, as used in the LAMP assay for HSV-1 and HSV-2 (Table 1). Secondly, we designed a primer set (primer set 2) for HSV-1 and HSV-2 (Table 2). PCR amplification was performed in 15-μl reaction mixtures containing 10 mM Tris, 50 mM KCl, 1.5 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate, a 500 nM concentration of each primer, and 0.5 unit of Taq DNA polymerase. Initial denaturation was done at 95°C for 3 min, followed by 40 cycles of denaturation (30 s at 95°C), annealing (1 min at 55°C and 50°C for primer set 1 and primer set 2, respectively), and extension (45 s at 72°C).
TABLE 2.
Primers used for PCR
| Virus | Primer | Size of PCR product (bp) | Positions | Sequence (5′-3′) |
|---|---|---|---|---|
| HSV-1 | HSV1-seqF | 511 | 9658-9680 | AGTTACCCCGCGTTTCCTGCCAA |
| HSV1-seqR | 10168-10145 | GTTTGTTAAATCCATGGGAGGGTC | ||
| HSV-2 | HSV2-seqF | 637 | 138010-138034 | GTGGCTCAATATTGTTATGCCTATC |
| HSV2-seqR | 138646-138626 | TCTGTGGGGGCAGACAGCGCT | ||
| VZV | VZV-seqF | 556 | 48461-48485 | GACGTTTTCCTCCAACTGTAAAGGT |
| VZV-seqR | 49016-48993 | ATTCCGGGATATTACGCTAGTGGA |
Moreover, in order to check the sensitivity of our PCR assay, we performed PCR amplification by using a commercial primer kit (Maxim Biotech. Inc., Calif.) which has often been used for the diagnosis of HSV in Japanese clinical laboratories (primer set 3). The sequences of these primers were 5′-CATCACCGACCCGGAGAGGGAC-3′ and 5′-GGGCCAGGCGCTTGTTGGTGTA-3′. PCR amplification with primer set 3 was performed in a 50-μl reaction mixture. Initial denaturation was done at 96°C for 1 min, followed by 35 cycles of denaturation (1 min at 94°C), annealing (1 min at 58°C), and extension (1 min at 72°C). Ten microliters of each reaction mixture was subjected to electrophoresis in a 2% agarose gel containing 0.5 μg of ethidium bromide per ml.
Quantification of HSV DNA.
The copy numbers in HSV-1 and HSV-2 DNA samples were analyzed by real-time PCR using a LightCycler (Roche Molecular Biochemicals). For each assay, 2 μl of an extracted DNA sample was added to 18 μl of PCR mixture in each reaction capillary, according to the manufacturer's instructions. The PCR mixture contained 1× FastStart DNA polymerase reaction buffer including deoxynucleoside triphosphates, Taq polymerase, and SYBR green I (Roche Molecular Diagnostics), 3 mM MgCl2, and a 0.5 μM concentration of each of the primers. The sequences of the primers for HSV-1 were 5′-TGTGGTCGTCGACGATTGCAGCAT-3′ and 5′-TGGGAGTGACCCGCGTGGTCGA-3′, and those for HSV-2 were 5′-CTACGACGCGTACCGGTCCGATG-3′ and 5′-GTGGTGCACGAACAGCGTGGTGA-3′. After denaturation for 10 min at 95°C, 40 cycles of amplification at 95°C for 0 s, 55°C for 10 s, and 72°C for 20 s were carried out. The amount of amplified product for the targeted nucleic acid was monitored by SYBR green I staining during each cycle of the PCR. Prediluted standard DNA templates comprising 103 to 107 copies of a quantified PCR product were applied to the real-time PCR analysis to make a standard curve. The copy numbers of HSV-1 and HSV-2 DNA were then determined by using the standard curve.
DNA sequencing of virus strains.
The nucleotide sequences of the LAMP target and primer regions in the HSV-1, HSV-2, and VZV strains were determined by a PCR-directed sequencing method. Briefly, double-stranded DNA templates for sequencing reactions were prepared by PCRs using the primer sets shown in Table 2 as described previously (18). PCR products from the HSV-1, HSV-2, and VZV strains were purified with a QIAquick PCR purification kit (QIAGEN, Hilden, Germany) and used as templates for DNA sequencing reactions. Nucleotide sequencing was performed by use of an autosequencer (ABI PRISM 3100 genetic analyzer) and a commercial kit (BigDye; Applied Biosystems, Foster City, Calif.).
Clinical samples.
We evaluated samples for 40 cases of genital herpes and 20 cases of ocular herpes. The genital samples were vaginal smears from patients suspected of having genital herpes. Ocular herpes samples were obtained from 5 patients with herpetic dendritic keratitis and 15 patients with uveitis and suspected ARN. Keratitis samples were corneal smears and uveitis samples were from aqueous (n = 6) and vitreous (n = 9) fluid. The smear samples were scraped with a cotton swab and placed in 0.5 ml of DMEM. Aqueous samples were obtained by anterior chamber paracentesis. Vitreous samples were obtained at the time of surgery. All samples were stored at −80°C until use. LAMP, PCR, and virus isolation were performed for all clinical samples. The sequences of the PCR primers are summarized in Table 2.
RESULTS
Specificity of LAMP assay.
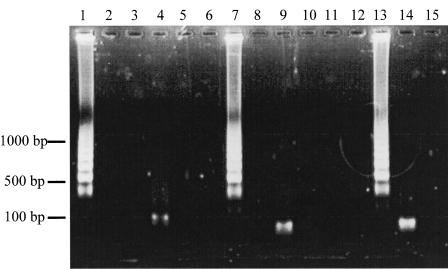
A successful LAMP reaction with virus-specific primers demonstrated many bands of different sizes upon agarose gel electrophoresis (Fig. 2). When the sample tube did not contain target DNA, no amplification was seen. To confirm that the products were amplified from the target region, we digested the products with a restriction enzyme, i.e., PstI for HSV-1, HaeIII for HSV-2, and AluI for VZV. The sizes of the fragments were in good agreement with those predicted theoretically from the expected DNA structures and showed that the reaction was specific.
FIG. 2.
Electrophoretic analysis of LAMP-amplified products. Lanes 1, 2, and 3, LAMP carried out with HSV-1 primers and genomic DNAs from HSV-1, HSV-2, and VZV, respectively; lane 4, LAMP product from lane 1 after digestion with PstI; lanes 5, 10, and 15, LAMP carried out in the absence of template DNA with the HSV-1, HSV-2, and VZV primers, respectively; lanes 6, 7, and 8, LAMP carried out with HSV-2 primers in the presence of genomic DNAs from HSV-1, HSV-2, and VZV, respectively; lane 9, LAMP product from lane 7 after digestion with HaeIII; lanes 11, 12, and 13, LAMP carried out with VZV primers in the presence of genomic DNAs from HSV-1, HSV-2, and VZV, respectively; lane 14, LAMP product from lane 13 after digestion with AluI. The positions of size markers are shown on the left.
Sequence variation of LAMP primer and target regions.
After PCR-directed sequencing of the LAMP target and primer regions, 3 HSV-1 strains, comprising 1 strain causing keratitis (KH15), 1 strain causing conjunctivitis (conj10), and 1 strain causing genitalitis (G1-4), of the 50 total strains had single nucleotide polymorphisms (Fig. 1). The polymorphism sites of the three strains were unique. The variations were in the F3 region of the G1-4 strain, the F1 region of the conj10 strain, and the B1 region of the KH15 strain. No HSV-2 or VZV strains had nucleotide polymorphisms. Thus, we speculated that our LAMP target and primer regions for HSV-1, HSV-2, and VZV had low levels of nucleotide sequence variation.
Sensitivity of LAMP assay for HSV-1 and HSV-2.
The sensitivity of the LAMP assay for HSV-1 and HSV-2 was determined by testing serial 10-fold dilutions of samples for which the copy numbers of virus DNA had previously been quantified by real-time PCR and comparing the results with those of PCR. The analyzed strains showed 100% nucleotide sequence homology in the LAMP primer region. In order to check the sensitivity of PCR accurately, we carried out a PCR assay using three primer sets. Primer set 1 consisted of B3 and F3 from the HSV-1 and HSV-2 LAMP assays, with which regions of low nucleotide sequence variation were proven in this study. The sensitivity of this primer set was much lower than those of primer set 2 and primer set 3 (data not shown). The PCR assay using primer set 2 or primer set 3 had a detection limit of 100 copies/tube. Therefore, we used primer set 2 for this study.
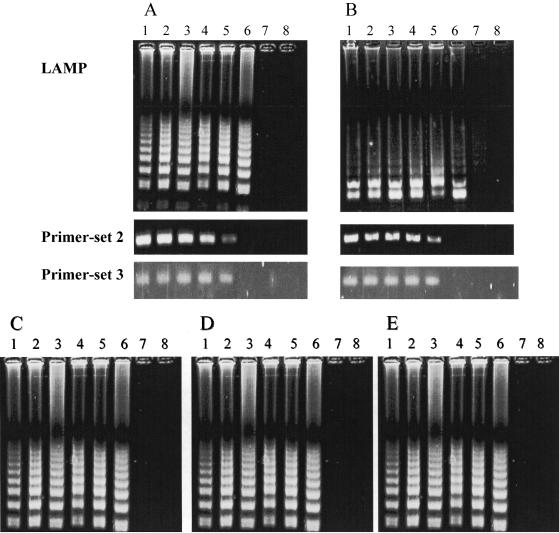
The HSV-1 and HSV-2 LAMP assay could detect as few as 10 copies/tube, which was a 10-fold higher sensitivity than that of the PCR assays with primer set 2 (Table 2) and primer set 3. We studied the influence of nucleotide polymorphisms in the primer region on the sensitivity of the LAMP assay by using three HSV-1 strains. The detection limit for all three strains was 10 copies/tube, and there were no differences in the sensitivity of the LAMP assay for HSV-1, regardless of the presence of single nucleotide variations (Fig. 3).
FIG. 3.
Comparative sensitivities of LAMP and PCR for the detection of HSV-1 (A) and HSV-2 (B). Amplification by LAMP shows a ladder-like pattern, whereas that by primer set 2 and primer set 3 shows single bands for the amplification products obtained by using the primer set shown in Table 2 and that from a commercial PCR kit, respectively. (C to E) Comparative sensitivities to single nucleotide polymorphisms of three HSV-1 strains. The electrophoretic profiles of the G1-4 strain (C), the conj10 strain (D), and the KH15 strain (E) are shown. Lanes: 1, 106 copies/tube; 2, 105 copies/tube; 3, 104 copies/tube; 4, 103 copies/tube; 5, 102 copies/tube; 6, 101 copies/tube; 7, 100 copies/tube; 8, negative control without target DNA.
Evaluation and comparison of LAMP and PCR for detection of DNA in clinical samples.
Previous reports have described clinical samples as containing some PCR inhibitors (8, 9, 15, 21); therefore, DNA preparation is required to obtain sensitive results for PCR assays. In order to clarify whether a DNA preparation procedure was necessary to obtain sensitive results with LAMP, we tested 40 genital and 20 ocular samples, with and without a DNA extraction procedure, by both the LAMP and PCR assays. For the assay of genital herpes samples, 30 samples subjected to DNA extraction were detected as HSV-1 or HSV-2 by both PCR and LAMP, indicating that the methods possessed equal sensitivities (Table 3). Of these 30 samples, HSV was isolated from 24 samples. Using direct sample fluid, we detected HSV-1 or HSV-2 DNA in 21 samples by the LAMP assay, but in just 1 sample by PCR, showing that the LAMP assay was less influenced by inhibitors than was PCR. For the assay of ocular herpes samples, virus DNA was detected by LAMP in 11 of 20 samples subjected to DNA extraction. However, the HSV genome was not detected in four LAMP-positive uveitis samples by PCR. With direct sample fluid, five cases were diagnosed as HSV infections by the LAMP assay, but HSV was not detected in the ocular samples by PCR (Table 3).
TABLE 3.
Comparison of LAMP assay and PCR assay by using clinical samples
| Condition (no. of samples) | Virus | No. of DNA samples with positive resulta
|
No. of direct samples with positive resultb
|
No. of isolated samples | ||
|---|---|---|---|---|---|---|
| LAMP | PCR | LAMP | PCR | |||
| Genitalitis (40) | HSV-1 | 17 | 17 | 12 | 1 | 15 |
| HSV-2 | 13 | 13 | 9 | 0 | 9 | |
| Keratitis (5) | HSV-1 | 4 | 4 | 2 | 0 | 2 |
| HSV-2 | 0 | 0 | 0 | 0 | 0 | |
| Uveitis (15) | HSV-1 | 2 | 1 | 1 | 0 | 0 |
| HSV-2 | 5 | 2 | 2 | 0 | 0 | |
Samples subjected to DNA extraction.
Direct sample fluid without DNA extraction.
DISCUSSION
We used a novel nucleic acid amplification method, LAMP, for the detection of HSV-1, HSV-2, and VZV. With the method used, viruses could be identified within 45 min of DNA extraction. The LAMP operation is quite simple; it starts with the mixing of buffer, primers, DNA lysates, and DNA polymerase in a tube, and then the mixture is incubated at 65°C for a certain period. There is no need for a thermal cycler because there is no heat denaturation step of the template DNAs with this method. The only equipment needed for the LAMP reaction is a regular laboratory water bath or a heat block that furnishes a constant temperature of 65°C. Moreover, the sensitivity of the LAMP assay for HSV-1 and HSV-2 was 10 times higher than that of PCR assays.
The nucleotide sequences of HSV-1 and HSV-2 have high homology, and regions of nucleotide sequence specificity between HSV-1 and HSV-2 are limited. We found sequence specificity for HSV-1 and HSV-2 in the UL1 to UL2 genes and the US4 gene, respectively; therefore, we designed primers to target these regions. The sequences of the HSV-1-specific forward primers (F3, FIP, and LPF) have high homology to HSV-2, but the reverse primers (B3, BIP, and BPF) are specific for HSV-1. All HSV-2-specific LAMP primers had a low homology with HSV-1. The virus specificities of these primers were confirmed in our study.
An evaluation of three strains with single nucleotide sequence variations showed that there was no difference in the sensitivity of the LAMP assay for these HSV-1 strains. Sakaoka et al. reported that the nucleotide diversity of HSV-1 was 0.0037 (3 or 4 nucleotides per 1,000 bases) between any two strains (16). The nucleotide diversity in the LAMP target and primer regions of 271 bp of 50 HSV-1 strains was 0.00044 (4 or 5 nucleotides per 10,000 bases). Thus, the LAMP target and primer regions of HSV-1 are regarded as having adequately low degrees of variation. Compared to HSV-1, HSV-2 and VZV have little nucleotide diversity among strains (2, 17, 19). Our sequencing study indicated that the targeted regions of HSV-2 and VZV have no nucleotide sequence variation and are also adequate for the LAMP assay.
In an evaluation of clinical samples, 30 of 40 patients suffering from genitalitis were identified as having HSV-1 or HSV-2 infection by a LAMP assay with samples subjected to DNA extraction. We could not diagnose whether the other 10 patients suffered from HSV infection or other diseases. In the case of keratitis, the diagnosis of herpes keratitis is relatively easy on the basis of its typical symptom, dendritic keratitis, so the frequency of HSV infection was high (four of five cases). However, the frequency of HSV detection in uveitis samples was low because the frequency of ARN during uveitis is low.
We paid attention to and evaluated the two methods by using clinical samples. We first evaluated the sensitivity of LAMP compared to that of PCR, using samples subjected to DNA extraction. Both LAMP and PCR showed the same sensitivity for identifying genital herpes and herpetic keratitis. However, three of seven LAMP-positive uveitis samples were negative in the PCR assay. This indicated that the sensitivity of LAMP is high enough to detect low DNA copy numbers and is therefore useful for small-volume samples such as those of vitreous and aqueous humor. Next, we evaluated the necessity of DNA extraction. Biological fluids, e.g., aqueous and vitreous humor, contain substances that inhibit PCR (8, 9, 15, 21). Therefore, samples for PCR assays require DNA extraction. Using direct sample fluid (in DMEM) with genital smear suspensions, we identified HSV-1 or HSV-2 in 21 samples by LAMP and in just 1 sample by PCR. Using direct ocular sample fluid, we found that 5 of the 11 samples that were positive after DNA extraction were also positive in the LAMP assay, but none were positive in the PCR assay. The inhibitory substances reduced the sensitivity of PCR, resulting in a PCR sensitivity which was lower than that required for virus isolation. These results indicate that the LAMP assay is more tolerant of substances contained in clinical specimens than is PCR.
In conclusion, the LAMP assay is a simple, rapid, and accurate nucleoside amplification method that is useful for the clinical diagnosis of HSV and VZV infection. Compared to PCR, the LAMP assay possesses advantages in its sensitivity and tolerance of inhibitory substances. These results suggest that the LAMP assay could be introduced into clinical laboratories and even private clinics.
REFERENCES
- 1.Arvin, A. M. 1996. Varicella-zoster virus, p. 2547-2585. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 2.Chiba, A., T. Suzutani, M. Saijo, S. Koyano, and M. Azuma. 1998. Analysis of nucleotide sequence variations in herpes simplex virus types 1 and 2 and varicella-zoster virus. Acta Virol. 42:401-407. [PubMed] [Google Scholar]
- 3.Hong, T. C., Q. L. Mai, D. V. Cuong, M. Parida, H. Minekawa, T. Notomi, F. Hasebe, and K. Morita. 2004. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 42:1956-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ichikawa, T., J. Sakai, Y. Yamauchi, H. Minoda, and M. Usui. 1997. A study of 44 patients with Kirisawa type uveitis. J. Jpn. Opthamol. 41:35. [PubMed] [Google Scholar]
- 5.Ihira, M., T. Yoshikawa, Y. Enomoto, S. Akimoto, M. Ohashi, S. Suga, N. Nishimura, T. Ozaki, Y. Nishiyama, T. Notomi, Y. Ohta, and Y. Asano. 2004. Rapid diagnosis of human herpesvirus 6 infection by a novel DNA amplification method, loop-mediated isothermal amplification. J. Clin. Microbiol. 42:140-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwamoto, T., T. Sonobe, and K. Hayashi. 2003. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J. Clin. Microbiol. 41:2616-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawana, T., T. Kawaguchi, and S. Sakamoto. 1976. Clinical and virological studies on genital herpes. Lancet ii:964. [DOI] [PubMed] [Google Scholar]
- 8.Khan, G., H. O. Kangro, P. J. Coates, and R. B. Heath. 1991. Inhibitory effects of urine on the polymerase chain reaction for cytomegalovirus DNA. J. Clin. Pathol. 44:360-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercier, B., C. Gaucher, O. Feugeas, and C. Mazurier. 1990. Direct PCR from whole blood, without DNA extraction. Nucleic Acids Res. 18:5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mori, Y., K. Nagamine, N. Tomita, and T. Notomi. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150-154. [DOI] [PubMed] [Google Scholar]
- 11.Nagamine, K., K. Watanabe, K. Ohtsuka, T. Hase, and T. Notomi. 2001. Loop-mediated isothermal amplification reaction using a nondenatured template. Clin. Chem. 47:1742-1743. [PubMed] [Google Scholar]
- 12.Nagamine, K., T. Hase, and T. Notomi. 2002. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell Probes 16:223-229. [DOI] [PubMed] [Google Scholar]
- 13.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parida, M., G. Posadas, S. Inoue, F. Hasebe, and K. Morita. 2004. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J. Clin. Microbiol. 42:257-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruano, G., E. M. Pagliaro, T. R. Schwartz, K. Lamy, D. Messina, R. E. Gaensslen, and H. C. Lee. 1992. Heat-soaked PCR: an efficient method for DNA amplification with applications to forensic analysis. BioTechniques 13:266-274. [PubMed] [Google Scholar]
- 16.Sakaoka, H., K. Kurita, Y. Iida, S. Takada, K. Umene, Y. T. Kim, C. S. Ren, and A. J. Nahmias. 1994. Quantitative analysis of genomic polymorphism of herpes simplex virus type 1 strains from six countries: studies of molecular evolution and molecular epidemiology of the virus. J. Gen. Virol. 75:513-527. [DOI] [PubMed] [Google Scholar]
- 17.Sakaoka, H., K. Kurita, T. Gouro, Y. Kumamoto, S. Sawada, M. Ihara, and T. Kawana. 1995. Analysis of genomic polymorphism among herpes simplex virus type 2 isolates from four areas of Japan and three other countries. J. Med. Virol. 45:259-272. [DOI] [PubMed] [Google Scholar]
- 18.Suzutani, T., K. Ishioka, E. De Clercq, K. Ishibashi, H. Kaneko, T. Kira, K. Hashimoto, M. Ogasawara, K. Ohtani, N. Wakamiya, and M. Saijo. 2003. Differential mutation patterns in thymidine kinase and DNA polymerase genes of herpes simplex virus type 1 clones passaged in the presence of acyclovir or penciclovir. Antimicrob. Agents Chemother. 47:1707-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takayama, M., N. Takayama, N. Inoue, and Y. Kameoka. 1996. Application of long PCR method of identification of variations in nucleotide sequences among varicella-zoster virus isolates. J. Clin. Microbiol. 34:2869-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitley, R. J. 1996. Herpes simplex viruses, p. 2297-2342. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 21.Wiedbrauk, D. L., J. C. Werner, and A. M. Drevon. 1995. Inhibition of PCR by aqueous and vitreous fluids. J. Clin. Microbiol. 33:2643-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshikawa, T., M. Ihira, S. Akimoto, C. Usui, F. Miyake, S. Suga, Y. Enomoto, R. Suzuki, Y. Nishiyama, and Y. Asano. 2004. Detection of human herpesvirus 7 DNA by loop-mediated isothermal amplification. J. Clin. Microbiol. 42:1348-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]